Abstract
In this study, cellular membrane fragments from SH-EP1-pCEP4-hα7 and α7 HEK-293 cell lines were used to synthesize cellular membrane affinity chromatography (CMAC) columns containing functional α7 nicotinic acetylcholine receptors, CMAC(α7 nAChR) columns. The synthesis of stable columns required the addition of cholesterol to the 2% cholate solubilization/immobilization (s/i) buffer and to the mobile phase. In addition, when membranes from the SH-EP1 cell line were used, L-α-phosphatidylserine and L-α-phosphatidylethanolamine also had to be added to the s/i buffer. A CMAC(α4β2 nAChR) column was prepared using membrane fragments from a SH-EP1-pCEP4-hα4β2 cell line, and this process required the addition of L-α-phosphatidylserine and L-α-phosphatidylethanolamine to the s/i buffer, but not cholesterol. The s/i buffers from the three columns were compared with the s/i buffer utilized in the preparation of a CMAC(α4β2 nAChR) column prepared using an α4β2 HEK-293 cell line, which required no additions to the 2% cholate s/i buffer. The data demonstrate that both cell type and receptor type affect the protocol required to produce a stable CMAC column and that, at the current time, the development of an optimum immobilization protocol is an empirical process. The results are also consistent with the observation that the α7 nAChR is localized in lipid rafts in both of these cell lines and that the cholate detergent removed cholesterol from these microdomains.
Neuronal nicotinic acetylcholine receptors (nAChRs) are ligand gated ion channels that are composed of five transmembrane subunits oriented around a central pore.1,2 To date, 12 different neuronal subunits have been identified, 9 α subunits (α2–α10) and 3 β subunits (β2–β4). These subunits combine to form a wide variety of homomeric αx subtypes where x = 7, 9, 10 and heteromeric αxβy subtypes where x = 2, 3, 4, 5, 6, and y = 2, 3, 4.
The homomeric α7 nAChR is found in the mammalian brain,3 and these receptors appear to be involved in learning and memory4 and to play a role in Alzheimer’s disease.5,6 The receptor is also involved in schizophrenia through the mediation of the release of GABA by hippocampal interneurons,7 and α7 nAChR agonists such as clozapine are currently used in the clinical treatment of this disease.8 In the peripheral nervous system, the α7 nAChR has been implicated in the initial nicotine-induced heart rate decrease.9 In addition, α7 nAChRs have been associated with the regulation of inflammation through the inhibition of cytokine synthesis in the cholinergic anti-inflammatory pathway.10
The majority of the studies on the characterization of the human α7 nAChR have been carried out using electrophysiological techniques on receptors heterologously expressed in Xenopus oocytes.11,12 While the experimental procedures associated with this technique are demanding and time-consuming, this approach continues to be the predominant method for characterization of neuronal nAChRs.
An alternative approach to the study of the binding of agonists and antagonists to nAChRs is cellular membrane affinity chromatography (CMAC).13,14 In this experimental approach, cellular membrane fragments obtained from cell lines expressing the target nAChRs are immobilized on the surface of an immobilized artificial membrane (IAM) liquid chromatography stationary phase and the resulting nAChR-IAM stationary phases used to create the desired CMAC column. The columns are then used in frontal affinity chromatography or nonlinear chromatography studies to characterize ligand–nAChR interactions.
This technique has been used to produce CMAC columns containing heteromeric αxβy subtypes, CMAC(αxβy nAChR) columns, containing the α3β2, α3β4, α4β2, and α4β4 nAChRs.15 The data from the studies with the CMAC(αxβy nAChR) columns have demonstrated that this approach can be used to directly determine Kd values within and between αxβy nAChR subtypes,15 to sort compounds by relative EC50 values16 and to identify and characterize noncompetitive inhibitors of the nAChR.14
To date, CMAC columns containing homomeric nAChRs, such as the α7 nAChR, have not been created. This article reports the synthesis and initial characterization of two CMAC(α7 nAChR) columns. The cellular membranes used in this study were obtained from an epithelial cell line, SH-EP1-pCEP4-hα7, and a human embryonic kidney cell line, KXa7-HEK293. There were clear differences in the stabilities of the two CMAC(α7 nAChR) columns, which appeared to be due to the cell type. However, both of the CMAC(α7 nAChR) columns were less stable than the previously described CMAC(αxβy nAChR) columns, even though one of the columns had been created using the same HEK293 cell type. Thus, it appeared that both cell type and receptor subtype played a role in the observed differences. Thus, this paper also reports the results of a comparative study using SH-EP1 and HEK293 cell lines that stably express either the α7 nAChR or α4β2 nAChR designed to examine the sources of these differences. The data indicate that both cell type and receptor subtype play a role in the observed stability, and the results demonstrate that the creation of CMAC columns must be undertaken with an understanding of the biological and pharmacological properties of the target and the expression system.
MATERIALS AND METHODS
Materials
Phenylmethylsulfonyl fluoride (PMSF), benzamidine, leupeptin, (±)-epibatidine (EB), (S)-nicotine tartrate (NIC), cholesterol, fluorescent isothiocyanate (FITC)-labeled-α-bungarotoxin (FITC-αBgt), cholic acid sodium salt, and ethylenediaminetetraacetic acid (EDTA) were purchased from Sigma Aldrich (St. Louis, MO). L-α-Phosphatidylserine and L-α-phosphatidylethanolamine were purchased from Avanti Lipids (Alabastar, AL). [3H]-EB (66.6 Ci/mmol) and [3H]-NIC (86.7 Ci/mmol) were purchased from NEN Life Science Products (Boston, MA). Dulbecco’s modified Eagle’s medium 1× (DMEM) and sodium pyruvate was purchased from Invitrogen Corp. (Carlsbad, CA). Horse serum, fetal bovine serum, and amphotericin B were purchased from Biosource International (Rockville, MD). Immobilized artificial membrane (IAM-PC) silica beads (12 μm particle size, 300 Å pore size) were from Regis Technologies Inc. (Morton Grove, IL). Nitrocellulose dialysis tubing (MW cutoff < 10 000 Da) was purchased from Pierce Chemical (Rockford, IL). Chromatographic glass columns HR 5/2 and a 50-mL superloop were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden).
Cell Line
The SH-EP1, SH-EP1-pCEP4-hα4β2 and SH-EP1-pCEP4-hα7 cell lines used in this study were kindly provided by R. Lukas (Division of Neurobiology, Barrow Neurological Institute, Phoenix, AZ), and the HEK-293 cell line was obtained from American Type Tissue Culture (Manassas, VA). The cell lines were maintained following a previously described procedure,17 in which the cells were grown in DMEM supplemented with 10% horse serum, 5% fetal bovine serum, 1% sodium pyruvate, and 8% amphotericin B.
Immobilization of Cellular Membrane Fragments
The membranes obtained from the SH-EP1, SH-EP1-pCEP4-hα7, SH-EP1-pCEP4-hα4β2, hα7HEK-293, hα4β2HEK-293, and HEK-293 cells were immobilized on the IAM stationary phase following a previously described method,15 which was modified as needed for the different cell lines and receptor subtypes. In brief, (10–30) × 106 cells were placed in 10 mL of Tris-HCl buffer (50 mM, pH 7.4) containing 5 mM EDTA, 3 mM benzamidine, and 0.2 mM PMSF. The suspension was homogenized for 3 × 15 s at the setting of 12.5 on a model PT-2100 homogenizer (Kinematica AG, Luzern, Switzerland). The homogenate was centrifuged at 800g for 4 min, and the pellet containing the nuclear proteins was discarded. The supernatant was centrifuged at 100000g for 20 min at 4 °C, and the resulting pellet containing the cellular membranes was collected and resuspended in 10 mL of one of the following solubilization buffers: (a) SH-EP1-pCEP4-hα7, SH-EP1. Tris-HCl buffer (50 mM pH 7.4), containing 2 mM MgCl2, 100 mM NaCl, 3 mM CaCl2, 5 mM KCl, 10 μM leupeptin, 100 nM cholesterol, 60 μM L-α-phosphatidylserine, 40 μM L-α-phosphatidylethanolamine, and 2% (w/v) cholate. (b) SH-EP1-pCEP4-hα4β2. Tris-HCl buffer (50 mM pH 7.4), containing 2 mM MgCl2, 100 mM NaCl, 3 mM CaCl2, 5 mM KCl, 10 μM leupeptin, 60 μM L-α-phosphatidylserine, 40 μM L-α-phosphatidylethanolamine, and 2% (w/v) cholate. (c) hα7 HEK-293, HEK-293. Tris-HCl buffer (50 mM pH 7.4), containing 2 mM MgCl2, 100 mM NaCl, 3 mM CaCl2, 5 mM KCl, 10 μM leupeptin, 100 nM cholesterol, 60 μM L-α-phosphatidylserine, 40 μM L-α-phosphatidylethanolamine, and 2% (w/v) cholate. (d) hα4β2 HEK-293. Tris-HCl buffer (50 mM pH 7.4), containing 2 mM MgCl2, 100 mM NaCl, 3 mM CaCl2, 5 mM KCl, 10 μM leupeptin, and 2% (w/v) cholate.
The resulting mixture rotated at 150 rpm using an orbit shaker (Lab-line model 3520, Melrose Park, IL) for 18 h at 4 °C and subsequently centrifuged at 80000g for 20 min, and the supernatant containing membrane–cholate solution was collected. The supernatant was mixed with 200 mg of the IAM stationary phase; the resulting mixture was rotated at room temperature for 1 h at 150 rpm using an orbit shaker and then dialyzed against 1 L of Tris HCl (50 mM, pH 7.4) containing 100 mM NaCl, 5 mM EDTA, 0.1 mM CaCl2, and 0.02 mM PMSF for 1 day at 4 °C. The resulting mixture was centrifuged for 3 min at 400g, and the supernatant was discarded. The pellet was washed with 5 mL of Tris-HCl (10 mM, pH 7.4) and centrifuged. This process was repeated until the supernatant was clear.
The IAM supports were collected and packed into HR 5/2 chromatographic glass columns to create the respective CMAC columns containing 18 mm × 5 mm I.D chromatographic beds.
Frontal Chromatographic Studies
(1) Chromatographic System
The CMAC columns were placed in a chromatographic system, which has been previously described.15 The mobile phase consisted of ammonium acetate (10 mM, pH 7.4), which contained 10 pM cholesterol when the CMAC(hα7-SH-EP1), CMAC(hα7 HEK-293), and CMAC(hα4β2-SH-EP1) columns were used. The mobile phase was delivered at 0.2 mL/min at room temperature, and the CMAC column was washed for 18 h between injections. Detection of the marker ligands [3H]-EB or [3H]-NIC was accomplished using an on-line scintillation detector (IN/US system, β-ram model 3, Tampa, FL) with a dwell time of 2 s using Laura lite 3.
(2) Chromatographic Studies
On the CMAC(α7 nAChR) columns, the marker ligand for the frontal chromatographic studies of NIC was [3H]-NIC (120 pM) and 10-mL solutions containing the marker ligand and a series of concentrations of NIC, 0.50, 1.0, 10, 5, 0.050, 0.100, and 0.500 nM, were applied to the columns. For studies involving EB, the concentration of the marker ligand ([3H]-EB) was 120 pM and the concentrations of EB were 60 pM, 100 pM, 1 nM, and 5 nM.
On the CMAC(α4β2 nAChR) columns, the marker ligand was [3H]-EB (30 pM) and the concentrations of displacers were EB 15, 30, 60, 120, and 180 pM; and NIC 0.1, 1, 5, 10, and 50 nM.
(3) Data Analysis
The data were analyzed as previously described.15
Confocal Microscopy and α-Bungarotoxin Binding
FITC-α-Bgt (1 mg/mL) was pumped through the CMAC columns created using the SH-EP1 and SH-EP1-pCEP4-hα7, hα7 HEK-293, and HEK-293 cell lines for 30 min using a peristaltic pump (Dynamax RP-1) at a flow rate of 50 μL/min, followed by water for 30 min. A small amount of stationary phase was collected and analyzed by confocal microscopy. The stationary phases were imaged with a Zeiss LSM-410 inverted confocal microscope (Carl Zeiss Inc., Jena, Germany) with excitation at 488 nm and collecting emission at greater than 515 nm simultaneously with transmitted image using a Zeiss Plan-Apochromat 63×/1.4 NA oil immersion lens. The images were processed by MetaMorph software (Universal Imaging Corp., Downingtown, PA).
RESULTS
Preparation of CMAC(α7 nAChR) Columns
In the previous studies on the synthesis and characterization of CMAC(αxβy nAChR) columns, (1–3) × 106 HEK-293 cells expressing the target αxβy nAChR were required to produce a viable column.15 Based on these results, the initial preparation of the CMAC(α7 nAChR) columns was carried out using membranes obtained from 3 × 106 cells from either the SH-EP1-hα7 cell line or the KXα7-HEK-293 cell line. However, no measurable binding activity was observed on either CMAC column. In order to determine if this was due to the level of expression of the α7 nAChRs in the respective cell lines, the number of cells used in the preparation of the CMAC columns was sequentially increased. Using [3H]-EB as the marker ligand, measurable binding activity was observed with columns prepared from (20–30) × 106 of SH-EP1-hα7 cells and 10 × 106 KXα7-HEK-293 cells.
In the protocol employed in the synthesis and characterization of CMAC(αxβy nAChR) columns, initial frontal displacement chromatography studies are used to establish the apparent affinity, Kd, of the immobilized nAChRs for [3H]-EB, established nAChR marker ligand. When the CMAC(α7 nAChR) column prepared using SH-EP1-hα7 cells was studied, the characteristic frontal chromatography curves with initial flat, breakthrough, and plateau regions were observed. However, although the data indicated that EB was specifically retained on the CMAC(α7 nAChR) column, the calculated Kd for EB, 80 nM, was significantly weaker than the Kd of 0.64 nM previously reported for the SH-EP1-hα7 cells.17 Further, the Kd, value for NIC could not be obtained due to a continuous decrease in the observed specific retention of [3H]-EB. The unstable behavior of the column suggested that the protocol used in the synthesis of the CMAC column had affected the stability of the membrane fragments, α7 nAChRs, or both.
In previous studies involving G-protein-coupled receptors (GPCRs), the addition of mixed egg phosphatidylcholine (PC) and phosphatidylserine (PS) lipids to the solubilization and chromatographic buffers was required to produce stable CMAC columns.18,19 This approach was used in this study, and the membranes from the SH-EP1-hα7 cells were stabilized by the addition of 100 nM cholesterol, 60 μM L-α-phosphatidylserine, and 30 μM L-α-phosphatidylethanolamine to the solubilization buffer. Once the protocol was developed for the SH-EP1-hα7 cells, the same approach was applied to the synthesis of the CMAC(α7 nAChR) from the KXα7HEK-293 cells. However, this cell line only required the addition of cholesterol to the solubilization buffer.
Characterization of CMAC(α7 nAChR) Columns
The activity and selectivity of the CMAC(α7 nAChR) columns were determined using frontal displacement studies employing both [3H]-NIC and [3H]-EB as the marker ligands and NIC and EB as the displacers, cf. Figure 1. Elution profiles containing front and plateau regions were observed, and the relationship between the concentrations of the displacer and the corresponding breakthrough volumes of the marker was used to calculate the Kd values for the displacer as previously described.15 However, the observed affinity of EB on the CMAC column prepared using the SH-EP1-hα7 cells, was still weaker than the expected value. This was addressed by the addition of 10 pM cholesterol to the chromatographic mobile phase. Under these conditions, the observed binding affinity for EB increased as the calculated Kd value decreased from 80 to 0.67 nM, Table 1. This value was consistent with the previously reported Kd value of 0.64 nM obtained using membranes obtained from SH-EP1-hα7 cells and competitive membrane binding techniques.17 This is not surprising as both methods used membranes obtained from the SH-EP1-hα7 cell line, the same marker, [3H]-EB, and the same general experimental approach, i.e., competitive binding.
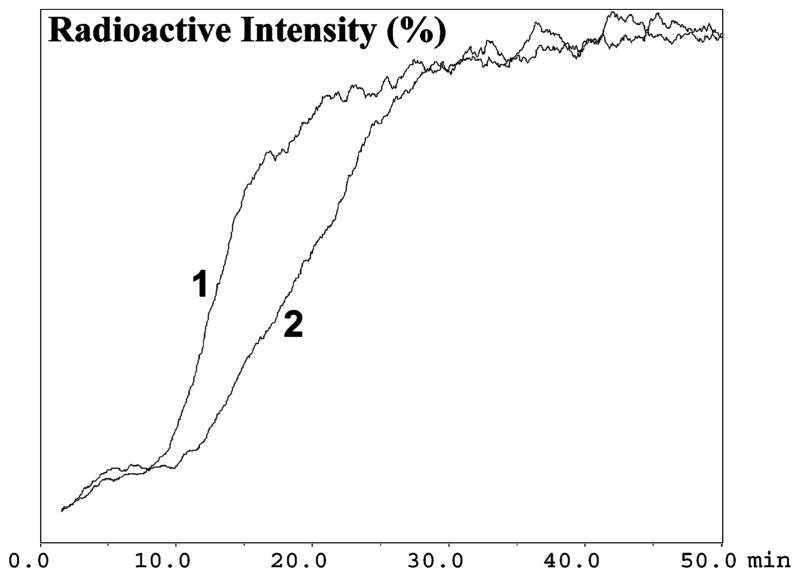
Figure 1.
Competitive displacement of [3H]-nicotine on the α7(+) nAChR-IAM, where chromatographic traces 1 and 2 represent the chromatography of 120 pM [3H]-nicotine alone and in the presence of 500 pM nicotine, respectively. Running buffer: ammonium acetate (10 mM, pH 7.4), 10 pM cholesterol at a flow rate of 0.2 mL/min.
Table 1.
Binding Affinities, Kd Values, Calculated by Frontal Affinity Chromatography on the α7 nAChR and α4β2 nAChR CMAC Columns Prepared in This Study
|
Kd (nM)
|
||
|---|---|---|
| cell type | epibatidine | nicotine |
| SH-EP1-hα7 | 0.67 | 7,610 |
| SH-EP1-hα4β2 | 0.46 | 12.80 |
| α7 HEK-293 | 0.82 | 5,590 |
| α4β2 HEK-293a | 0.01 | 16.40 |
Data previously reported.
The Kd values for EB and NIC were determined on both CMAC(α7 nAChR) columns, Table 1. There was no significant difference between the Kd values for EB determined on the two columns. There was also no significant difference between the calculated Kd values for NIC, which were ~7 μM. The Kd values for the binding of NIC to the human α7 nAChR used in this study have not been reported, and no direct comparison can be made with the data from membrane binding studies. However, in a study using α7 nAChR immunopurified from chick retina and [125I]-α-bungarotoxin as the marker, NIC displayed a Ki value of 1.2 μM.11 Thus, the results indicate that the CMAC(α7 nAChR) columns developed in this study can be used to determine the binding affinities of compounds at the α7 nAChR.
Preparation and Characterization of CMAC(α4β2 nAChR) Columns
Following the protocol developed for the immobilization of the membrane fragments from the SH-EP1-hα7 cell line, (20–30) × 106 SH-EP1-hα4β2 cells were used in the synthesis of the CMAC(α4β2 nAChR) column. A systematic examination of the solubilization and immobilization parameters indicated that while L-α-phosphatidylserine and L-α-phosphatidylethanolamine were necessary in the solubilization buffer, it was not necessary to add cholesterol to this buffer. The synthesis of the CMAC-(α4β2 nAChR) column from the hα4β2HEK-293 cell line has been previously reported15 and was accomplished using membranes from 1 × 106 to 3 × 106 cells and did not require the addition of lipids to the solubilization buffer.
The nAChR binding affinities on the CMAC(α4β2 nAChR) columns were characterized using EB and NIC, Table 1. On the CMAC(α4β2 nAChR) column created using the SH-EP1-hα4β2 cell line, the Kd value for EB was 0.46 nM and the Kd value for NIC was 12.8 nM. These values were consistent with the Kd values of 0.49 and 76 nM, respectively, determined using competitive binding experiments with SH-EP1-hα4β2 membranes and [3H]-EB as the marker ligand.17 On the previously reported CMAC-(α4β2 nAChR) column created using α4β2HEK-293 cells, the Kd value for EB was 0.01 nM and the Kd value for NIC was 16.4 nM. These values were consistent with the Kd values of 0.06 and 10 nM, respectively, determined using competitive binding experiments with α4β2HEK-293 membranes and [3H]-EB as the marker ligand.20 The data indicate that the immobilized α4β2 nAChRs retained their ability to bind ligands and that there were no significant differences between the observed affinities on the two CMAC(α4β2 nAChR) columns.
Confocal Microscopy
In the previous studies of CMAC(αxβy nAChR) columns, the presence of the cellular membranes and immobilized receptors on the surface of the silica beads was not visualized, in part due to the lack of an appropriate fluorescent marker. However, the expression of functional hα7 nAChRs on the cellular surface of macrophages has been recently demonstrated using a fluorescently labeled isothiocyanate derivative of α-bungarotoxin (FITC-α-Bgt) and fluorescent confocal microscopy.10 This approach was employed in this study using the IAM stationary phases used to create the CMAC(α7 nAChR) columns from the SH-EP1-hα7 and α7HEK-293 cell lines. In order to ensure that nonspecific binding on the cellular surfaces did not produce false positive staining, membranes obtained from nontransfected SH-EP1 and HEK-293 cell lines were also immobilized on the IAM stationary phase using the same conditions employed with the respective transfected cell line.
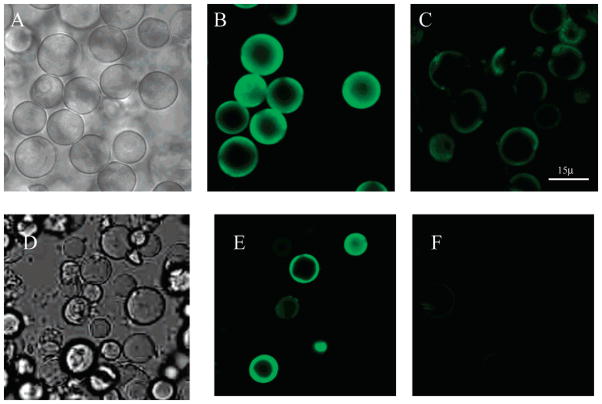
The 12-μm silica beads, which comprise the backbone of the IAM stationary phase, were clearly evident when the IAM phase coated with membranes from the SH-EP1-hα7 was imaged without fluorescent light, Figure 2A. Similar results were obtained with the other three phases (data not shown). Under fluorescent conditions, the measured emission at λ > 515 nm from the IAM phases containing membranes from the SH-EP1-hα7 and α7 HEK-293 cell lines, Figure 2B, E, were significantly greater than the emissions from the corresponding IAM phase containing membranes from the respective nontransfected cell lines, Figure 2C, F. The results indicate that the membrane fragments from the SH-EP1-hα7 and α7HEK-293 cell lines coated the IAM phase and retained the ability to bind a specific α7 nAChR marker.
Figure 2.
FITC-labeled α-bungarotoxin (FITC-α-Bgt) (1 mg/mL) pumped through an hα7(+) nAChR-SH-EP1 IAM column and α7(+)-nAChR-HEK-293 IAM column during 30 min using a peristaltic pump (Dynamax RP-1) at a flow rate of 50 μL/min followed by water for 30 min. The transmitted light images are displayed in (A) and (D), respectively, and the fluorescent images are displayed in (B) and (E). FITC-α-Bgt was also pumped through the hα7(−) nAChR-SH-EP1 IAM column and the α7(−) nAChR-HEK-293 IAM column. The fluorescent images are displayed in (C) and (F), respectively. A small amount of stationary phase was collected and analyzed by confocal microscopy. The stationary phases were imaged with a Zeiss LSM-410 inverted confocal microscope with excitation at 488 nm and collecting emission at greater than 515 nm simultaneously with transmitted image using a Zeiss Plan-Apochromat 63x/1.4 NA oil immersion lens. The images were processed by MetaMorph software.
It is of interest to note that a comparison of the IAM beads coated with the SH-EP1-hα7 membrane fragments imaged without fluorescent light (Figure 2A) with the number of particles observed under fluorescent imaging (Figure 2B) indicated that functional hα7 nAChRs were present in ~60% of the IAM particles. The observed surface coverage on the IAM particles can be attributed to the immobilization technique. After detergent solubilization, no further attempts were made to purify the homogenized membranes. Thus, it is likely that membranes fractions that did not contain the expressed hα7 nAChRs were immobilized on the surface of the IAMs, thereby reducing the area available for the immobilization of membrane fractions containing active receptors. Further, the level of expression obtained with the SH-EP1 cells was only 80%.16
DISCUSSION
In previous studies, membrane fragments from HEK-293 cell lines expressing heteromeric αxβy nAChRs were used to create CMAC(αxβy nAChR) columns. In these studies, the membrane fragments were solubilized using a 2% cholate buffer and then immobilized on IAM beads by removing the detergent with dialysis. This protocol did not work when applied to the immobilization of the cellular membranes obtained from the SH-EP1-hα7 cells, and lipids had to be added to the solubilization buffer in order to stabilize the membrane fragments and retain α7 nAChR function. Since both the cell line and the receptor subtype differed between the two experiments, it was not immediately clear which factor was responsible for the necessary changes in the solubilization/immobilization buffer. In order to examine this question, CMAC columns were produced using membranes obtained from α7 HEK-293, SH-EP1-hα7, and SH-EP1-hα4β2 cell lines and the data from the three columns were compared to the previous results obtained using the α4β2 HEK-293 cell line.15 The data are presented in Table 2. It is important to note that once the immobilization processes and the mobile-phase conditions had been optimized, the observed affinities of the immobilized receptor subtypes were essentially equivalent and independent of the source of the cellular membrane, Table 1. Thus, the answers lie in the immobilization process.
Table 2.
Composition of the Solubilization/Immobilization Buffers Used in the Preparation of CMAC Columnsa
The solubilization/immobilization buffer used with both of the SH-EP1 cell lines required the addition of PS and PE lipids. In contrast, these lipids were not required when HEK-293 cells were the source of the membrane fragments. The data suggest that a key factor is the composition of the cellular membrane.
The comparison of the results from the point of view of the nAChR subtypes adds another dimension to this analysis. With the α7 nAChRs, it was necessary to add cholesterol to both the solubilization/immobilization buffers and mobile phase irrespective of the source of the cellular membranes, while this was not required with the α4β2 subtypes. The observation that the addition of cholesterol was essential to achieve a stable α7 nAChR suggests that this subtype is located in an environment, or microdomain, different from the α4β2 nAChRs within the cellular membranes.
Membrane microdomains composed of sphingolipids, cholesterol, glycolipids, and proteins have been identified and shown to provide an ordered lipid environment within the membrane and to play key roles in signal transduction, transcellular transport, and cholesterol homostasis.21–23 These microdomains are a group of related structures, lipid rafts, which have been classified on the basis of the presence or absence of caveolin within the structure, and designated as caveolae or lipid rafts, respectively. However, the biological significance of this designation has not been firmly established.
Proteomic studies of lipid rafts and caveolae have demonstrated that these structures are highly complex and diverse.23–25 It has also been shown that detergent extraction removes some lipids and proteins from rafts and that the extractability of proteins from rafts can be used to probe the variability in lipid raft structure.23,26 These rafts can also be disrupted by the methyl-β-cyclodextrin-mediated extraction of cholesterol.22
A number of GPCRs, ion channels, and enzymes have been found in caveolae and lipid rafts.27 The association of lipid rafts and α7 nAChR has also been demonstrated in somatic spines of ciliary neurons28 and in PC-12 cells.29 The latter studies with the PC-12 cell line are particularly relevant to the present work. The PC-12 cells have been shown to express both homomeric (α7) and heteromeric (α3, α5, β2, β4) nAChR subunits, and the presence of functional α7 nAChR was also demonstrated. In addition, caveolin of any subtype was not detected in the cell line. When the lipid rafts were prepared using sucrose gradient centrifugation, only the α7 nAChR was found in the rafts. In addition, cholesterol depletion disrupted the receptor’s activity. The authors concluded that the α7 nAChRs were localized to the lipid rafts while the other receptor subunits were located in other microdomains of the plasma membrane.
In this study, the synthesis of stable CMAC(α7 nAChR) columns from membrane fragments obtained from both the SH-EP1 and HEK-293 cell lines required the addition of cholesterol to the solubilization/immobilization buffers and to the mobile phase. This is consistent with the observation that the α7 nAChR is localized in lipid rafts in both of these cell lines and that the cholate detergent removed cholesterol from these microdomains. Indeed, cholate has been shown to effectively remove cholesterol during membrane solubilization and would be expected to affect these domains.30
The fact that the synthesis of stable CMAC(α4β2 nAChR) columns from membrane fragments obtained from both the SH-EP1 and HEK-293 cell lines did not require the addition of cholesterol to the solubilization/immobilization buffers is also consistent with the previous observation that heteromeric subunits of the nAChR are not located within lipid rafts.
The results of this study also demonstrate that, since lipid rafts exist within a membrane environment, disruption of this environment will also destabilize the receptor. Thus, it is likely that the PS and PE lipids were extracted during cholate solubilization. This disrupted the membrane structure of the SH-EP1 cells requiring the addition of these lipids to the solubilization/immobilization buffers in order to stabilize the membrane fragments. It is of interest that this did not occur during the solubilization of the HEK-293 membrane fragments indicating that the HEK-293 cell line has a different membrane architecture and suggesting that it might be preferred over the SH-EP1 cell line in the development of CMAC columns.
Thus, it appears that both cell type and receptor affect the protocol required to produce a stable CMAC column and that these requirements reflect the membrane localization of the receptor. This observation is consistent with a previous study on the development of a CMAC column from membrane fragments obtained from a HEK-293 cell line expressing the β2-adrenergic receptor (β2-AR).19 In this study, the preparation of a stable CMAC column required the addition of mixed egg PC to the solubilization/immobilization and chromatographic buffers, Table 2. The β2-AR is a GPCR that has been identified in caveolin-3 containing lipid rafts in neonatal rat cardiomyocytes,31 while a second GPCR, the D1 dopamine receptor, localized in caveolin-2 containing lipid rafts in the HEK-293 cell line.28 In addition to cholesterol, the cholate detergent effectively removes phosphatidyl lipids from cellular membranes30 and the required addition of PC to the solubilization/immobilization and chromatographic buffers suggests that, in the HEK-293 cell line used in the study, the β2-AR was localized to a lipid raft containing a caveolin subtype. It is of interest to note that the same protocol was required when the cholate detergent was replaced by CHAPS, which has a detergent extractability profile that is similar to cholate.30
The results with the β2-AR and the nAChRs discussed in this study indicate that, at the current time, the development of an optimum immobilization protocol is an empirical process. This is emphasized by the development of a CMAC column containing the P2Y1 receptor, a GPCR, which was produced using cellular membrane fragments obtained from the 1321N1 astrocytoma cell line.32 In this case, the detergent was n-octyl β-D-glucopyranoside and it was not necessary to add lipids to the solubilization/immobilization buffer. This may be due to the cell type, the membrane localization of the P2Y1 receptor, or the fact that n-octyl β-D-glucopyranoside has a different detergent extractability profile than cholate and CHAPS. These factors are under investigation, and the results will be reported elsewhere.
While the empirical nature of the process may require added experimentation, the resulting CMAC columns are an interesting and useful addition to receptor–ligand studies. The optimized CMAC(α7 nAChR) columns produced in this study were viable for over 1 month and have been used in a variety of studies. These results will also be published elsewhere.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging.
References
- 1.Holladay MW, Dart MJ, Lynch JK. J Med Chem. 1997;40:4169–4194. doi: 10.1021/jm970377o. [DOI] [PubMed] [Google Scholar]
- 2.Hucho F, Tsetlin VI, Machold J. Eur J Biochem. 1996;239:539–557. doi: 10.1111/j.1432-1033.1996.0539u.x. [DOI] [PubMed] [Google Scholar]
- 3.Lukas RJ, Changeux JP, Le Novere N, Albuquerque EX, Balfour DJ, Berg DK, Bertrand D, Chiappinelli VA, Clarke PB, Collins AC, Dani JA, Grady SR, Kellar K, Lindstrom JM, Marks MJ, Quik M, Taylor PW, Wonnacott S. Pharmacol Rev. 1999;51:397–401. [PubMed] [Google Scholar]
- 4.Radcliffe KA, Dani JA. J Neurosc. 1998;18:7075–7083. doi: 10.1523/JNEUROSCI.18-18-07075.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kihara T, Shimohama S, Sawada H, Honda K, Nakamizo T, Shibasaki H, Kume T, Akaike A. J Biol Chem. 2002;276:13541–13546. doi: 10.1074/jbc.M008035200. [DOI] [PubMed] [Google Scholar]
- 6.Picciotto MR, Zoli M. J Neurobiol. 2002;53:641–655. doi: 10.1002/neu.10102. [DOI] [PubMed] [Google Scholar]
- 7.Martin LF, Kem WR, Freedman R. Psychopharmacology. 2004:54–64. doi: 10.1007/s00213-003-1750-1. [DOI] [PubMed] [Google Scholar]
- 8.Simosky JK, Stevens KE, Adler LE, Freedman R. Psychopharmacology. 2003;165:386–396. doi: 10.1007/s00213-002-1285-x. [DOI] [PubMed] [Google Scholar]
- 9.Ji S, Tosaka T, Whitfield BH, Katchman AN, Kandil A, Knollmann BC, Ebert SN. J Pharm Exp Ther. 2002;301:893–899. doi: 10.1124/jpet.301.3.893. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Yu M, Ochani M, Amella CA, Tanovic M, Susaria S, Li JH, Wang H, Yang H, Ulloa L, Al-Abed Y, Czura CJ, Tracey KJ. Nature. 2003;421:384–388. doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]
- 11.Gerzanich V, Peng X, Wang F, Wells G, Anand R, Fletcher S, Lindstrom J. Mol Pharm. 1995;48:774–782. [PubMed] [Google Scholar]
- 12.Papke RL, Sanberg PR, Shytle RD. J Pharm Exp Ther. 2001;297:646–656. [PubMed] [Google Scholar]
- 13.Moaddel R, Jozwiak K, Wainer IW. Med Res Rev. 2007;27:723–753. doi: 10.1002/med.20091. [DOI] [PubMed] [Google Scholar]
- 14.Jozwiak K, Haginaka J, Moaddel R, Wainer IW. Anal Chem. 2002;74:4618–4624. doi: 10.1021/ac0202029. [DOI] [PubMed] [Google Scholar]
- 15.Moaddel R, Jozwiak K, Whittington K, Wainer IW. Anal Chem. 2005;77:895–901. doi: 10.1021/ac048826x. [DOI] [PubMed] [Google Scholar]
- 16.Moaddel R, Jozwiak K, Yamaguchi R, Cobello C, Whittington K, Sarkar T, Basak S, Wainer IW. J Chromatogr, B. 2004;813:235. doi: 10.1016/j.jchromb.2004.09.042. [DOI] [PubMed] [Google Scholar]
- 17.Peng JH, Fryer JK, Hurst RS, Schroeder KM, George AA, Morrissy S, Groppi VE, Leonard SS, Lukas RJ. J Pharm Exp Ther. 2005;313:24–35. doi: 10.1124/jpet.104.079004. [DOI] [PubMed] [Google Scholar]
- 18.Beigi F, Wainer IW. Anal Chem. 2004;75:4480–4485. doi: 10.1021/ac034385q. [DOI] [PubMed] [Google Scholar]
- 19.Beigi F, Chakir K, Xiao RP, Wainer IW. Anal Chem. 2004;76:7187–7193. doi: 10.1021/ac048910c. [DOI] [PubMed] [Google Scholar]
- 20.Zhang YX, Xiao YX, Kellar KJ, Wainer IW. Anal Biochem. 1998;264:22–25. doi: 10.1006/abio.1998.2828. [DOI] [PubMed] [Google Scholar]
- 21.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 22.Tsui-Pierchala BA, Encinas M, Milbrandt J, Johnson EM., Jr Trends Neurosci. 2002;25:412–417. doi: 10.1016/s0166-2236(02)02215-4. [DOI] [PubMed] [Google Scholar]
- 23.Sprenger RR, Horrevoets AJ. Methods Mol Biol. 2007;357:199–213. doi: 10.1385/1-59745-214-9:199. [DOI] [PubMed] [Google Scholar]
- 24.Li N, Shaw AR, Zhang N, Mak A, Li L. Proteomics. 2004;4:3156–3166. doi: 10.1002/pmic.200400832. [DOI] [PubMed] [Google Scholar]
- 25.Foster JL, De Hoog CL, Mann M. Proc Natl Acad Sci USA. 2003;100:5813–5818. doi: 10.1073/pnas.0631608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madore N, Smith KL, Graham HC, Jen A, Brady K, Hall S, Morris R. Eur Mol Biol Org. 1999;18:6917–6926. doi: 10.1093/emboj/18.24.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu P, Yang Z, Jones JE, Wang Z, Owens SA, Mueller SC, Felder RA, Jose PA. Kidney Int. 2004;66:2167–2180. doi: 10.1111/j.1523-1755.2004.66007.x. [DOI] [PubMed] [Google Scholar]
- 28.Bruses JL, Chauvet N, Rutishauser U. J Neurol. 2001;21:504–512. doi: 10.1523/JNEUROSCI.21-02-00504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshikawa J, Toya Y, Fujita T, Egawa M, Kawabe J, Umemura S, Ishikawa Y. Am J Physiol Cell Physiol. 2003;285:C567–C574. doi: 10.1152/ajpcell.00422.2002. [DOI] [PubMed] [Google Scholar]
- 30.Banerjee P, Joo JB, Buse JT, Dawson G. Chem Phys Lipids. 1995;77:65–78. doi: 10.1016/0009-3084(95)02455-r. [DOI] [PubMed] [Google Scholar]
- 31.Ostrom RS, Gregorian C, Drenan RM, Xiang Y, Regan JW, Insel PA. J Biol Chem. 2001;276:42063–42069. doi: 10.1074/jbc.M105348200. [DOI] [PubMed] [Google Scholar]
- 32.Moaddel R, Calleri E, Massolini G, Frazier C, Wainer IW. Anal Biochem. 2007;364:216–218. doi: 10.1016/j.ab.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]