Abstract
ADME/Tox (absorption, distribution, metabolism, elimination and toxicity) technology is traditionally associated as a tool in the drug discovery process which is often used to predict the efficiency of drug adsorption, distribution, metabolic pathways, and elimination. For the past four years we have been involved in an effort to evaluate readily available Food and Drug Administration (FDA) consumer drug profiles and pharmacological data. Portable Document Format (PDF) data from drug profiles available on the FDA Drug Information website were used to create a searchable FDA Consumer Drug Database© using Bio-Rad’s KnowItAll® platform which includes ADME/Tox in silico predictors. 14 pertinent pharmaceutical and pharmacological properties were collected for 75 structurally diverse consumer prescription drugs, and for several drugs, not all properties were completely populated. The major objective of this investigation was to validate the platforms prediction models for plasma protein binding (PPB) and bioavailability (BIO).
Keywords: FDA consumer drug database©, KnowItAll®, ADME/Tox, (quantitative) structure activity relationship (Q)SAR, predictor tools, chemical informatics
Introduction
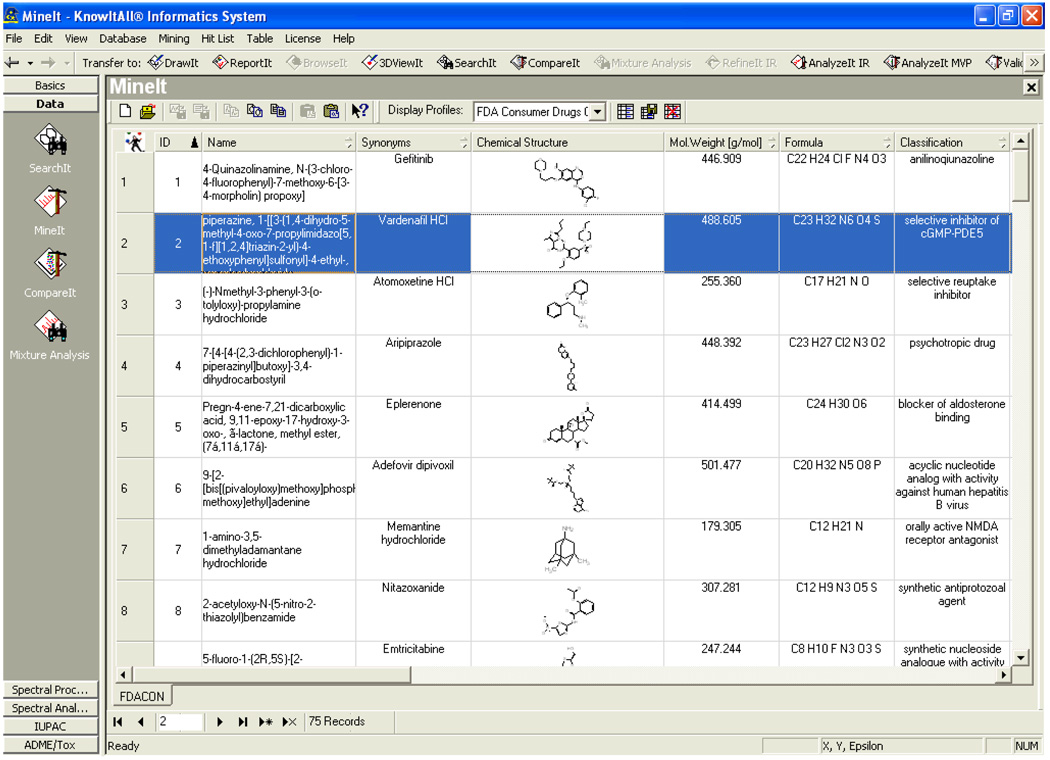
Earlier this year, in an article [1] in Pharmaceutical Reviews we outlined the steps taken to build the FDA Consumer Drug Database© [2] (shown in Figure 1.) containing 75 structurally diverse prescription drugs. The chemical, pharmacokinetic, and toxicological data for each drug were extracted from their web-accessible FDA portable document format (PDF) consumer drug information files [3–5]. Additionally, further useful information gleaned from documents submitted to the NIH Molecular Libraries-Small Molecule Repository [6] and the University of Alberta’s DrugBank database [7] were incorporated to populate fields in this newly designed searchable drug information database [2]. The database was created using our KnowItAll™ Informatics System–ADME/Tox Edition, which hosts predictors covering 14 in silico ADME/Tox properties and was designed by Bio-Rad Laboratories, Inc., Philadelphia, PA [8, 9]. These parameters are common to traditional (Q)SAR modeling and each parameter value was then extracted (when reported) from a drug’s profile. The fourteen pharmaceutical and pharmacological properties were Oncogenicity, Teratogenicity, Mutagenicity, Human Intestinal Absorption, Plasma Protein Binding, Water Solubility, Volume of Distribution, Elimination Half Time, Rate of Absorption, Blood Brain Barrier, NeuroToxicity, pKa, Bioavailability, and log P. As explained in our earlier article [1], the published FDA drug profiles [3–5] come from different pharmaceutical companies and there are differences in the reporting of a set of properties provided for each drug profile. Hence for some of the drugs, before using their reported information in the pharmaceutical and pharmacological data content section of their FDA files, data normalizations were sometimes needed [1]. The KnowItAll® system also contains a chemical informatics platform used to record experimental data [8–13] and has an ability to validate the predictors as well as the potential to build a consensus model [14–17].
Figure 1.
FDA Consumer Drug Database© [2] created with the KnowItAll® platform [8, 9]. It includes chemical structure, chemical name, synonym, drug classification, and other pharmacological properties, for each of the 75 drugs studied.
During the past decade, (Q)SAR studies have proved to be pliable encapsulated tools used for modeling non-congeneric data in drug discovery, toxicity prediction, lead optimization, and in the formulation of regulations [18, 19]. The acceptance or rejection of such predictions is dependent on the ability to estimate accuracy [20]. One objective now is to use the available ChemSilico predictor tools [21] in our KnowItAll® ADME/Tox edition [8, 9], to try to predict a drug’s pharmacokinetic, or toxicological property on the basis of chemical structure. The next step is to take this predicted data and compare it with the drug’s empirical data (available within the data published by the FDA) [3–5] to demonstrate and further support the accuracy of the KnowItAll® predictor tools.
Materials and Methods
The three dimensional chemical structure for each of the following 75 consumer drugs, (limited to orally administered drugs and pro-drugs with a chemical structure): Iressa®, Levitra®, Strattera®, Abilify®, Inspra®, Hepsera®, Namenda®, Alinia®, Emtriva®, Emend®, Tindamax®, Sensipar®, Pletal®, Viagra®, Zavesca®, Orfadin®, Zyvox®, Uroxatral®, Zelnorm®, Avodart®, Frova®, Provigil®, Aromasin®, Detrol®, Thalomid®, Atacand®, Zonegran®, Micardis®, Maxalt®, Xeloda®, Arava®, Gleevec®, Cialis®, Reyataz®, 5FU from of Xeloda®, Candesartan, Hectorol®, Singulair®, Spectracef®, Cefditoren (active metabolite of Spectracef®), Sanctura®, Colazal®, active metabolite of Arava®, Zetia®, Hepsera® M1, Spiriva®, Starlix®, Tasmar®, Xifaxan®, Crestor®, Nolvadex®, Detrol® M1, Mobic®, Sustiva®, Aciphex®, Ziagen®, Protonix®, Celebrex®, Agenerase®, Trileptal®, Oxcarbazepine MHD (active metabolite of Trileptal®), Exelon®, Temodar®, Keppra®, Tequin®, Avandia®, Actos®, Avelox®, Tamiflu®, Oseltamivir Carboxylate (active metabolite of Tamiflu®), Lopinavir®, Ritonavir®, Benicar®, Rapamune®, and Ketek®, were drawn using the drawing application DrawIt™ available in the KnowItAll® Cheminformatics Edition. For prodrugs, chemical structures of both the prodrugs and their corresponding real drugs were drawn and dealt with separately (for example Detrol® and Detrol M1). For the study of the computational prediction of the properties, the comparison of the predicted data with the empirical data from FDA drug profiles [3–5] and the prediction of missing data were conducted using the ChemSilico prediction and analyzing tools [21] available in the KnowItAll® ADME/Tox edition.
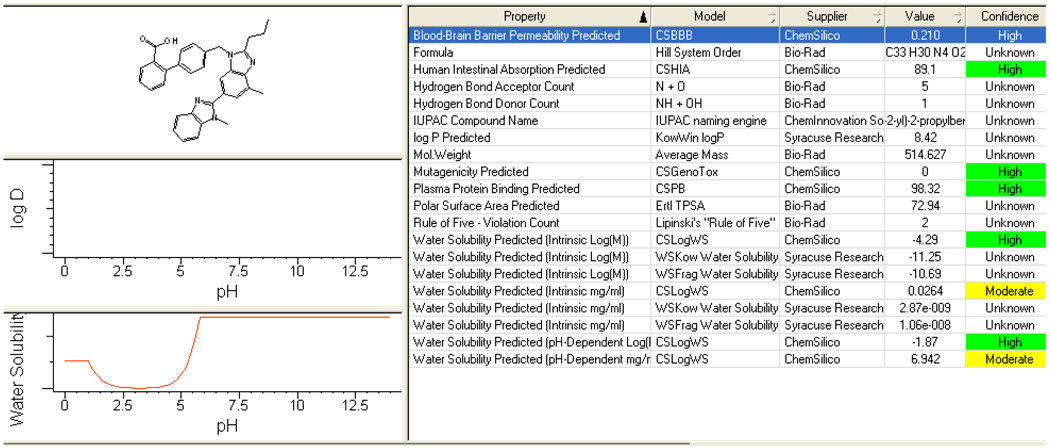
To investigate structure activity relationships (SARs), two of the most common biological properties – bioavailability (BIO) and human plasma protein binding (PPB) - were extracted for each of the 75 drugs from its FDA drug profile. Then, 7 major functional groups were considered: carbamoyl, carboxylic acid, ketone, sulfamoyl, sulfide, sulfone and sulfinyl. The 7 functional groups were further divided into 2 subset categories: (1) sulfur-containing functional groups (sulfide, sulfamoyl, sulfone, and sulfinyl); and (2) carbonyl-containing functional groups (ketone, carbamoyl, and carboxylic acid). In July 2009, results obtained from the first subset were presented and published in the very selective peer-reviewed International Conference on Bioinformatics, Computational Biology, Genomics, and Chemoinformatics [22]. Now in this paper, results from the second subset of categories for carbonyl-containing functional groups are reported. Each carbonyl-containing functional group was first drawn using the DrawIt™ tool in KnowItAll®, then the SearchIt™ tool was utilized to search the FDA Consumer Drug Database© for the maximum hits for that functional group, sorting among the 75 drugs. However SearchIt™ results reported some drugs several times (within the 3 carbonyl-containing functional groups), not because they contain more than one functional group, but simply because of a common chemical structural motif, such as C=O, shared between the 3 carbonyl-containing functional groups. Hence, a visual sorting was then done. On completion, the ChemSilico predictions were made for each of the 75 drugs using the KnowItAll® predictor ProfileIt™ as represented in the Figure 2 below, and the BIO and PPB data were extracted from these predictions. Each set of empirical and predicted data was then examined, and the FDA drug profiles of these drugs were reanalyzed to find explanations for those drugs for which the ChemSilico predictions differed from the empirical data by more than a 10% difference.
Figure 2.
ProfileIt™ prediction results for Telmisartan® an angiotensin II receptor antagonist (ARB) used in the management of hypertension. Bioavailability (BIO) and Plasma Protein Binding (PPB) data were extracted from these ChemSilico prediction results.
Discussion
Table 1 below lists the groupings obtained using the SearchIt™ tool, and tabulates the data extracted from the ProfileIt™ predictor tool in the KnowItAll® ADME/Tox edition. This table also contains relevant information harvested from the drug’s web-accessible profiles [3–5]. ChemSilico’s published guidelines [23] for human plasma protein binding predictions are: <45% low binding, 45–70% medium binding, and >70% high binding. Of the 49 drugs containing carbonyl groups considered, 14 had predictions for plasma protein binding that differed from FDA profile data by more than 10%. FDA drug profile values for Zelnorm®, Detrol®, Cialis®, and Avandia® were all in the high-binding range; and although predicted values were lower by more than 10%, all predictions were still within the upper range. In addition, both Zelnorm® and Detrol® were found experimentally to bind primarily to α1-acid glycoprotein. Because drug manufacturers determine binding by in vitro methods using purified protein, the profile data for these two drugs may be overstated since α1-acid glycoprotein is present in the plasma in fairly low concentrations (0.4–1.0%). With regard to the prediction for Colazal®, which was extremely low (62.33% compared to a profile value of >99%), the data are not physiologically relevant since this drug is administered in a hard gelatin capsule designed to be delivered intact to the colon where it is enzymatically cleaved to form the active metabolites. These then act locally on the colonic mucosa with minimal absorption into the systemic circulation. The plasma protein binding prediction for Ketek® was low when it was considered as a ketone but high as a carboxylic acid. This, in itself, however, does not explain the discrepancy since 3 other drugs had both carboxylic acid and ketone properties, yet all 3 had plasma protein binding predictions that were comparable to profile values in both categories. Of the drugs with predicted binding that was higher than values reported in FDA profiles, Provigil® and Thalomid® are racemic compounds. Chirality is an important consideration in pharmacology because enantiomers can display different pharmacokinetic properties [24]. For the remaining 6 drugs with plasma protein binding predictions that differed from experimental values, no specific information was found in their drug profiles that might explain the discrepancies. However, many physiological factors influence in vivo binding, and it is not known which of these are included in the ChemSilico model. ChemSilico binding predictions are based on over 500 topological structure and E-state descriptors, 158 of which are proprietary. Also, their plasma protein binding model uses single-activity values [25] (i.e., drug is not concentration dependent), which may or may not be true of a specific drug. Most drugs bind primarily to albumin or α1-acid glycoprotein but may also bind to other plasma proteins, such as lipoproteins and immunoglobulins. The drug concentration, protein concentration, and association constant all influence the fraction of drug bound. Also, some drugs bind to or penetrate erythrocytes, but this is not always indicated in drug information sheets. In addition, many substances besides drugs bind to plasma proteins, such as free fatty acids, bilirubin, various hormones, and tryptophan. Competition for saturable binding sites could result in reduced drug binding, especially at high drug concentrations.
Table 1.
The FDA empirical values, and the ChemSilico predicted values for the Bioavailability (BIO) and Plasma Protein Binding (PPB) parameters for the Sulfur- and Carbonyl-containing drugs within our database. Red: percent difference greater than 10. Yellow background: values in different PPB ranges.
| Sulfide | |||||||
|---|---|---|---|---|---|---|---|
| # | Brand Name | Classification | Potency | Bioavailability (%) | Plasma Protein Binding (PPB)(%) | ||
| FDA data | ChemSilico Prediction | FDA data | ChemSilico Prediction | ||||
| 8 | Alinia | A synthetic antiprotozoal agent | Treatment of intestinal protozoan infections | N/A | 92.9 | 99 | 72.15 |
| 9 | Emtriva | A synthetic nucleoside analog with activity against HIV-1 reverse transcriptase | anti-HIV-1 agent | 93 | 86.4 | <4 | 11.69 |
| 38 | Singulair | A selective and orally active leukotriene receptor antagonist that inhibits CysLT1 receptor | Long-term treatment of asthma | 64 | 82.7 | 99 | 97.65 |
| 39 | Spectracef | A semi-synthetic cephalosporin antibiotic | Treatment of bacterial infections | 14 | 18.4 | N/A | 64.79 |
| 40 | A.M. of Spectracef | A semi-synthetic cephalosporin antibiotic | Treatment of bacterial infections | N/A | 78.9 | 88 | 46.24 |
| 46 | Spiriva | An anticholinergic with specificity for muscarinic receptors | Treatment of COPD | 19.5 | 10.9 | 72 | 68.32 |
| 53 | Mobic | A member of the enolic acid group of nonsteroidal anti-inflammatory drugs | Treatment of arthritis | 89 | 93 | ~99.4 | 97.45 |
| 66 | Avandia | An antidiabetic agent | Treatment of Type 2 diabetes mellitus | 99 | 92.6 | 99.8 | 84.47 |
| 67 | Actos | An antidiabetic agent | Treatment of Type 2 diabetes mellitus | N/A | 93.7 | >99 | 96.24 |
| 71 | Lopinavir | An inhibitor of the HIV protease (Kaletra) | Treatment of HIV infection | N/A | 66.4 | 98.5 | 86.99 |
| 72 | Ritonavir | An inhibitor of the CYP3A-mediated metabolism of lopinavir (Kaletra) | Elevator of plasma levels of lopinavir | N/A | 41.9 | N/A | 91.79 |
| Sulfamoyl | |||||||
| # | Brand Name | Classification | Potency | Bioavailability (%) | Plasma Protein Binding (PPB)(%) | ||
| FDA data | ChemSilico Prediction | FDA data | ChemSilico Prediction | ||||
| 2 | Levitra | A selective inhibitor of cGMP-specific PDE5 | Treatment of erectile dysfunction | 15 | 79.7 | 95 | 94.33 |
| 14 | Viagra | A selective inhibitor of cGMP-specific PDE5 | Treatment of erectile dysfunction | 40 | 78.6 | 95 | 89.07 |
| 27 | Zonegran | An antiseizure drug | Treatment of partial seizures in adults | N/A | 91.9 | 40 | 84.17 |
| 50 | Crestor | A synthetic lipid-lowering agent | Treatment of high cholesterol | N/A | N/A | 88 | N/A |
| 53 | Mobic | A member of the enolic acid group of nonsteroidal anti-inflammatory drugs | Treatment of arthritis | 89 | 93 | ~99.4 | 97.45 |
| 58 | Celebrex | A nonsteroidal anti-inflammatory drug | Treatment of arthritis | N/A | 92 | ~97 | 96.21 |
| 59 | Agenerase | An inhibitor of the HIV protease | Treatment of HIV infection | N/A | 44.9 | 90 | 88.49 |
| Sulfone | |||||||
| # | Brand Name | Classification | Potency | Bioavailability (%) | Plasma Protein Binding (PPB)(%) | ||
| FDA data | ChemSilico Prediction | FDA data | ChemSilico Prediction | ||||
| 11 | Tindamax | A synthetic antiprotozoal and antibacterial agent | Treatment of protozoal or bacterial infection | N/A | 97.9 | 12 | 29.81 |
| 32 | Gleevec | A protein-tyrosine kinase inhibitor | Treatment Philadelphia chromosome positive CML | 98 | 87.3 | 95 | 69.07 |
| 34 | Reyataz | An azapeptide inhibitor of HIV-1 protease | Treatment of HIV infection | N/A | 28.9 | 86 | 83.1 |
| 56 | Ziagen | A synthetic carbocyclic nucleotide analogue with inhibitory activity against HIV | Treatment of HIV infection | 83 | 81.2 | 50 | 36.37 |
| Sulfinyl | |||||||
| # | Brand Name | Classification | Potency | Bioavailability (%) | Plasma Protein Binding (PPB)(%) | ||
| FDA data | ChemSilico Prediction | FDA data | ChemSilico Prediction | ||||
| 22 | Provigil | A wakefulness-promoting agent | Treatment of excessive sleepiness | N/A | 97.7 | ~60 | 72.02 |
| 55 | Aciphex | A substituted benzinidazole that inhibits gastric acid secretion | Treatment of duodenal ulcers and GERD | 52 | 87.2 | 96.3 | 86.63 |
| 57 | Protanix | A inhibitor of gastric acid secretion | Treatment of GERD | 77 | N/A | 98 | N/A |
| Carbamoyl | |||||||
| # | Brand Name | Classification | Potency | Bioavailability (%) | Plasma Protein Binding (PPB)(%) | ||
| FDA data | ChemSilico Prediction | FDA data | ChemSilico Prediction | ||||
| 2 | Levitra | A selective inhibitor of cGMP-specific PDE5 | Treatment of erectile dysfunction | 15 | 79.7 | 95 | 94.33 |
| 4 | Abilify | A partial agonist at dopamine D2 & serotonin 5-HT1A receptors and antagonist at serotonin%-TH2A receptor | Psychotropic drug | 87 | 89.6 | 99 | 95.84 |
| 8 | Alinia | A synthetic antiprotozoal agent | Treatment of intestinal protozoan infections | N/A | 92.2 | 99 | 72.15 |
| 13 | Pletal | A quinolinone derivative that inhibits cellular phosphodiesterase | Treatment of intermittent claudification | N/A | 90.7 | 96.5 | 99.44 |
| 14 | Viagra | A selective inhibitor of cGMP-specific PDE5 | Treatment of erectile dysfunction | 40 | 78.6 | 95 | 89.07 |
| 17 | Zyvox | A synthetic antibacterial agent of the oxazolidinone class | Treatment of bacterial infection | 100 | 93.1 | 31 | 47.83 |
| 18 | Uroxatral | A selective antagonist of post-synaptic alpha1-adrenoreceptors | A reduction in symptoms of BPH | 49 | 89.6 | 86 | 86.98 |
| 20 | Avodart | A synthetic 4-azasteroid compound, selectiove inhibitor of both type 1 and type 2 insoforms of 5AR | Treatment of enlarging prostate | 60 | 75.1 | 99 | 98.88 |
| 21 | Frova | A selective 5-hydroxy-tryptamine1 (5-HT1B/1D) receptor subtype agonist | Treatment of migraine and cluster headaches | 20(M) & 30 (F) | 70.7 | 15 | 21.46 |
| 22 | Provigil | A wakefulness-promoting agent | Treatment of excessive sleepiness | N/A | 92.7 | ~60 | 72.02 |
| 25 | Thalomid | An immunomodulatory agent | Treatment of skin conditions from leprosy | N/A | 91.4 | 55(+R)/66(−S) | 83.55 |
| 31 | Arava | A pyrimidine synthesis inhibitor | Treatment of rheumatoid arthritis | N/A | 94.5 | N/A | 91.37 |
| 32 | Gleevec | A protein-tyrosine kinase inhibitor | Treatment Philadelphia chromosome positive CML | 98 | 87.3 | 95 | 69.07 |
| 33 | Cialis | A selective inhibitor of cyclic guanosine monophosphate (cGMP)-specific phosphidiesterase type 5 (PDE5) | Treatment of erectile dysfuction | N/A | 79.7 | 94 | 76.54 |
| 34 | Reyataz | An azapeptide inhibitor of HIV-1 protease | Treatment of HIV infection | N/A | 79.7 | 86 | 76.54 |
| 35 | A.M. of Xeloda | A fluoropyrimidine carbamate with anti-neoplastic activity | Treatment for some types of cancer | N/A | 71.4 | <60 | 15.58 |
| 39 | Spectracef | A semi-synthetic cephalosporin antibiotic | Treatment of bacterial infections | 14 | 18.4 | N/A | 64.79 |
| 40 | A.M. of Spectracef | A semi-synthetic cephalosporin antibiotic | Treatment of bacterial infections | 14 | 78.9 | 88 | 46.24 |
| 42 | Colazal | A mesalamine (5-ASA) producing prodrug, anti-inflammatory drug | Treatment of active ulcerative colitis | N/A | 4.9 | >99 | 62.33 |
| 43 | A.M. of Arava | A pyrimidine synthesis inhibitor | Treatment of rheumatoid arthritis | N/A | 96.3 | >99.3 | 66.44 |
| 44 | Zetia | A selective inhibitor of the intestinal absorption of cholesterol and related phytosterols | Treatment of high blood cholesterol | N/A | 90.2 | >90 | 86.32 |
| 47 | Starlix | An antidiabetic agent used in the management of Type 2 diabetes mellitus | Treatment of Type 2 diabetes | 73 | 84.8 | 98 | 90.03 |
| 49 | Xifaxan | A semi-synthetic, nonsystemic antibiotic | Treatment of travelers' diarrhea | N/A | 61.4 | N/A | 83.72 |
| 53 | Mobic | A member of the enolic acid group of nonsteroidal anti-inflammatory drugs | Treatment of arthritis | 89 | 93 | ~99.4 | 97.45 |
| 63 | Temodar | An imidazotetrazine derivative | Treatment of recurrent malignant brain tumors | N/A | 85.1 | 15 | 21.49 |
| 64 | Keppra | An antiepileptic drug | Treatment of partial-onset or myoclonic seizures | 100 | 85.1 | <10 | 79.38 |
| 66 | Avandia | An antidiabetic agent | Treatment of Type 2 diabetes mellitus | 99 | 92.6 | 99.8 | 84.47 |
| 67 | Actos | An antidiabetic agent | Treatment of Type 2 diabetes mellitus | N/A | 93.7 | >99 | 96.24 |
| 69 | Tamiflu | An ethyl ester prodrug requiring ester hydrolysis for conversion to the active form, oseltamivir carboxylate | Treatment of the flu | N/A | 91.1 | 42 | 42.97 |
| 70 | A.M. of Tamiflu | The active metabolite of tamiflu | Treatment of the flu | 75 | 78.8 | 3 | 7.46 |
| 72 | Ritonavir | An inhibitor of the CYP3A-mediated metabolism of lopinavir (Kaletra) | Elevator of plasma levels of lopinavir | N/A | 41.9 | 98.5 | 91.79 |
| 73 | Benicar | A selective AT1 subtype angiotensin II receptor antagonist | Treatment of hypertension | 26 | 23.1 | 99 | 90.19 |
| Carboxylic Acid | |||||||
| # | Brand Name | Classification | Potency | Bioavailability (%) | Plasma Protein Binding (PPB)(%) | ||
| FDA data | ChemSilico Prediction | FDA data | ChemSilico Prediction | ||||
| 14 | Viagra | A selective inhibitor of cGMP-specific PDE5 | Treatment of erectile dysfunction | 40 | 78.6 | 95 | 89.07 |
| 19 | Zelnorm | A 5-HT4 receptor partial antagonist that binds with high affinity at human 5-HT4 receptors | Treatment of constipation and IBS | 10 | 77.1 | 98 | 74.44 |
| 21 | Frova | A selective 5-hydroxy-tryptamine1 (5-HT1B/1D) receptor subtype agonist | Treatment of migraine and cluster headaches | 20(M) & 30 (F) | 70.7 | 15 | 21.46 |
| 24 | Detrol | A competitive muscarinic receptor antagonist | Treatnment of an overactive bladder | >77 | 75 | 96.3 | 84.7 |
| 28 | Micardis | A nonpeptide angiotensin II receptor (type AT1) antagonist | Treatment of hypertension | 42(40mg) & 58(160mg) | 89.1 | >99.5 | 98.32 |
| 29 | Maxalt | A selective 5-hydrotryptamine1B/1D (5-HT 1B/1D) receptor agonist | Treatment of migraine headaches | 45 | 91.9 | 14 | 12.68 |
| 36 | A.M. of Atacand | A selective AT1 subtype angiotensin II receptor antagonist | Treatment of hypertension | 15 | 87 | >99 | 96.08 |
| 38 | Singulair | A selective and orally active leukotriene receptor antagonist that inhibits CysLT1 receptor | Long-term treatment of asthma | 64 | 82.7 | 99 | 97.65 |
| 47 | Starlix | An antidiabetic agent used in the management of Type 2 diabetes mellitus | Treatment of Type 2 diabetes | 73 | 84.8 | 98 | 90.03 |
| 51 | Nolvadex | A nonsteroidal antiestrogen | Treatment of breast cancer | N/A | 87.2 | N/A | 94.22 |
| 52 | A.M. of Detrol | A competitive muscarinic receptor antagonist | Treatnment of an overactive bladder | >77 | 85.1 | 64 | 79.38 |
| 62 | Exelon | A reversible cholinesterase inhibitor | Treatment of Alzheimer disease | 40 | 95 | 40 | 52.97 |
| 65 | Tequin | A synthetic broad-spectrum 8-methoxyfluoroquinolone antibacterial agent | Treatment of bacterial infection | 96 | 76.7 | 20 | 28.12 |
| 66 | Avandia | An antidiabetic agent | Treatment of Type 2 diabetes mellitus | 99 | 92.6 | 99.8 | 84.47 |
| 68 | Avelox | A synthetic broad spectrum antibacterial agent | Treatment of bacterial infection | 90 | 90.5 | 40 | 45.21 |
| 70 | A.M. of Tamiflu | The active metabolite of tamiflu | Treatment of the flu | 75 | 78.8 | 3 | 7.46 |
| 73 | Benicar | A selective AT1 subtype angiotensin II receptor antagonist | Treatment of hypertension | 26 | 80.1 | 99 | 94.69 |
| 75 | Ketek | A semi-synthetic antibacterial in the ketolide class | Treatment of bacterial infection | 57 | 23.1 | 65 | 90.19 |
| Ketone | |||||||
| # | Brand Name | Classification | Potency | Bioavailability (%) | Plasma Protein Binding (PPB)(%) | ||
| FDA data | ChemSilico Prediction | FDA data | ChemSilico Prediction | ||||
| 5 | Inspra | A blocker of aldosterone binding at the mineralocorticoid receptor | Treatment of HBP and heart failure | N/A | 90.7 | 50 | 43.74 |
| 16 | Orfadin | A synthetic reversible inhibitor of 4-hydroxyphenylpyruvate dioxygenase | Treatment of hereditary tyrosinemia type 1 | 90 | 82 | N/A | 73.15 |
| 23 | Aromasin | An irreversible, steroidal aromatase inactivator | Treatment of hormone-dependent breast cancer | >42 | 83.2 | 90 | 87.87 |
| 48 | Tasmar | An inhibitor of catechol-O-methyltransferase (COMT) | Treatment of Parkinson's disease | 65 | 61.4 | >99.9 | 90.55 |
| 49 | Xifaxan | A semi-synthetic, nonsystemic antibiotic | Treatment of travelers' diarrhea | N/A | 61.4 | N/A | 83.72 |
| 60 | Trileptal | An antiepileptic drug | Treatment of certain types of seizures | N/A | 94 | 40 | 48.2 |
| 65 | Tequin | A synthetic broad-spectrum 8-methoxyfluoroquinolone antibacterial agent | Treatment of bacterial infection | 96 | 76.7 | 20 | 28.12 |
| 68 | Avelox | A synthetic broad spectrum antibacterial agent | Treatment of bacterial infection | 90 | 90.5 | 40 | 45.21 |
| 73 | Benicar | A selective AT1 subtype angiotensin II receptor antagonist | Treatment of hypertension | 26 | 23.1 | 99 | 90.19 |
| 75 | Ketek | A semisynthetic antibacterial in the ketolide class | Treatment of bacterial infection | 57 | 55.1 | 65 | 45.79 |
 : percent difference greater than 10.
: percent difference greater than 10.  : values in different PPB ranges (Low: <45%, Middle: 45–70%. & High: >70%)
: values in different PPB ranges (Low: <45%, Middle: 45–70%. & High: >70%)
Bioavailability predictions for 4 of the drugs were low. For Keppra®, its drug profile value of 100% is unrealistic. 100% is the value used for an IV dose, and it would be extremely unlikely that all of an oral dose would reach the systemic blood. In fact, the FDA profile indicates it is “almost completely absorbed” (emphasis added). Tequin® is a racemic compound; ChemSilico predictions may not take this into account. In the case of Ketek®, the low prediction refers to its carboxylic acid properties. When considered as a ketone, the predicted bioavailability is comparable to empirical data. For Gleevec® there was no obvious explanation for the low prediction; however, as is the case with the majority of consumer drugs, including all four listed above, a number of excipients are present in the formulation that increase solubility and thus may increase absorption. Bioavailability predictions for 14 of the drugs were high. Drug profiles for Levitra®, Viagra®, Uroxatral®, Zelnorm®, Avodart®, Frova®, Micardis®, Maxalt®, and Exelon® listed volumes of distribution (VD) over 100 liters, indicating extensive extravascular binding which may not be reflected in ChemSilico predictions. In addition to having a high VD, Frova® and Micardis® are racemic mixtures with differing bioavailabilities for individual enantiomers. Although Singulair® and the active metabolite of Atacand® are also racemic, their drug profiles give only one value for bioavailability. There was no VD reported for Aromasin®; however, its FDA profile indicated it is “distributed extensively into tissues”. In the case of Starlix® and Benicar®, the VD indicates distribution to interstitial fluid. In addition to being an important plasma protein, albumin is also present in interstitial fluids in concentrations up to 60% that of plasma; and Starlix®, in particular, is reported to have a high affinity for albumin. A number of factors can complicate the predictability of drug behavior, and it should be noted that specific data that might help explain discrepancies between predicted and clinically-derived values are not always available. In some instances, it is recommended that the drug be administered with food which can delay gastric emptying, change gastrointestinal pH, increase splanchnic blood flow, and have physical and chemical interactions with the drug itself. Many drugs are administered as film-coated tablets and/or contain excipients expressly included to slow their dissolution, with an increase in first-pass effect (high pre-systemic elimination). Such drugs often exhibit greater variability among individuals in terms of bioavailability. Clinical assessment of bioavailability is most often determined from measurement of drug concentrations in samples taken from venous blood, the composition of which can differ greatly from arterial blood. In addition, values for bioavailability are often obtained from small clinical studies, which may poorly reflect the general population. The bioavailability value of 60% for Avodart® was determined using only 5 subjects, and values ranged from 40–94% [3–5]. Finally, some orally administered drugs also contain substances used simply as fillers that are perceived to be inert. This may, however, not always be the case. For example, calcium salts used as fillers were reported to interfere with the absorption of tetracycline [26].
Conclusions
The KnowItAll® platform [8–12] offers a fully integrated environment that is designed so that the user can transfer data from application to application without having to leave the main interface or open another program, which makes study set up easy and intuitive. The browseable high-quality mass of data [3–5] can be mined, analyzed, and interpreted using KnowItAll®. The FDA Consumer Drug Database© [1, 2] proved to be a valuable resource of available structurally diverse drug data to generate predictive computational models for the interpretation of ADME/Tox properties based on chemical structure. Due to the dynamic and complex process of metabolism, prediction of the biological properties was found to be very complicated. Some of the difficulties in the current project also arose due to the differences in the reporting of the physico-chemical data that were used to correlate to the SAR patterns [1]. This research has clearly demonstrated that possible reasons for the outliers could be sought by evaluating consistencies in physiological properties (such as BIO and PPB) among the drugs sharing the same functional group.
Supplementary Material
Acknowledgements
This research was supported by grant number 2 P2O RR016472-09 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). This IDeA Network of Biomedical Research Excellence (INBRE) grant to the state of Delaware was obtained under the leadership of the University of Delaware, and the authors sincerely appreciate their efforts.
Footnotes
Fumie Koyoshi a Wesley College Biology major completed this project during an INBRE supported Undergraduate Research Assistantship in the Directed Research Program in Chemistry at Wesley. Initial results of this project when presented as a poster in the Division of Chemical Education (CHED) - Medicinal Chemistry Section, at the 235th National American Chemical Society (ACS) Meeting, New Orleans, LA, April 6–8, 2008; earned a Certificate of Recognition from the ACS-CHED-Medicinal Chemistry Section. On graduation from Wesley, Fumie joined the University of Pennsylvania Hospital, School of Medical Technology, Philadelphia, PA.
References
- 1.D’Souza MJ, Kyoshi F. Extracting Relevant Information from FDA Drug Files to Create a Structurally Diverse Drug Database Using KnowItAll®. Pharmaceutical Reviews. 2009;7(3) ISSN 1918-5561. [PMC free article] [PubMed] [Google Scholar]
- 2.D’Souza MJ. HaveItAll - ADME/Tox Experimental Databases Datasheet. Bio-Rad Laboratories; 2008. FDA Consumer Drug database – 2007. Bulletin # INF-96199. [Google Scholar]
- 3.Food and Drug Administration (FDA) profiles. Retrieved from http://www.fda.gov/cder/drug/default.htm.
- 4.Drugs@FDA. Retrieved from http://www.accessdata.fda.gov/Scripts/cder/DrugsatFDA/
- 5.FDA Safety Information and Adverse Reporting program (MedWatch) Retrieved from http://www.fda.gov/medwatch/
- 6.NIH Molecular Libraries-Small Molecule Repository. Retrieved from http://mlsmr.glpg.com/MLSMR_HomePage/submitcompounds.html.
- 7.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: A Comprehensive Resource for In Silico Drug Discovery and Exploration. Nucleic Acid Research. 2006;34(Database Issue):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.KnowItAll® Informatics System Desktop Solutions. Retrieved from http://www.knowitall.com/
- 9.KnowItAll® U System. Retrieved from http://www.knowitallu.com/
- 10.D’Souza MJ. KnowItAll® - Software Reviews. Chemistry World. 2005;2(9):70–71. [Google Scholar]
- 11.D’Souza MJ. KnowItAll® U System - Software Reviews. Chemistry World. 2007;4(11):70–72. [Google Scholar]
- 12.Anand V, Gera M, Kumar V, Karwasara P, Kataria M, Kukkar V. Comparative Evaluation of Freely Available Chemical Structure Drawing Software. Pharmaceutical Reviews. 2008;6(2) ISSN 1918-5561. [Google Scholar]
- 13.Scandone M, Kernan D. Data Management in the Spectral Laboratory. International Labmate. 2004:25–26. [Google Scholar]
- 14.Banik GM. In Silico ADME/Tox Prediction: The More, the Merrier. Current Drug Discovery. 2004:31–34. [Google Scholar]
- 15.D’Souza ML, Abshear T, Banik GM, Nedwed K, Peng C. A Model Validation and Consensus Building Environment. SAR and QSAR in Environmental Research. 2006;17(3):311–321. doi: 10.1080/10659360600787551. [DOI] [PubMed] [Google Scholar]
- 16.Bidault Y. A Flexible Approach for Optimizing In Silico ADME/Tox Characterization of Lead Candidates. Expert Opinion on Drug Metabolism and Toxicity. 2006;2(1):157–168. doi: 10.1517/17425255.2.1.157. [DOI] [PubMed] [Google Scholar]
- 17.Dearden JC. In Silico Predictions of ADMET Properties: How Far Have We Come? Expert Opinion on Drug Metabolism and Toxicity. 2007;3(5):635–639. doi: 10.1517/17425255.3.5.635. [DOI] [PubMed] [Google Scholar]
- 18.Swaan PW, Ekins S. Reengineering the Pharmaceutical Industry by Crash-Testing Molecules. Drug Discovery Today. 2005;10(17):1191–1200. doi: 10.1016/S1359-6446(05)03557-9. [DOI] [PubMed] [Google Scholar]
- 19.Johnson DE, Rodgers AD. Computational Toxicology: Heading Toward More Relevance in Drug Discovery and Development. Current Opinion in Drug Discovery and Development. 2006;9:29–37. [PubMed] [Google Scholar]
- 20.Franklin RB. In Silico Studies in ADME/Tox: Caveat Emptor. Current Computer-Aided Drug Design. 2009;5(2):128–138. [Google Scholar]
- 21.Dearden J, Worth A. In Silico prediction of physicochemical properties. JRC Scientific and Technical Reports. :1–68. EUR 23051, EN-2007. [Google Scholar]
- 22.D’Souza MJ, Koyoshi F, Everett LM. Structure Activity Relationship (SAR) Patterns Observed Within a Series of Unrelated Common Consumer Drugs. Bioinformatics, Computational Biology Genomics, and Chemoinformatics. 2009:1–6. [Google Scholar]
- 23. Retrieved from http://www.chemsilico.com/CS_methods/methods_descriptors.html.
- 24.Crossley R. The Relevance of Chirality to the Study of Biological Activity. Tetrahedron. 1992;48(38):8155–8178. [Google Scholar]
- 25. Retrieved from http://www.chemsilico.com/CS_prPB/PBintro.html.
- 26.Dave RH. Drug Topics. 2008. Oct 24, retrieved from http://drugtopics.modernmedicine.com.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.