Abstract
There are several factors like angiogenesis, lymphangiogenesis, genetic alterations, mutational factors that are involved in malignant transformation of potentially malignant oral lesions (PMOLs) to oral squamous cell carcinoma (OSCC). Fibroblast growth factor-2 (FGF-2) is one of the prototypes of the large family of growth factors that bind heparin. FGF-2 induces angiogenesis and its receptors may play a role in synthesis of collagen. FGFs are involved in transmission of signals between the epithelium and connective tissue, and influence growth and differentiation of a wide variety of tissue including epithelia. The present study was undertaken to analyze expression of FGF-2 and its receptors FGFR-2 and FGFR-3 in 72 PMOLs, 108 OSCC and 52 healthy controls, and their role in risk assessment for malignant transformation of Leukoplakia (LKP) and Oral submucous fibrosis (OSMF) to OSCC. Immunohistochemistry was performed using antibodies against FGF-2, FGFR-2 and FGFR-3. IHC results were validated by Real Time PCR. Expression of FGF-2, FGFR-2 and FGFR-3 was upregulated from PMOLs to OSCC. While 90% (9/10) of PMOLs which showed malignant transformation (transformed) expressed FGF-2, only 24.19% cases (15/62) of PMOLs which were not transformed (untransformed) to OSCC expressed FGF-2. Similarly, FGFR-2 expression was seen in 16/62 (25.81%) of untransformed PMOLs and 8/10 (80%) cases of transformed PMOLs. FGFR-3 expression was observed in 23/62 (37.10%) cases of untransformed PMOLs and 6/10 (60%) cases of transformed PMOLs. A significant association of FGF-2 and FGFR-2 expression with malignant transformation from PMOLs to OSCC was observed both at phenotypic and molecular level. The results suggest that FGF-2 and FGFR-2 may be useful as biomarkers of malignant transformation in patients with OSMF and LKP.
Introduction
Oral squamous cell carcinoma (OSCC) is a multifactorial and complex disease. Several factors including angiogenesis, lymphangiogenesis, alterations in expression or structure of tumor suppressor genes and oncogenes and their proteins, etc are involved in malignant transformation of potentially malignant oral lesions (PMOLs) to OSCC [1, 2].
The malignant transformation rates of PMOLs show a great variation; for example, 10–20% of hyperkeratosis or epithelial hyperplasia or epithelial dysplasia may transform to cancer and the estimated annual rate is 1.4–7% [3, 4]. Lumerman in 1995 reported that 6.6%–36% of epithelial dysplasias may transform to invasive SCC [5]. The malignant transformation rate of leukoplakia (LKP) has been reported to range from 0.13 to 17.5% [6,7], 1.1% in Oral lichen planus [8] and 2.3–7.6% in oral submucous fibrosis (OSMF), during 10–17 years of follow-up [9,10]. In a study from Taiwan [11], the malignant transformation rate in an average follow-up period of 42.6 months ranged from 1.9 to 5.4% for various types of PMOLs.
At present, histologic assessment of epithelial dysplasia is the gold standard for determining the malignant transformation risk of PMOLs. However, the problem of inter and intra observer variation is always there in the histopathologic assessment of presence and severity of epithelial dysplasia [12, 13]. In spite of tremendous progress in the field of molecular biology, there is as yet no single marker or set of markers that reliably predict malignant transformation of LKP in individual patients [14]. Therefore, objective biomarkers which do not require the ability to recognize morphologic changes are needed to evaluate the risk of malignant transformation of PMOLs.
FGF-2 is one of the prototypes of a large family of growth factors which is expressed in several tissues and has a wide scope of biologic activities [15]. It binds to low affinity heparin sulfate proteoglycans that are involved in the interaction with high affinity receptors that in turn mediate the cellular response to FGF-2 [16, 17]. The FGF receptor family consists of four members (FGFR-1(flg), FGFR-2 (bek), FGFR-3 and FGFR-4) that have 55–72% amino acid homology [18].
FGF-2 induces angiogenesis [19, 20, 21] and its receptors may play a role in synthesis of collagen. It is involved in the transmission of signals between the epithelium and connective tissue, and influences growth and differentiation of a wide variety of tissue including epithelia [22].
Studies have reported FGF-2 overexpression in high grade malignant tumours and malignant transformation of normal cells transfected with FGF-2 gene [23, 24]. FGF-2 is involved in the invasion of cancer cells and the proliferation of fibroblasts around cancer cells in an autocrine or paracrine fashion [25]. The study of expression of this factor in head and neck carcinomas (HNC) has yielded controversial results [26, 27, 28, 29]. It has been suggested that deployment of FGFR-specific tyrosine kinase inhibitors as single agent or in combination with EGFR inhibitors may be one of the effective therapeutic strategies in Head and Neck squamous cell carcinoma [30].
The aim of present study was to analyze the expression of FGF-2 and its receptors FGFR-2 and FGFR-3 in PMOLs and OSCC and their role in risk assessment for malignant transformation of LKP and OSMF to OSCC.
Materials and Methods
Subjects and sample collection
Tissue biopsies were obtained from cases of 72 PMOLs which included 43 cases of LKP and 29 cases of OSMF, 108 OSCC and 52 healthy controls from the Departments of Oral and Maxillofacial Surgery and Otorhinolaryngology, King George’s Medical University Lucknow after obtaining the Institutional Ethical approval and informed written consent from patients during the year 2007 to 2012. Healthy oral tissues were obtained from patients undergoing cosmetic surgery, who otherwise did not have any infective or inflammatory oral lesion. A small part of the tissue was snap frozen for molecular work and was stored at -80°C. The inclusion criteria followed for PMOLs patients was as described by Ho et al. (2009) [31]. Relevant clinical and demographic details of each patient were recorded on a structured proforma.
Histopathological examination
All tissues were fixed in 10% neutral buffered formalin and processed for histopathological examination as per standard procedure. 5μm thick sections were cut and stained with haematoxylin and eosin (H&E). Sections were reviewed by two independent pathologists and histological diagnosis was made as per WHO criteria and following the classification by Warnakulasuriya et al 2007 [32] where the clinically diagnosed cases of leukoplakia on histopathology of tissue biopsies, are divided into leukoplakia with dysplasia and leukoplakia without dysplasia. Among 29 cases of the OSMF group in the present study, 11 were without dysplasia, 4 showed mild dysplasia, 5 with moderate dysplasia and 9 cases showed severe dysplasia. Among 43 cases in the Leukoplakia group, 10 cases showed no dysplasia, 3 had mild dysplasia, 10 with moderate dysplasia and 20 cases showed severe dysplasia.
The patients were followed up every 2 months in the first year, every 3 months in the second year, and every 4 to 6 months thereafter. The patients were followed to a maximum of 5 years. Between the follow ups some patients with leukoplakic lesions and OSMF without dysplasia, underwent treatment at an early stage and were discontinued from the follow-up.
Immunohistochemistry
Sections were deparaffinized in xylene followed by hydration in descending ethanol grades. Endogenous peroxidase was blocked in 3% H2O2 in methanol for 30 min. Antigen retrieval was performed by heating specimens for 15 min at 95°C in citrate buffer (pH 6.0) using an EZ antigen retriever system (BioGenex, USA). Sections were then incubated with power block (BioGenex, USA) for 10 min to reduce nonspecific antibody binding. The sections were incubated overnight at 4°C with primary antibodies. Mouse monoclonal antibodies against human FGF-2, FGFR-2 and FGFR-3 (Santa Cruz Biotechnology Inc., Santa Cruz, CA) were used. Primary antibodies were detected using super sensitive polymer-HRP IHC detection system (BioGenex, USA). After thorough washing with Tris buffered saline (TBS; pH 7.4) sections were treated with super enhancer for 20 min at room temperature followed by incubation with poly-HRP reagent for 30 min at room temperature. After three washes with TBS, DAB substrate (3, 3’-diaminobenzidine tetra hydrochloride) was applied to the sections for 5–10 min in the dark. Sections were counterstained with hematoxylin, dehydrated with ascending ethanol grades and xylene and mounted permanently with DPX. Negative control sections were processed by omitting primary antibody and breast carcinoma tissue was used as positive control.
Evaluation of staining
The level of expression was assessed by semiquantitative scoring which included the overall percentage area of the lesion stained positive (0–100%), and the staining intensity (0–3). In all the cases, the expression in epithelium, endothelial cells and stroma was analyzed. Grading for percentage area positivity was done as follows: <10% = 0, 10–25% = 1, 25–50% = 2, 50–75% = 3, >75 = 4. To evaluate the intensity, grading was done as; 0 = none, 1 = mild, 2 = moderate, 3 = strong staining. The percentage score (0–4) was multiplied by the intensity score (0–3) and a final score was assigned, 0–4 as negative staining, 5–12 as positive staining [33]. At least five best fields were taken for interpreting results of percentage area.
Quantitative real-time PCR (qPCR) for FGF-2, FGFR-2 and FGFR-3 genes
Total RNA extraction
RNA was extracted from frozen tissue samples with Trizol reagent (Invitrogen, Carlsbad, CA). RNA purification was done by DNase1 treatment (Invitrogen, Amplification grade). In brief, 1μg of total RNA sample was treated with 10X DNase I reaction buffer and DNase I (1U/10μl) and incubated for 15 min at room temperature followed by inactivation of DNase I with 25mM EDTA at 65°C for 10 min. RNA was quantified by Qubit 2.0 fluorometer (Molecular Probes, Invitrogen, USA).
cDNA synthesis
250 ng of the total RNA was subjected to reverse transcription using random hexamer primers with Gene AMP RNA PCR kit (Applied Biosystems, Foster city, CA) for cDNA synthesis, as per manufacturer’s instructions. Briefly, the 20μl reaction was performed in 3 steps. Step 1 at 25°C for 10 min, step 2 at 37°C for 2 hrs and finally step 3 at 85°C for 5min. cDNA was stored at -20°C for real time PCR.
Quanitative real time PCR (qPCR)
qPCR was performed using StepOne Real-time PCR system (Applied Biosystems, USA) in the presence of SYBR Green fluorescent dye according to the manufacturer’s instructions. Briefly, 20μl of the reaction mixture consisting of reverse transcribed cDNA, 2X SYBR Green master mix containing dNTPs, ROX dye and 10μM of forward and reverse primers was dispensed into a fast optical 48-well real time PCR reaction plate (Applied Biosystems, USA). The PCR primers for FGF-2 [34], FGFR-2 [34], and FGFR-3 [30] genes were selected from a published article and synthesized by MWG, India. Primer sequences were rechecked using Primer Express software 3.0 (Applied Biosystems, USA) and checked for homology by Blast sequence analysis (National Centre for Biotechnology Information). Following primers sequences were used: β-actin (endogenous control): forward 5’-GAGACCTTCAACACCCCAGCC-3’; reverse 5’-AGACGCAGGATGGCATGGG-3’, FGF-2: forward5’-ATGGCAGCCGGGAGCATCACCCACG-3’; reverse: 5’TCAGCTCTTCGCAGACATTGGAAG-3’, FGFR-2: forward 5’-TCCACATGGAGATATGGAACAGGA-3’; reverse 5’-GGAGCTATTTATCCCCGAGTG-3’ FGFR-3 forward 5’- CCATCGGGCATTGACAAGGAC-3’, reverse 5’- GCATCGTCTTTCAGCATCTTCAC-3’. Thermal cycle conditions consisted of on initial denaturation incubation at 95°C for 10min followed by 40 cycle of 15sec incubation at 95°C and 60sec incubation at 60°C followed by the thermal dissociation (melt curve) protocol for fluorescence detection. Gene expression level was determined using the 2-ΔΔCt method using beta-actin as an endogenous control. A negative control without a template was run in parallel to assess the overall specificity of the reaction. All reactions were run in replicates. Data are presented as “relative gene expression”.
Statistical analysis
Statistical analysis was performed using version 17.0 SPSS software for windows (SPSS, INC, Chicago, IL). For assessing proportional data, chi-square test was carried out. Correlation between FGF-2, FGFR-2 & FGFR-3 was determined by Spearman correlation coefficient. Logistic regression was applied to evaluate hazard ratios for the malignant transformed PMOLs. Odds ratios (OR) with 95% confidence interval (95% CI) was calculated and p-values were reported. For all the tests, a p-value < 0.05 was considered statistically significant.
Results
Clinicopathological features
The study population comprised of 172 (74.14%) males and 60 (25.86%) females with a median age of 42 years (range 8 to 85 years). In PMOL group dysplasia was present in 51 out of 72 cases. In OSCC group, histologically 8 cases were poorly differentiated, 25 cases were moderately differentiated and 75 cases were well differentiated. 62 tumors were clinical stage I or II, and 46 tumors were clinical stage III or IV. Lymph node involvement was found in 41 cases of OSCC.
Immunohistochemical expression of FGF-2 and its receptors (FGFR-2 and FGFR-3) in PMOLs and OSCC
Immunhistochemical expression of FGF-2, FGFR-2 and FGFR-3 is shown in Table 1. Expression of FGF-2 was cytoplasmic in basal, parabasal layers in tissues with lining epithelium, tumor cells and also in stroma as shown in “Fig 1A, 1B, 1C and 1D”. FGF-2 positivity was observed in 49.08% (53/108) OSCC cases, 31.95% (23/72) PMOLs and in 26.92% (14/ 52) control cases. Expression of FGFR-2 was found to be cytoplasmic in full thickness of epithelium, tumor cells and stromal cells as shown in “Fig 2A, 2B and 2C”. FGFR-2 expression was observed in 57.41% (62/108) cases of OSCC, 33.33% (24/72) cases of PMOLs and 15.38% (8/52) cases of controls. Cytoplasmic FGFR-3 expression was observed in upper and midstratum layer of lining epithelium, stromal fibroblast cells, tumor cells and seldom in endothelium of tumor stroma as shown in “Fig 3A, 3B and 3C”. 75% cases (81/108) of OSCC, 40.28% cases (29/72) of PMOLs and 36.54% (19/52) controls stained positive for FGFR-3.
Table 1. Immunohistochemical expression of FGF-2, FGFR-2 and FGFR-3 in OSCC and PMOLs.
| Groups | FGF2Negative | FGF-2 Positive | P -value | FGFR-2Negative | FGFR-2 Positive | P -value | FGFR-3Negative | FGFR-3 Positive | P -value |
|---|---|---|---|---|---|---|---|---|---|
| N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | ||||
| Controls | 38(73.0) | 14(26.92) | 44(84.62) | 8(15.38) | 33(63.46) | 19(36.54) | |||
| PMOLs | 49(68.0) | 23(31.95) | 0.445 | 48(66.67) | 24(33.33) | 0.024 | 43(59.72) | 29(40.28) | 0.428 |
| OSMF | 20(68.9) | 9(31.03) | 0.694 | 22(75.86) | 7(24.14) | 0.331 | 18(62.07) | 11(37.93) | 0.282 |
| LKP | 29(67.4) | 14(32.56) | 0.402 | 26(60.47) | 17(39.53) | 0.008 | 25(58.14) | 18(41.86) | 0.296 |
| OSCC | 55(50.92) | 53(49.08) | 0.008 | 46(42.59) | 62(57.41) | 0.000 | 27(25) | 81(75) | 0.002 |
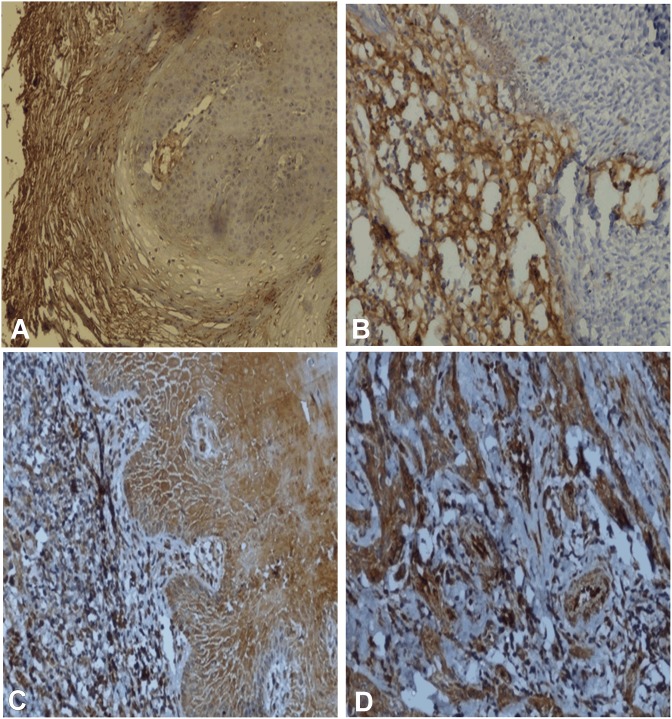
Fig 1. Immunohistochemical staining of FGF-2.
(A-D). A, FGF-2 positivity in transformed case of Leukoplakia with dysplasia X 100. B, FGF-2 positivity in transformed cases of OSMF in fibrosis area X 100. C, FGF-2 positivity in epithelium of OSCC X 400. D, FGF-2 positivity in tumor cells OSCC X 400.
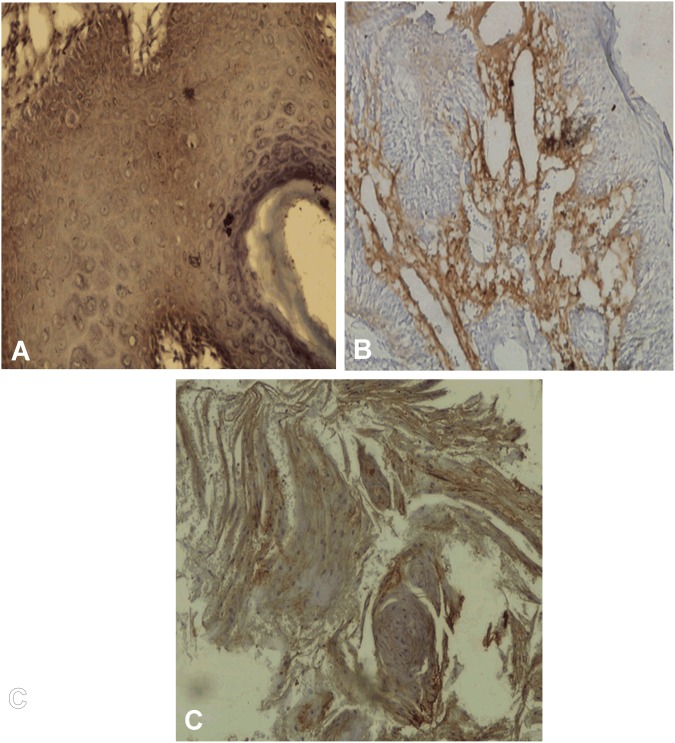
Fig 2. Immunohistochemical staining of FGFR-2 (A-C).
A, FGFR-2 positivity in transformed cases of Leukoplakia with dysplasia X 200. B, FGFR-2 positivity in transformed case of OSMF in fibrosis area X 100. C, FGFR-2 positivity in OSCC X 100.
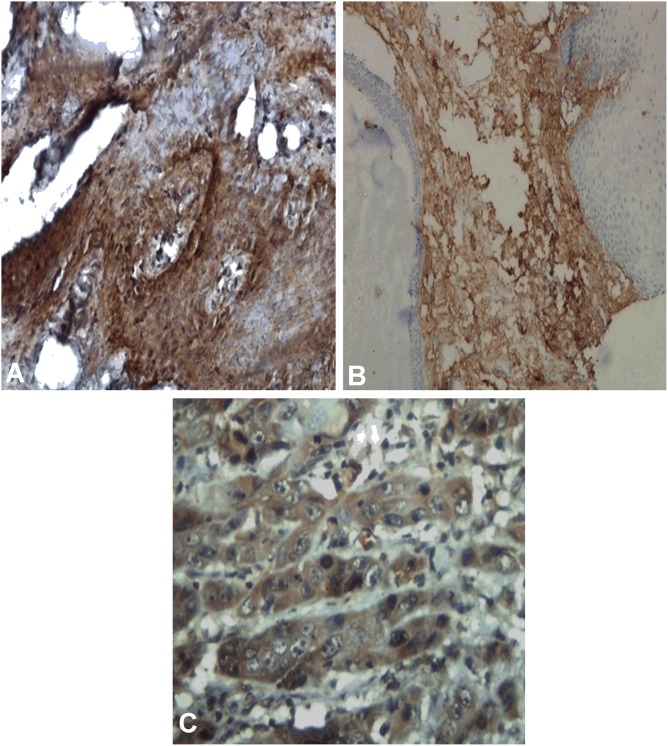
Fig 3. Immunohistochemical staining of FGFR-3 (A-C).
A, FGFR-3 positivity in epithelium of Leukoplakia with dysplasia in transformed case X 200. B, FGFR-3 positivity in transformed case of subepithelial fibrosis area of OSMF X 100. C, FGFR-3 positivity in tumor cells of OSCC X 400.
Association of FGF-2, FGFR-2 and FGFR-3 with clinicopathological parameters in PMOLs and OSCC
Association of FGF-2, FGFR-2 and FGFR-3 with clinicopathological parameters in PMOLs and OSCC is shown in Tables 2 and 3. FGF-2 expression was significantly associated with sex (p<0.013) and tumor differentiation (p<0.0001) in OSCC patients. While, FGFR-2 was not associated with any clinicopathological features in oral cancer, FGFR- 3 expression was significantly higher (p<0.043) in stages I-II. No association of FGF-2, FGFR-2 and FGFR-3 was found with clinicopathological parameters in PMOLs.
Table 2. Relation of FGF-2, FGFR-2 and FGFR-3 expression with clinico-pathological parameters in PMOLs.
| Variables | FGF-2positive | FGF-2 negative | P-value | FGFR-2positive | FGFR-2negative | P-value | FGFR-3positive | FGFR-3negative | P-value |
|---|---|---|---|---|---|---|---|---|---|
| Age | |||||||||
| <42 | 16 | 35 | 0.582 | 15 | 36 | 0.271 | 21 | 30 | 0.809 |
| ≥42 | 8 | 13 | 9 | 12 | 8 | 13 | |||
| Sex | |||||||||
| Male | 17 | 37 | 0.564 | 20 | 34 | 0.248 | 21 | 33 | 0.677 |
| Female | 7 | 11 | 4 | 14 | 8 | 10 | |||
| Dysplasia | |||||||||
| Present | 19 | 32 | 0.271 | 17 | 34 | 1.000 | 18 | 33 | 0.179 |
| Absent | 5 | 16 | 7 | 14 | 11 | 10 | |||
| Tobacco chewing habit | |||||||||
| Present | 16 | 32 | 1.000 | 16 | 32 | 1.000 | 20 | 28 | 0.734 |
| Absent | 8 | 16 | 8 | 16 | 9 | 15 | |||
| Alcohol | |||||||||
| Present | 8 | 20 | 0.494 | 10 | 18 | 0.732 | 9 | 19 | 0.262 |
| Absent | 16 | 28 | 14 | 30 | 20 | 24 | |||
| Smoking | |||||||||
| Present | 12 | 24 | 1.000 | 10 | 26 | 0.317 | 12 | 24 | 0.230 |
| Absent | 12 | 24 | 14 | 22 | 17 | 19 |
Table 3. Association of FGF-2, FGFR-2 and FGFR-3 with clinico-pathological features in OSCC.
| Variables | FGF-2 positive | FGF-2Negative | P-value | FGFR-2positive | FGFR-2negative | P-value | FGFR-3positive | FGFR-3negative | P-value |
|---|---|---|---|---|---|---|---|---|---|
| Age | |||||||||
| <42 | 14 | 16 | 0.756 | 19 | 11 | 0.440 | 26 | 4 | 0.082 |
| ≥42 | 39 | 39 | 43 | 35 | 55 | 23 | |||
| Sex | |||||||||
| Male | 49 | 41 | 0.013 | 55 | 35 | 0.080 | 69 | 21 | 0.371 |
| Female | 4 | 14 | 7 | 11 | 12 | 6 | |||
| Lymph node | |||||||||
| Positive | 16 | 25 | 0.102 | 20 | 21 | 0.156 | 27 | 14 | 0.086 |
| Negative | 37 | 30 | 42 | 25 | 54 | 13 | |||
| Differentiation | |||||||||
| WD | 32 | 43 | 0.000 | 40 | 35 | 0.069 | 52 | 23 | 0.077 |
| MD | 19 | 6 | 19 | 6 | 23 | 2 | |||
| WD | 2 | 6 | 3 | 5 | 6 | 2 | |||
| Tumor stage | |||||||||
| Stage I-II | 31 | 31 | 0.823 | 31 | 31 | 0.071 | 42 | 20 | 0.043 |
| StageIII-IV | 22 | 24 | 31 | 15 | 39 | 7 | |||
| Tobacco chewing habit | |||||||||
| Present | 43 | 41 | 0.410 | 50 | 34 | 0.405 | 61 | 23 | 0.285 |
| Absent | 10 | 14 | 12 | 12 | 20 | 4 | |||
| Alcohol | |||||||||
| Present | 28 | 24 | 0.339 | 27 | 25 | 0.267 | 37 | 15 | 0.374 |
| Absent | 25 | 31 | 35 | 21 | 44 | 12 | |||
| Smoking | |||||||||
| Present | 46 | 44 | 0.344 | 50 | 40 | 0.384 | 67 | 23 | 0.766 |
| Absent | 7 | 11 | 12 | 6 | 14 | 4 |
Quantitative real time PCR for FGF-2, FGFR-2 and FGFR-3
Real time PCR was performed to validate the results of immunohistochemistry as shown in Table 4, “Fig 4A”. The same tissue specimens that overexpresed FGF-2 and receptors by IHC were subjected to PCR.
Table 4. Gene expression profile of FGF-2, FGFR-2 & FGFR-3 by Quantitative Real—time PCR.
| Gene | Controls(N = 11) | LKP(N = 8) | OSCC(N = 11) | OSMF(N = 29) |
|---|---|---|---|---|
| FGF-2 | ||||
| Fold Change | 1 | 21.75 | 28.62 | 6.79 |
| SE | 0.2730769 | 0.45445 | 0.6142065 | 0.3886392 |
| P-value | 0.002 | 0.000 | 0.001 | |
| FGFR-2 | ||||
| Fold Change | 1 | 16.79 | 20.21 | 9.72 |
| SE | 0.3936878 | 0.6452975 | 0.496684 | 0.9335131 |
| P-value | 0.065 | 0.000 | 0.322 | |
| FGFR-3 | ||||
| Fold Change | 1 | 4.45 | 7.05 | 3.95 |
| SE | 0.333856 | 0.7594367 | 0.6345427 | 0.3587593 |
| P-value | 0.008 | 0.000 | 0.005 |
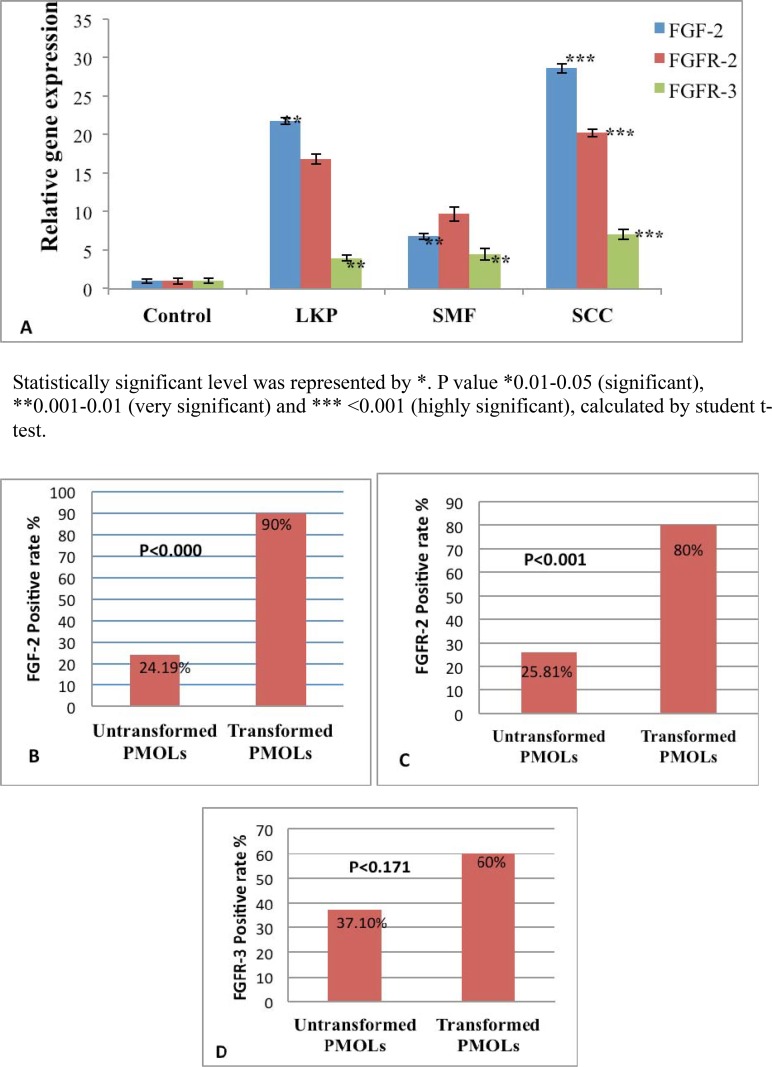
Fig 4. Real time PCR results of FGF-2, FGFR-2 and FGFR-3 and frequency of FGF-2, FGFR-2 and FGFR-3 malignant transformation rate (A-D).
Bar diagram showing fold change expression of FGF-2, FGFR-2 and FGFR-3 by Real Time. A, PCR in OSMF, LKP, OSCC and healthy controls. B, Frequency of FGF-2 malignant transformation rate in PMOLs. C, Frequency of FGFR-2 malignant transformation rate in PMOLs. D, Frequency of FGFR-3 malignant transformation rate in PMOLs.
We found that OSCC patients had significantly higher level of FGF-2, FGFR-2 and FGFR-3 expression followed by LKP and OSMF. In OSCC patients relative gene expression of FGF-2 was 28.62 fold higher, FGFR-2 was 20.21 fold higher and FGFR-3 was 7.05 fold higher as compared to healthy controls.
Correlation between FGF-2, FGFR-2 and FGFR-3 in PMOLs and OSCC
In PMOLs, FGF-2 was significantly correlated with FGFR-2 (rs = 0.313, p<0.008). FGFR-2 showed significant correlation with FGFR-3 (rs = 0.260, p<0.027). In OSCC, FGF-2 was significantly correlated with FGFR-2 (rs = 0.321, p<0.001) and FGFR-3 (rs = 0.353, p<0.000). As in PMOLs, FGFR-2 showed significant correlation with FGFR-3 (rs = 0.368, p<0.000).
Malignant transformation (from PMOLs to OSCC) and association with FGF-2, FGFR-2 and FGFR-2
Malignant transformation of PMOLs to OSCC was observed in 13.89% (10/72) cases (3 OSMF and 7 LKP cases) as shown in Table 5. Some cases of both OSMF (n = 3) and LKP (n = 4) groups, with either no dysplasia or mild dysplasia regressed after the course of treatment.
Table 5. Patients characteristic of malignant transformed and untransformed cases in PMOLS.
| Patients Characteristics | Untransformed PMOLs (N = 62) | Malignant transformed PMOLs (N = 10) | P-value |
|---|---|---|---|
| Age (Years) | |||
| Mean ± SD | 37.81± 11.98 | 43.60± 18.65 | 0.383 |
| Range | 20–80 | 22–75 | |
| Sex N (%) | |||
| Female | 17 (27.42%) | 1 (10%) | 0.238 |
| Male | 45 (72.58%) | 9 (90%) | |
| Dysplasia | |||
| Present | 41 (66.13%) | 10 (100%) | 0.029 |
| Mild | 7 | 0 | |
| Moderate | 15 | 0 | |
| Severe | 19 | 10 | |
| Absent | 21 (33.87%) | 0 (0%) | |
| Tobacco chewing habit | |||
| Present | 40 (64.52%) | 8 (80%) | 0.335 |
| Absent | 22 (35.48%) | 2 (20%) | |
| Alcohol | |||
| Present | 24 (38.71%) | 4 (40%) | 0.938 |
| Absent | 38 (61.29%) | 6 (60%) | |
| Smoke | |||
| Present | 29 (46.77%) | 7 (70%) | 0.173 |
| Absent | 33 (53.23%) | 3 (30%) | |
| Follow—up | |||
| Mean | 26.21 | 29.41 | 0.749 |
| Range(months) | 3–55 | 10–60 | |
| FGF-2 expression | |||
| Negative | 47 (75.81%) | 1 (10%) | 0.000 |
| Positive | 15 (24.19%) | 9 (90%) | |
| FGFR-2 expression | |||
| Negative | 46 (74.19%) | 2 (20%) | 0.001 |
| Positive | 16 (25.81%) | 8 (80%) | |
| FGFR-3 expression | |||
| Negative | 39 (62.90%) | 4 (40%) | 0.171 |
| Positive | 23 (37.10%) | 6 (60%) |
FGF-2 expression was seen in 15/62 cases (24.19%) of untransformed PMOLs and 9/10 cases (90%) of transformed PMOLs as shown in “Fig 4B”. FGFR-2 expression was observed in 16/62 (25.81%) of untranformed PMOLs and 8/10 (80%) transformed PMOLs as shown in “Fig 4C”. FGFR-3 expression was observed in 23/62 (37.10%) cases of untransformed PMOLs and 6/10 cases (60%) in transformed PMOLs as shown in “Fig 4D”. The characteristics of individual malignant transformed cases are described in Table 6. FGF-2 and FGFR-2 expression was found to be significantly associated with malignant transformation of PMOLs to OSCC both at phenotypic (FGF-2 p<0.000, FGFR-2 p<0.001), and molecular level (FGF-2 p<0.019, FGFR-2 p<0.025). No association of FGFR-3 was found with malignant transformation.
Table 6. Characteristics of the malignant transformed cases.
| CaseNo. | Age | Sex | Diagnosis | Site of lesion | Tobacco chewing habit | Alcohol | Smoking | dysplasia | FGF-2 | FGFR-2 | FGFR-3 | Follow- up in months |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 25 | M | SMF | BM | P | A | A | P | A | A | P | 40 |
| 2 | 22 | M | SMF | BM | P | A | P | P | P | P | P | 60 |
| 3 | 60 | M | SMF | BM | P | A | A | P | P | P | A | 18 |
| 4 | 34 | M | LKP | T | A | P | P | P | P | P | P | 34 |
| 5 | 55 | F | LKP | BM | P | A | P | P | P | P | P | 39 |
| 6 | 65 | M | LKP | BM | P | A | A | P | P | P | A | 29 |
| 7 | 30 | M | LKP | G | P | P | P | P | P | P | A | 22 |
| 8 | 40 | M | LKP | BM | P | P | P | P | P | A | A | 23 |
| 9 | 75 | M | LKP | BM | P | P | P | P | P | P | P | 10 |
| 10 | 30 | M | LKP | BM | A | A | P | P | p | p | P | 18 |
M, Male; F, Female; BM, Buccal mucosa; T, Tongue; G, Gingival buccal sulcus. P, Present; A, Absent.
Logistic regression analysis of malignant transformation risk in PMOLs
To evaluate the risk of malignant transformation of PMOLs we analyzed clinicopathological parameters and FGF-2 and FGFR-2 expression by logistic regression. In the univariate regression analysis, FGF-2 and FGFR-2 expression was associated with 28.2-fold (95% CI, (3.29–241.72); P <0.002) and 11.5-fold (95% CI, 2.20–59.91; P <0.004) increased risk of malignant transformation, respectively as shown in Table 7. To further assess the influence of each factor, we did multivariate regression analysis. Both these factors retained statistical significance. The OR for transformation was 19.91 for FGF-2 (95% CI, 12.20–180.32; P <0.008) and 7.22 for FGFR-2 (95% CI, 1.20–43.31; P <0.031). Interestingly, when co-expression of FGF-2 and FGFR-2 was considered as cofactor, the risk of malignant transformation was considerably higher in PMOLs showing co-expression as compared to PMOLS without co-expression of FGF-2and FGFR-2 (OR, 4.19; 95% CI, 1.81–9.71; P < 0.001).
Table 7. Logistic regression analysis of variables in PMOLs malignant transformation.
| Variables | OR (95% CI) | P-value |
|---|---|---|
| Univariate analysis | ||
| Age | 1.03 (.98–1.08) | 0.201 |
| Sex | 0.29 (.03–2.50) | 0.262 |
| Tobacco | 2.20 (.42–11.27) | 0.344 |
| Alcohol | 1.05 (.27–4.13) | 0.938 |
| Smoking | 2.65 (.62–11.22) | 0.184 |
| FGF-2 expression | 28.20 (3.29–241.72) | 0.002 |
| FGFR-2 expression | 11.5 (2.20–59.91) | 0.004 |
| FGFR-3 expression | 2.54 (.64–9.97) | 0.180 |
| Multivariate analysis | ||
| FGF-2 expression | 19.91 (2.20–180.32) | 0.008 |
| FGFR-2 expression | 7.22 (1.20–43.31) | 0.031 |
| Coexpression of FGF-2/FGFR-2 | 4.19 (1.81–9.71) | 0.001 |
OR, Odds ratio; 95%CI, 95% confidence interval.
Discussion
The transformation of oral precancerous lesions to cancer is a known fact. However it is difficult to predict which precancerous lesions will progress into malignancy. The present study evaluated the expression of FGF-2 and its receptors FGFR-2 and FGFR-3 in PMOLs and OSCC and their role in assessment for malignant transformation of PMOLs to OSCC.
FGF-2 is involved in induction of angiogenesis in several cancers; phaeochromocytoma, renal cell carcinoma, astrocytoma, bladder carcinoma, hepatocellular carcinoma, and prostate cancer [35] and also in the signals between the epithelium and connective tissue, influencing growth and differentiation. We observed expression of FGF-2 and its receptors FGFR-2 and FGFR-3 in neoplastic progression from normal through stages of epithelial dysplasia to oral squamous cell carcinoma. FGF-2 immunohistochemical expression was observed in 49.08% (53/108) cases of OSCC, 31.95% (23/72) cases of PMOLs and 26.92% (14/52) of controls. The expression of FGF-2 was cytoplasmic in basal and parabasal layers with lining squamous epithelium, tumor cells and also in the tumor stroma. Our findings are similar to that of Raimondi et al. (2006) [36] who have reported FGF-2 immunostaining only in cytoplasm of the basal layers of the epithelium in hamster cheek pouch model of oral cancer. Higher expression of FGF-2 in breast cancer stroma as compared to normal breast stroma has been reported [37].
FGF-2, besides being expressed in cancer and precancer, was also found in 26.92% cases of healthy controls in the present study. Wakulich et al (2002) [28] noticed weak to intense staining of FGF-2 in basal and parabasal layers in normal epithelium of 78.57% (11/ 14) cases. They found no statistically significant difference in staining intensity between normal and carcinomatous tissue. Further the presence of FGF-2 expression in basal and parabasal layers suggested that this growth factor was involved in proliferation but not in differentiation of normal oral keratinocytes.
FGF-2 expression was significantly high (60.38%) in well differentiated tumors in the present study. Janot et al. (1995) [26] and Wakulich et al. (2002) [28] have also reported increased FGF-2 staining in well differentiated tumors suggesting that it may be involved in mitosis seen in dysplasia, carcinoma in situ and OSCC.
In the present study, FGFR-2 expression was observed in 57.41% (62/108) cases of OSCC, 33.33% (24/72) cases of PMOLs and 15.38% (8/52) cases of controls. Wakulich et al. (2002) [28] have also reported generalized increase in staining intensity at all levels in dysplasias and carcinomatous tissue. While we observed FGFR-2 expression in normal epithelium in only 8 control cases, Dellacono et al. (1997) [27] found positive FGFR-2 staining in normal epithelium as well as in patches in OSCC. Forootan et al. (2000) [38] have reported increased FGFR-2 expression in superficial layers of normal epithelium and in areas of keratinization in squamous cell carcinomas. Recently, overexpression of FGFR-2 in breast cancer cell lines was reported to lead to constitutive FGFR-2 activation. Inhibition of FGFR-2 signaling in these cells induced apoptosis [38]. Thus, constitutive FGFR-2 signaling due to FGFR-2 overexpression can lead to protection from apoptosis which is one of the hallmarks of cancer [39].
Cytoplasmic FGFR-3 expression was observed in upper and middle layers of lining epithelium, stromal fibroblast cells, tumor cells and sometimes in the endothelium of tumor stroma. We found FGFR-3 positivity in 75% cases (81/108) of OSCC, 40.28% cases (29/72) of oral PMOLs and 36.54% cases (19/52) of controls. Our findings of FGFR-3 expression in full thickness of normal epithelium and its significant expression (p<0.008) in carcinomas are supported by the results of Raimondi et al. (2006) [36] in experimental model of hamster cheek pouch. The authors found FGFR-3 expression in normal mucosa as well as its significant expression in carcinoma, predominantly in the basal layers. The presence of FGFR-2 and FGFR-3 receptors in epithelial cells, fibroblasts and endothelia are an evidence to their participation in normal epithelial growth and in the maintenance of connective tissue structures and of vascular network [36].
FGFR-3 has been shown to play an important role in bladder cancer growth and is suggested as a candidate for targeted therapy [40]. In human bladder cancer, the FGFR-3 mutations were observed to be strongly associated with non invasive, low grade and stage. A two pathway model of carcinogenesis (favorable and unfavorable) has been proposed in urinary bladder cancer [41,42]. The favorable pathway is characterized by mutations in FGFR-3 and a clinically unfavorable pathway characterized by mutations in p53. Further the detection of FGFR-3 mutations in urine from patients with FGFR-3 mutations in primary tumor indicating tumor recurrence has also been reported. Thus identification of FGFR-3 mutations may be a potential biomarker of prognosis and recurrence [43,44]. This needs to be studied in oral cancer.
In the present study, we found that the FGFR-3 expression was significantly associated with tumor stage. FGFR-3 expression was observed in 51.85% of cases in stages I and II. Unlike our results, other workers have observed more pronounced staining in stages III and IV [27].
Over expression of a gene can be caused by its amplification or aberrant transcriptional regulation. Increased mRNA levels of FGF-2 and its receptors were observed in oral cancer and precancer patients as compared to normal controls in the present study. Relative gene expression of FGF-2 was 28.62 fold higher in OSCC, 21.75 fold in Leukoplakia and 6.79 fold higher in OSMF as compared to healthy controls. FGFR-2 relative gene expression was 20.21 fold higher in OSCC, 16.79 fold higher in LKP and 9.72 fold higher in OSMF as compared to healthy controls. Relative gene expression of FGFR-3 was 7.05 fold higher in OSCC, 4.45 fold higher in LKP and 3.95 fold higher in OSMF as compared to healthy controls. Elevated levels of FGFRs have been found in numerous cancers such as cancer of brain, head and neck, lung, stomach, breast, prostate and in sarcomas and multiple myeloma. However, an elevated level of a protein in cancer cell does not necessarily mean that this protein plays a role in carcinogenesis. Further it is not always clear that the FGFR alterations found in human cancers are “drivers” or “passengers” [39].
Several alterations, most often leading to increased FGFR signaling have been associated with human carcinogenesis and development of a malignant phenotype [39]. FGFs/FGFRs are key regulators of mesenchymal-epithelial communication. In adults, FGFR signaling continues to regulate tissue homeostasis and is also involved in processes such as tissue repair, angiogenesis and inflammation. In angiogenesis and neovascularization, FGFR signaling is mainly thought to play an indirect role by influencing other growth factors such as VEGF and hepatocyte growth factor [45].
FGFRs have also been observed in several cancers suggesting a tumor suppressor role of FGFR signaling in these cases. However, it is currently not well understood how FGFR-2 signaling seems to exhibit tumor suppressing effects in some cells and oncogenic effects in others. The basic mechanisms of FGFR signaling have been discussed in an excellent review on roles of fibroblast growth factor receptors in carcinogenesis by Haugsten et al. (2010) [39].
Imbalanced FGFR signaling could contribute to carcinogenesis and could thus be a potent therapeutic target in several human cancers. Several promising FGFR tyrosine kinase inhibitors and FGFR-blocking antibodies have been developed and some of them are in early phases of clinical trials [46, 47].
We found malignant conversion of PMOLs (OSMF, LKP) to OSCC in 13.89% (10/72) cases with mean follow up of 29.41 months. All the precancer cases 7 LKP (16.28%) and 3 OSMF (10.34%) which transformed into malignancy, had associated epithelial severe dysplasia. This number is an under-representation of the population because our study was restricted to only biopsy proven cases of pre-cancerous lesions, that too only in two categories i.e. LKP and OSMF. Further, the cases of OSMF are also limited, as all clinical cases which report to us, have not been included in the study. Only those cases of OSMF who underwent surgery as a part of their treatment were included in our study. FGF-2 was positive in 90% (9/10) and FGFR-2 was positive in 80% (8/10) of transformed cases, a finding of statistical significance (FGF-2 p<.0001; FGFR-2 p<.001). Myoken et al. (1994) [48] in their immunohistochemical study, reported stronger expression of FGF-2 in OSCCs as compared to normal tissue, suggesting the possibility of its involvement in malignant transformation and self proliferation of cells. Fibroblasts transfected with FGF-2 cDNA underwent malignant transformation thereby acquiring self proliferative ability.
OSMF is an established precancerous lesion prevalent in India [49]. It is caused mainly due to the habit of chewing betel quid which subjects the oral mucosa to direct and frequent contact with chemical carcinogens. One third of OSMF cases slowly progress into squamous cell carcinoma. In experimental carcinogenesis model of oral cancer, when chemical cancerization solution is spread over the mucosa of hamster cheek pouch (one of the most widely accepted animal model of oral cancer), the first change observed is marked desmoplasia or fibrosis underlying the cancerized epithelium, even before the abnormal morphologic epithelial lesions occur. Fibrosis or desmoplasia in the oral cancer may be either a stromal component of the tumor itself or it may be associated with preceding OSMF [36].
The principal cells in OSMF implicated as a source of extracellular matrix in the areas of fibrosis are fibroblasts. Accumulation of connective tissue matrix is secondary to factors such as cytokines and growth factors. In the present study, 3 out of 29 (10.34%) cases of OSMF progressed into OSCC. In a report from southern India, 40% of oral cancer patients had OSMF. Incidence of 7.6% oral cancer in OSMF patients was reported by authors in a median follow-up period of 10-year. We however, had a follow up period of 5 years in this study. Increased FGF-2 expression can be explained due to an initial injury phase because of areca consumption, followed by cellular activation by chemotactic cytokines and other growth factors with eventual fibrosis occurring as a result of molecular alteration at the cellular level [50].
The univariate and multivariate analysis with logistic regression showed statistically significant expression of only FGF-2 and FGFR-2 and their combined co-expression in cases which transformed from precancer into malignancy in the present study. Tumor derived FGF-2 may promote cancer progression by elevating proteolytic enzymes and by paracrine stimulation of vascular endothelial cell growth [51].
Shi et al. (2010) [52] have reported immunohistochemical expression of 2 other modulators of angiogenesis; podoplanin and ABCG2 as markers for evaluating risk of malignant transformation of oral lichen planus. We however, did not have any case of oral lichen planus in our study.
In the present study, presence of dysplasia was found to be a significant clinico-pathologic predictor for malignant transformation in precancer [53]. Age, sex, tobacco, smoking, and alcohol intake were not predictors of malignant transformation. Our findings are in close conformity with previous reports [52, 54].
Our data showed up-regulated FGF-2, FGFR-2 and FGFR-3 expression both at phenotypic and molecular level from PMOLs to OSCC with statistically significant co-expression of FGF2 and FGFR2. Therefore, expression of FGF-2 and FGFR-2 may serve as an adjunct to histopathologic assessment of epithelial dysplasia for evaluating progression and malignant transformation in PMOLs. However, the study is limited by the overall numbers of converted patients. Further studies need to be done in a large cohort to draw a strong conclusion.
Acknowledgments
The authors wish to thank Indian Council of Medical Research, GOI (Project No. 5/13/56/2008-NCDIII) and Council of Science and Technology (UP, India) for financial support. We are grateful to all participants for their cooperation during the course of the study.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by Indian council of Medical Research, New Delhi, India-110029 (Grant No. 5/13/56/2008-NCDIII, www.icmr.nic.in); and Council of Science and Technology (Uttar Pradesh), India (Grant No. CST/SERPD/D-1888(ii), www.cstup.gov.in). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jorden RCK, Daley TD. Oral squamous cell carcinoma: new insights, J Can Dent Assoc. 1997; 63:517–518. [PubMed] [Google Scholar]
- 2. Sarnath D, Bhoite LT, Deo MG. Molecular lesions in human oral cancer: the Indian scene. Eur J Cancer B Oral Oncol. 1993; 29B: 107–112. [DOI] [PubMed] [Google Scholar]
- 3. Schepman KP, van der Meij EH, Smeele LE, van der Waal I. Malignant transformation of oral leukoplakia: a follow-up study of a hospital-based population of 166 patients with oral leukoplakia from The Netherlands. Oral Oncol. 1998; 34:270–275. [PubMed] [Google Scholar]
- 4. Silverman S Jr, Gorsky M, Lozada F. Oral leukoplakia and malignant transformation. A follow-up study of 257 patients. Cancer. 1984;53:563–568. [DOI] [PubMed] [Google Scholar]
- 5. Lumerman H, Freedman P, Kerpel S. Oral epithelial dysplasia and the development of invasive squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1995;79:321–329. [DOI] [PubMed] [Google Scholar]
- 6. Amagasa T, Yamashiro M, Uzawa N. Oral premalignant lesions: from a clinical perspective. Int J Clin Oncol. 2011;16:5–14. 10.1007/s10147-010-0157-3 [DOI] [PubMed] [Google Scholar]
- 7. Brouns E, Baart J, Karagozoglu Kh, Aartman I, Bloemena E, van der Waal I. Malignant transformation of oral leukoplakia in a well-defined cohort of 144 patients. Oral Dis. 2014. April;20(3):e19–24. 10.1111/odi.12095 [DOI] [PubMed] [Google Scholar]
- 8. Fang M, Zhang W, Chen Y, He Z. Malignant transformation of oral lichen planus: a retrospective study of 23 cases. Quintessence Int. 2009;40:235–242. [PubMed] [Google Scholar]
- 9. Murti PR, Bhonsle RB, Pindborg JJ, Daftary DK, Gupta PC, Mehta FS. Malignant transformation rate in oral submucous fibrosis over a 17-year period. Community Dent Oral Epidemiol. 1985;13:340–341. [DOI] [PubMed] [Google Scholar]
- 10. Pindborg JJ, Murti PR, Bhonsle RB, Gupta PC, Daftary DK, Mehta FS. Oral submucous fibrosis as a precancerous condition. Scand J Dent Res. 1984;92:224–229. [DOI] [PubMed] [Google Scholar]
- 11. Hsue SS, Wang WC, Chen CH, Lin CC, Chen YK, Lin LM. Malignant transformation in 1458 patients with potentially malignant oral mucosal disorders: a follow-up study based in a Taiwanese hospital. J Oral Pathol Med. 2007;36:25–29. [DOI] [PubMed] [Google Scholar]
- 12. Karabulut A, Reibel J, Therkildsen MH, Praetorius F, Neilson HW, Dabelsteen E. Observer variability in the histologic assessment of oral premalignant lesions. J Pathol Med. 1995; 24:198–200. [DOI] [PubMed] [Google Scholar]
- 13. Abbey LM, Kaugars GE, Gunsolley JC, Burns JC, Page DG, Svirsky JA, et al. Intraexaminer and interexaminer reliability in diagnosis of oral epithelial dysplasia. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1995; 80:188–191. [DOI] [PubMed] [Google Scholar]
- 14. Smith J, Rattay T, McConkey C, Helliwell T, Mehanna H. Biomarkers in dysplasia of the oral cavity: a systematic review. Oral Oncol. 2009;45:647–653. 10.1016/j.oraloncology.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 15. Yoshitake Y, Nishikawa K. Distribution of fibroblast growth factors in cultured tumor cells and their transplants. In Vito Cell Dev Biol. 1992; 28A: 419–428. [DOI] [PubMed] [Google Scholar]
- 16. Klagsbrun M, Baird A. A dual receptor system is required for basic fibroblast growth factor activity. Cell. 1991; 67: 229–231. [DOI] [PubMed] [Google Scholar]
- 17. Coutts JC, Gallagher JT. Receptor for fibroblast growth factor. Immunol Cell Biol 1995; 37:584–9. [DOI] [PubMed] [Google Scholar]
- 18. Givol D, Yayon A. Complexity of FGF receptors: genetic basis for structural diversity and functional specificity. FASEB J. 1992;6: 3362–3369. [PubMed] [Google Scholar]
- 19. Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005; 16: 233–247. [DOI] [PubMed] [Google Scholar]
- 20. Yamanaka Y, Fries H, Buchler M, Beger HG, Uchida E, Onda M, et al. Overexpression of acidic and basic fibroblast growth factors in human pancreatic cancer correlates with advanced tumor stage. Cancer Res. 1993; 53: 5289–5296. [PubMed] [Google Scholar]
- 21. Fukui S, Nawashiro H, Otani N, Ooigawa H, Nomura N, Yano A, et al. Nuclear accumulation of basic fibroblast growth factor in human astrocytic tumors. Cancer. 2003; 97: 3061–3067. [DOI] [PubMed] [Google Scholar]
- 22. Bikfalvi A, Klein S, Pintucci G, Rifkin DB. Biological roles of fibroblast growth factor-2. Endocr Rev. 1997; 18:26–45. [DOI] [PubMed] [Google Scholar]
- 23. Jaye M, Lyall RM, Mudd R, Schlessinger J, Sarver N. Expression of acidic fibroblast growth factor cDNA confers growth advantage and tumorigenesis to Swiss 3T3 cells. EMBO J. 1988; 7: 963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sasda R, Kurokawa T, Iwane M, Igarashi K. Transformation of mouse BALB/c 3T3 cells with human basic fibroblast growth factor cDNA. Mol Cell Biol. 1988;8: 588–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012; 18(7):1855–62. 10.1158/1078-0432.CCR-11-0699 [DOI] [PubMed] [Google Scholar]
- 26. Janot F, el Naggar AK, Morrison RS, Liu TJ, Taylor DL, Clayman GL. Expression of basic fibroblast growth factor in squamous cell carcinoma of the head and neck is associated with degree of histologic differentiation. Int J Cancer. 1995; 64: 117–123. [DOI] [PubMed] [Google Scholar]
- 27. Dellacono FR, Spiro J, Eisma R, Kreutzer D. Expression of basic fibroblast growth factor and its receptors by head and neck squamous carcinoma tumor and vascular endothelial cells. Am J Surg. 1997;174: 540–544. [DOI] [PubMed] [Google Scholar]
- 28. Wakulich C, Jackson-Boeters L, Daley TD, Wysocki GP. Immunohistochemical localization of growth factors fibroblast growth factor-1 and factors fibroblast growth factor-2 and factors fibroblast growth factor-3 in normal oral epithelium, epithelial dysplasias, and squamous cell carcinoma. Oral surg Oral Med Oral Pathol Oral Radiol Endod. 2002; 93:573–579. [DOI] [PubMed] [Google Scholar]
- 29. Shareef KN, Majeed AH. Immunohistochemical expression of Basic fibroblast growth factor-2 and Heparanase in oral squamous cell carcinoma. J Bagh College Dentistry 2013; 25(1):94–98. [Google Scholar]
- 30. Marshall ME, Hinz TK, Kono SA, Singleton KR, Bichon B, Ware KE, et al. Fibroblast growth factor receptors are components of autocrine signaling networks in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2011;17: 5016–5025. 10.1158/1078-0432.CCR-11-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ho P-S, Chen P-L, Warnakulasuriya S, Shieh T-Y, Chen Y-K, Huang I-Y. Malignant transformation of oral potentially malignant disorders in males: a retrospective cohort study. BMC Cancer. 2009;9: 260–267. 10.1186/1471-2407-9-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Warnakulasuriya S, Johnson NW, van der Waal I. Nomenclature and classification of potentially malignant disorders of the oral mucosa. J Oral Pathol Med. 2007; 36(10):575–80. [DOI] [PubMed] [Google Scholar]
- 33. Karavasilis V, Malamou-Mitsi V, Briasoulis E, Tsanou E, Kitsou E, Kalofonos H. Angiogenesis in cancer of unknown primary: Clinicopathological study of CD34, VEGF and TSP-1. BMC Cancer. 2005; 5:25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hase T, Kawashiri S, Tanaka A, Nozaki S, Noguchi N, Kato K, et al. Fibroblast growth factor-2 accelerates invasion of oral squamous cell carcinoma. Oral Science international. 2006; 1–9. [DOI] [PubMed] [Google Scholar]
- 35. Giri D, Ropiquet F, Ittmann M. Alerations in expression of basic fibroblast growth factor (FGF-2) and its receptor FGFR-1 in human prostate cancer. Clin Cancer Res. 1999;5: 1063–1071. [PubMed] [Google Scholar]
- 36. Raimondi AR, Molinolo AA, Itoiz ME. Fibroblast growth factor-2 expression during experimental oral carcinogenesis. Its possible role in the induction of pre-malignant fibrosis. J Oral Pathol Med. 2006;35: 212–217. [DOI] [PubMed] [Google Scholar]
- 37. Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14: 518–527. 10.1038/nm1764 [DOI] [PubMed] [Google Scholar]
- 38. Forootan SS, Ke Y, Jones AS, Helliwell TR. Basic fibroblast growth factor and angiogenesis in squamous cell carcinoma of the tongue. Oral Oncol. 2000;36: 437–443. [DOI] [PubMed] [Google Scholar]
- 39. Haugsten EM, Wiedlocha A, Olsnes S, Wesche J. Roles of fibroblast growth factor receptors in carcinogenesis. Mol Cancer Res. 2010; 8:1439–1452. 10.1158/1541-7786.MCR-10-0168 [DOI] [PubMed] [Google Scholar]
- 40. Hernandez S, Lopez-Knowles E, Lloreta J, Kogevinas M, Amoros A, Tardon A, et al. Prospective study of FGFR3 mutations as a prognostic factor in non muscle invasive urothelial bladder carcinomas. J Clin Oncol. 2006; 24:3664–71. [DOI] [PubMed] [Google Scholar]
- 41. Cheng L, Zhang S, Davidson DD, MacLennan GT, Koch MO, Montironi R, et al. Molecular determinants of tumor recurrence in the urinary bladder. Future Oncol. 2009; 5:843–857. 10.2217/fon.09.50 [DOI] [PubMed] [Google Scholar]
- 42. Castillo-Martin M, Domingo-Domenech J, Karni-Schmidt O, Matos T, Cordon-Cardo C. Molecular pathways of urothelial development and bladder tumorigenesis. Urol Oncol 2010;28:401–408 10.1016/j.urolonc.2009.04.019 [DOI] [PubMed] [Google Scholar]
- 43. Miyake M, Sugano K, Sugino H, Imai K, Matsumoto E, Maeda K, et al. Fibroblast growth factor receptor 3 mutation in voided urine is a useful diagnostic marker and significant indicator of tumor recurrence in non-muscle invasive bladder cancer. Cancer Sci. 2010;101:250–258. 10.1111/j.1349-7006.2009.01334.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zuiverloon TCM, van der Aa MNM, Van der Kwast TH, Steyerberg EW, Lingsma HF, Bangma CH, et al. Fibroblast growth factor receptor 3 mutation analysis on voided urine for surveillance of patients with low-grade non-muscle-invasive bladder cancer. Clin Cancer Res. 2010;16: 3011–3018. 10.1158/1078-0432.CCR-09-3013 [DOI] [PubMed] [Google Scholar]
- 45. Murakami M, Simons M. Fibroblast growth factor regulation of neovascularization. Curr Opin Hematol. 2008;15: 215–220. 10.1097/MOH.0b013e3282f97d98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Turner N, Grose R. Fibroblast growth factor signaling: from development to cancer. Nat Rev Cancer. 2010; 10:116–129. 10.1038/nrc2780 [DOI] [PubMed] [Google Scholar]
- 47. Knights V, Cook SJ. De-regulated FGF receptors as therapeutic targets in cancer. Pharmacol Ther. 2010;125: 105–117. 10.1016/j.pharmthera.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 48. Myoken Y, Okamoto T, Sato JD, Takada K. Immunohistochemical localization of fibroblast growth factor -1 (FGF-1) and FGF-2 in oral squamous cell carcinoma (SCC). J Oral Parhol Med. 1994;23: 451–456. [DOI] [PubMed] [Google Scholar]
- 49. Mehrotra D, Pradhan R, Gupta S. Retrospective comparison of surgical treatment modalities in 100 patients with oral submucous fibrosis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;107:e1–10. [DOI] [PubMed] [Google Scholar]
- 50. Bishen KA, Radhakrishnan R, Satyamoorthy K. The role of basic fibroblast growth factor in oral submucous fibrosis pathogenesis. J Oral Pathol Med. 2008;37: 402–411. 10.1111/j.1600-0714.2008.00649.x [DOI] [PubMed] [Google Scholar]
- 51. Liu W, Bao ZX, Shi LJ, Tang GY, Zhou ZT. Malignant transformation of oral epithelial dysplasia: clinicopathological risk factors and outcome analysis in a retrospective cohort of 138 cases. Histopathology. 2011;59:733–740. 10.1111/j.1365-2559.2011.03938.x [DOI] [PubMed] [Google Scholar]
- 52. Shi P, Liu W, Zhou ZT, He QB, Jiang WW. Podoplanin and ABCG2: Malignant Transformation Risk Markers for Oral Lichen Planus. Cancer Epidemiol Biomarkers Prev. 2010;19(3): 844–9. 10.1158/1055-9965.EPI-09-0699 [DOI] [PubMed] [Google Scholar]
- 53. Liu W, Shi L-J, Wu L, Feng J-Q, Yang X, et al. (2012) Oral Cancer Development in Patients with Leukoplakia—Clinicopathological Factors Affecting Outcome. PLoS ONE 7(4): e34773 10.1371/journal.pone.0034773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Murti PR, Daftary DK, Bhonsle RB, Gupta PC, Mehta FS, Pindborg JJ. Malignant potential of oral lichen planus: observations in 722 patients from India. J Oral Pathol. 1986;15:71–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.