Abstract
Under basal conditions histone deacetylases (HDACs) and their associated co-repressor complexes serve as molecular ‘brake pads’ to prevent the gene expression required for long-term memory formation. Following a learning event, HDACs and their co-repressor complexes are removed from a subset of specific gene promoters, allowing the histone acetylation and active gene expression required for long-term memory formation. Inhibition of HDACs increases histone acetylation, extends gene expression profiles, and allows for the formation of persistent long-term memories for training events that are otherwise forgotten. We propose that emotionally salient experiences have utilized this system to form strong and persistent memories for behaviorally significant events. Consequently, the presence or absence of HDACs at a selection of specific gene promoters could serve as a critical barrier for permitting the formation of long-term memories.
Keywords: chromatin, epigenetics, gene expression, glucocorticoids, histone acetylation, histone deacetylases (HDACs)
Introduction
Emotionally arousing events often lead to the formation of strong and persistent memories that can last a lifetime. There is extensive research supporting the role of adrenal stress hormones in enhancing memory for emotionally salient events (reviewed in McGaugh, 2004), but the molecular events underlying their persistence are largely unexplored. Here we propose that behaviorally important events remove the molecular ‘brake pads’ that normally prevent the formation of long-term memories and that misregulation of this system may result in the formation of abnormally strong memories, such as in post-traumatic stress disorder or addiction.
The consolidation of long-term memories requires the coordinated expression of specific profiles of gene targets (reviewed in Alberini, 2009). Many recent studies have focused on epigenetic mechanisms of transcriptional regulation in controlling the gene expression underlying longterm memory formation. In a broad sense, epigenetics refers to the regulation of gene expression via chromatin structure that is independent of changes in DNA sequence (Day and Sweatt, 2011; Wood, 2011). There are five major mechanisms by which chromatin structure is regulated in the service of controlling gene expression: histone modification, DNA methylation, nucleosome remodeling, histone variants, and miRNAs. This review will focus on histone modification mechanisms, as this is the best-studied chromatin regulatory mechanism implicated in memory processes to date and has recently been implicated in regulating behavioral adaptations to emotional stimuli (Renthal et al., 2007; Roozendaal et al., 2010; Fukada et al., 2012).
For transcription to occur, both transcription factors and RNA polymerase must gain access to the DNA. Within eukaryotic cells DNA is highly compacted (~10,000-fold) into a structure called chromatin. The primary repeating unit of chromatin is termed the nucleosome, with each nucleosome consisting of 147 base pairs of DNA wrapped around a histone octamer. The histone octamer can vary in composition, but canonically includes two of each of the following core histone proteins: H2A, H2B, H3 and H4. Each histone contains a histone tail that interacts with the negatively charged phosphate backbone of DNA. Histone tails can be post-translationally modified by the addition or removal of specific chemical groups (phosphorylated, acetylated, methylated, etc.). These histone tail modifications can alter histone-DNA interactions, recruit DNA or histone-interacting proteins, and either promote or repress transcription (Kouzarides, 2007; Barrett and Wood, 2008). Specific histone modifications may alter gene expression by interacting with additional epigenetic mechanisms of gene regulation. For example, DNA methylation and histone acetylation appear to work in concert to regulate long-term changes in behavior (Weaver et al., 2004; Miller and Sweatt, 2007).
This dynamic state of chromatin ultimately regulates gene expression profiles that in turn dictate cellular function. In general, histone acetylation weakens the interactions between histone tails and the negatively charged DNA, allowing greater access for the transcriptional machinery and hence promoting transcription. In contrast, histone deacetylation is usually associated with repressive chromatin states and decreased transcription (Kouzarides, 2007). The interplay between histone acetylation and deacetylation is one of the most widely studied forms of epigenetic regulation with regard to learning and memory. In particular, the two classes of enzymes that control the addition and removal of acetyl groups on histone tail lysine residues are histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. The role of HATs in learning and memory has been reviewed extensively elsewhere (Barrett et al., 2008) and consequently, this review will focus on the role of HDACs as negative regulators of long-term memory formation. Specifically, we propose that HDACs serve as molecular ‘break pads’ that prevent the formation of long-term memory and it is the removal of these ‘break pads’ during emotional, stressful or salient events that triggers the gene expression required for long-term memory formation.
Histone deacetylases
Histone deacetylases (HDACs) are a family of enzymes that are categorized into five main subtypes (I, IIa, IIb, III and IV) based on their homology to yeast transcriptional regulators, subcellular localization and enzymatic activity (reviewed in Bolden et al., 2006). Both class I and II HDACs remove acetyl groups from the ε amino groups of lysine residues via a charge-relay system that requires both Zn 2+ and additional cofactors (Finnin et al., 1999). In contrast, the class III HDACs (sirtuin family) are structurally and functionally distinct from class I and II and depend on nicotinamide adenine dinucleotide (NAD + ) for deacetylase activity. In addition to targeting acetyl groups on specific histone tail lysine residues, class I, IIa and IIb HDACs also remove acetyl groups from a variety of non-histone targets including cytoskeletal proteins, molecular chaperones, and nuclear import factors (Glozak et al., 2005). The first non-histone target of HDAC-mediated deacetylation was the tumor suppressor and DNA binding transcription factor p53 (Juan et al., 2000). Since that original discovery, HDACs have been found to remove acetyl groups from a variety of transcription factors including Yin Yang 1, STAT3, MyoD, NF-kB and c-MYC. Deacetylation also regulates several other non-transcription factor proteins including importin- α (nuclear import), Ku70 (DNA repair), Hsp90 (molecular chaperone), and α-tubulin (cytoskeleton) (reviewed in Glozak et al., 2005). Overall, the acetylation/deacetylation of non-histone targets seems to have a diverse and protein specific functional impact. For example, depending on the transcription factor target, increased acetylation has been associated with both increased and decreased DNA binding affinity, protein stability and the stability of protein - protein interactions (reviewed in Glozak et al., 2005).
Class I HDACs (1, 2, 3 and 8) are mainly localized to the nucleus (reviewed in de Ruijter et al., 2003). HDACs 1, 2 and 3 are highly expressed throughout many brain areas including the amygdala, hippocampus and cortex. Brain expression of HDAC 8 is relatively low, with the exception of moderate expression in the hippocampus (Broide et al., 2007). HDAC1 and 2 are found in at least three distinct complexes: Sin3, nucleosome remodeling and histone deacetylation (NURD) and CoREST (reviewed in Ng and Bird, 2000; de Ruijter et al., 2003). The HDAC3 complex includes either the co-repressors silencing mediator for retinoid co-repressor (SMRT) or nuclear receptor co-repressor (N-CoR) as well as class IIa HDACs 4, 5 and 7 (Guenther et al., 2000; Li et al., 2000; Fischle et al., 2001, 2002; Zhang et al., 2002). Both SMRT and N-CoR contain a deacetylase-activating domain (DAD) that includes a conserved switching-defective protein 3, adapter 2, nuclear receptor co-repressor, transcription factor (SANT) motif required for HDAC3 enzymatic activity. Importantly, the DAD domain of SMRT is required for both HDAC3 de acetylase activity and repression by SMRT (Guenther et al., 2001) and recently the specificity of the HDAC3/SMRTDAD domain interaction was further demonstrated by the crystal structure of the co-repressor complex (Watson et al., 2012).
Class II HDACs are subdivided into IIa (HDAC 4, 5, 7 and 9) and IIb (HDAC 6 and 10). HDACs 4, 5, 7 and 9 shuttle between the nucleus and the cytoplasm, while the class IIb HDACs are found mainly within the cytoplasm (Jepsen and Rosenfeld, 2002; Chawla et al., 2003; reviewed in de Ruijter et al., 2003). HDACs 4 and 5 are highly expressed throughout the brain, while expression of HDACs 7 and 9 is comparably low. As described above, HDACs 4, 5 and 7 interact with HDAC3 and the NCoR/SMRT complex. Recently, Lahm and colleagues (2007) showed that class IIa HDACs may not actually possess true deacetylase activity and instead express their enzymatic activity through interactions with class I HDACs. Specifically, the authors examined the deacetylase activity of HDAC3 (class I) and 4 (class IIa) on histone H4-derived peptides with a single acetyl-lysine. The class I and IIa HDACs differ by a single amino acid (tyrosine vs. histidine) within the catalytic domain, which is predicted to disrupt enzymatic activity in the class IIa HDACs. Mutation of this critical tyrosine within the HDAC3 (class I) catalytic site eliminates its deacetylase activity without disrupting its presence in the co-repressor complex. In contrast, a similar mutation in HDAC4 had no effect on the complex’s deacetylase activity. This in combination with the lack of deacetylase activity of purified recombinant HDAC4 suggests the de acetylase activity of HDAC4, and perhaps other class IIa HDACs, actually comes from their association with class I HDACs. In further support of this hypothesis, substituting the histidine for tyrosine within the catalytic domain of HDAC 4, 5 and 7 creates gain-of-function mutants with potent deacetylase activity (Lahm et al., 2007). These findings suggest that class IIa HDACs may not be enzymatically active but instead rely on class I HDACs for histone deacetylase activity. It is currently unclear, however, whether these studies extend beyond the role for class IIa HDACs on traditional isolated histone substrates. It remains an open possibility that class IIa HDACs may act on other acetylated histone sites or nonhistone targets.
Class IIb HDAC6 possess several unique structural domains including two independent catalytic domains and acts in the cytoplasm to deacetylate α-tubulin and alter microtubule stability (reviewed in Valenzuela-Fernández et al., 2008). Over-expression of HDAC6 decreases acetylation, resulting in an increase in cell motility leading to microtubule depolymerization (Hubbert et al., 2002). HDAC6 is moderately expressed in the cortex, hippocampus and pons (Broide et al., 2007), suggesting a potential role for the regulation of microtubule-dependent transport and cytoskeletal dynamics in neurons (see the section on HDAC6 below). The other class IIb HDAC (HDAC10) is only moderately expressed within the brain (Broide et al., 2007) and has been found to interact with both class I and class IIa HDACs (reviewed in de Ruijter et al., 2003).
The sirtuin family (class III) is composed of seven different NAD + -dependent HDACs that differ both structurally and functionally from other HDACs. There are seven mammalian sirtuins (1–7) that all share a conserved catalytic core domain that utilizes NAD + as a cofactor for catalytic reactions, including both deacetylases and mono-ADP-ribosyl transferase activity (Michishita et al., 2005; reviewed in Michán and Sinclair, 2007). Sirtuins vary in the type of enzymatic activity they possess, with SIRT1 demonstrating the most robust deacetylation activity (Michishita et al., 2005). Cellular localization of sirtuins varies with SIRT1, 6 and 7 being primarily locali zed within the nucleus and SIRT 2, 3, 4 and 5 within the cytoplasm (Michishita et al., 2005). Sirtuins target both histones and other proteins to regulate stress, metabolism and cell survival (reviewed in Sauve et al., 2006); however, even though all seven sirtuins are expressed in the brain, little is known about their specific roles in normal brain function (Michishita et al., 2005). To date, only SIRT1 and 2 have been characterized in the brain (see the section on SIRT1 below), with SIRT1 expression occurring mainly in neuronal cell bodies (Michán and Sinclair, 2007) and SIRT2 within oligodendrocytes and Schwann cells in the olfactory bulb (Yu et al., 2005). SIRT1 germline knockout mice or transgenic mice expressing SIRT1 lacking the catalytic domain possess abnormalities in the central nervous system including impaired retinal and heart development, somatotropic signaling and dwarfism (Cheng et al., 2003; Cohen et al., 2009). The role of SIRT1 in learning and memory is discussed in the sections below.
The final class of HDACs (class IV) contains only HDAC11, which shares homology within the catalytic domain to both class I and II HDACs (Gao et al., 2002). HDAC11 is expressed highly throughout most regions of the brain (Broide et al., 2007) and co-immunopreciptiates with HDAC6 (Gao et al., 2002), suggesting a potential role in deacetylating non-histone targets. Identifying potential HDAC11 targets as well as its function within the central nervous system remain an open areas of research.
HDACs regulate gene expression
Transcription is a complex and highly orchestrated process required for basic cellular function. Originally, the presence or absence of co-activator or co-repressor complexes was viewed as a ‘switch’ between a transcriptionally active or repressed state (reviewed in Perissi et al., 2010). In this model, HATs are associated with active genes and HDACs with inactive genes. This model has been further refined to incorporate the finding that in addition to co-activators, transcriptional activation requires the cyclic recruitment of HDACs and nucleosome remodeling complexes. Meétivier and colleagues (2003) demonstrated the orchestrated cycling of a host of activation factors including HATs, transcription factors, RNA polymerase and nucleosome remodeling complexes in vitro. Furthermore, following initiation and elongation, HDACs and chromatin remodeling machinery cycle on to the promoter to silence or reduce gene expression to basal levels. The authors propose that active promoters continuously exchange co-activator and co-repressor complexes, with the removal of co-repressors signaling transcriptional activation (Meétivier et al., 2003; Perissi et al., 2010). In the case of the N-CoR/SMRT/HDAC3 complex, this removal appears to be mediated by transducin β-like protein 1(TBLR1). TBLR1 is required for both N-CoR/SMRT/HDAC3-mediated repression and recruiting the ubiquitin/19S proteosomal machinery, the latter resulting in the dismissal and/ or degradation of the N-CoR/SMRT/HDAC3 complex and the subsequent activation of gene expression (Perissi et al., 2004, 2008). The continual presence of TBLR1 within the co-repressor complex suggests that the complex is primed for removal (Perissi et al., 2010), but it is unclear which signaling pathways directly orchestrate the removal/addition of different complexes at specific promoters at distinct time points.
The dynamic interplay between HDACs and HATs is supported by Wang and colleagues’demonstration of the presence of both HATs and HDACs at active promoter and enhancer regions in human CD+ T lymphocytes (Wang et al., 2009). In the resting state, the presence of HATs such as CBP, p300, PCAF, MOF and Tip60 near the transcription start site correlated with the presence of RNA Pol II, H3 and H4 acetylation and active gene expression. Surprisingly, a similar set of correlations was found for HDAC1, 2, 3 and 6, and HDACs were not enriched at inactive genes. When cells were treated with the HDAC inhibitor Trichostatin A (TSA) and butyrate, both H3K9 and H4K16 acetylation levels were increased in active genes within as little as 10 min. Given that both HATs and HDACs are found at active genes (and not inactive ones), the increase in acetylation following global HDAC inhibition indicates that active genes are in a constant cycle between acetylation and deacetylation and that blocking HDACs simply shifts this dynamic interplay in favor of histone acetylation (Wang et al., 2009). Recently, similar cyclic states of DNA methylation/demethylation at active promoters were demonstrated in vitro, indicating a potential synergy between the dynamic cycling of HDACs/HATs and DNA methyltransferases/demethylases (Kangaspeska et al., 2008; Meétivier et al., 2008).
If HDACs and HATs continually cycle at active promoters, HDAC inhibitors would be predicted to shift this dynamic in favor of HAT activity and prolong transcriptional activation by preventing HDAC re-engagement at active promoters. Delivery of HDAC inhibitors in vivo during a learning event increases H3 and H4 histone acetylation levels (Levenson et al., 2004; Vecsey et al., 2007; Federman et al., 2009; Malvaez et al., 2010). These global increases in histone acetylation could be predicted to lead to a general increase in gene expression; however, HDAC inhibition induces changes in expression of only a small subset of actively transcribed genes in vitro in carcinoma (8–10%; Glaser et al., 2003), lymphoid (2%; Van Lint et al., 1996), and hepatoma (~4%; Chiba et al., 2004) cell lines. Similarly, in vivo changes in gene expression following learning and HDAC inhibition seem to be limited to a subset of specific activity-dependent neuronal genes (Vecsey et al., 2007; McQuown et al., 2011). More specific inhibitors of individual HDAC enzymes are predicted to impact an even smaller subset of actively transcribed genes. Following a learning event and HDAC inhibition, expression of the orphan nuclear receptors Nr4a1 and Nr4a2 were significantly increased over vehicle-treated controls two hours post training (Vecsey et al., 2007). Since there were no changes in expression of either gene in animals given HDAC inhibitors without training and with normal expression of these genes peaks prior to two hours post training, these findings indicate that HDAC inhibitors prolong the expression of a specific subset of activity-regulated genes (Vecsey et al., 2007). The prolonged expression following HDAC inhibition as well as the memory-enhancing effects of HDAC inhibitors (see below) depend on the interaction between phosphorylated cyclic AMP-response element binding (CREB) and CREB Binding Protein (CPB) in the hippocampus, indicating that both CREB and CPB are critical for mediating increases in hippocampal gene expression following HDAC inhibition (Vecsey et al., 2007; see also McQuown et al., 2010; Barrett et al., 2011; Haettig et al., 2011).
Similar to the effects observed with general HDAC inhibitors, HDAC3 focal deletions in the dorsal hippocampus of HDAC3-FLOX mice led to prolonged expression profiles for both Nr4a2 and c-fos in a CREB/CBP-dependent manner. This prolonged expression appears to be critical for memory formation, since blocking Nr4a2 expression within the hippocampus prior to subthreshold object location training blocked the memory enhancement observed in the vehicle-treated controls (McQuown et al., 2011), see the HDAC3 section below. The findings from general HDAC inhibitors as well as manipulations of a single HDAC (HDAC3) indicate that endogenous HDACs negatively regulate the expression of specific target genes required for long-term memory formation.
Another potentially critical function of HDACs may be to keep a subset of promoters in a ‘primed’ state. These promoters contain the H3K4 methylation mark associated with gene activation but are maintained in a silent state, presumably by the presence of HDACs that maintain low levels of acetylation and interfere with RNA Pol II binding (Wang et al., 2009). Fass et al. (2003) found that in PC12 cells the pre-initiation complex appears to be at least partly pre-assembled and poised to initiate transcription at both the c-fos and Nr4a1 promo ters, and that HDAC inhibition rapidly triggers the expression of both genes. Under normal conditions, HDACs may serve to prevent the complete formation of the pre-initiation complex and hence halt transcriptional activation. The role of HDAC inhibition may be more complex, however, as both Icer and Nor-1 transcription were decreased following HDAC inhibition in PC12 cells, indicating a potential role for HDACs in modulating trans criptional repression, perhaps by acting on non-histone targets (Fass et al., 2003). These findings indicate that HDACs may serve to negatively regulate gene expression by inhibiting the complete formation or activation of the initiation complex.
A similar ‘priming’-like mechanism was recently shown to mediate the rapid transcription of immediate early genes (IEGs; Saha et al., 2011). For a subset of IEGs, RNA polymerase II (RNA Pol II) transcribes approximately 20–50 nucleotides and then pauses until a sufficient activity-dependent stimulus releases the inhibition (Zeitlinger et al., 2007; Nechaev and Adelman, 2008). This promoter-proximal RNA Pol II stalling appears to occur at a subset of ‘rapidly’ engaged IEGs and allows for their immediate induction in response to neuronal activity. For example, in neuronal cultures RNA Pol II stalling was found at the c-fos promoter and following TTX withdrawal and c-fos transcription was rapidly engaged. In this study, both Nr4a1 and Nr4a2 were categorized as ‘delayed’ IEGs indicating that their expression is slower to engage than the more ‘rapid’ IEGs such as c-fos. Like most ‘delayed’ IEGS, RNA Pol II stalling did not appear to occur at the Nr4a1 promoter; however, RNA Pol II was bound at the Nr4a2 promoter indicating the presence of a pre-initiation complex under basal conditions (Saha et al., 2011). Furthermore, in non-neuronal cells HDAC corepressor complexes have been found to mediate basal state repression, specifically that of activity-regulated genes, with RNA Pol II stalling at their promoters. For example, in macrophages RNA Pol II stalling occurs at Toll-like receptor inducible genes. Under basal resting conditions, NCoR/HDAC3 and CoREST keep these genes switched off. Upon lipopolysaccharide stimulation, the co-repressor complexes are rapidly removed, histone acetylation increases, and RNA Pol II elongation progresses (Hargreaves et al., 2009). Together these findings indicate a potential role for HDACs in preventing both rapid and delayed neuronal IEG induction, potentially by maintaining a closed chromatin state prior to releasing a stalled RNA Pol II (rapid IEGs) or in preventing the formation of the pre-initiation complex or RNA Pol II binding (delayed IEGs). In either or both cases, HDACs could serve a critical role in negatively regulating transcriptional activation and HDAC inhibitors would be predicted to dramatically shift these kinetics. To date, the role of HDACs in regulating RNA Pol II stalling or the induction kinetics of neuronal gene expression in vivo remains unexplored.
HDAC inhibition in learning and memory
To date there is a large body of evidence to support the role of histone tail-modifying enzymes in learning and memory. Learning events in a variety of tasks, brain regions and species lead to increased histone acetylation and gene expression (Swank and Sweat, 2001; Guan et al., 2002; Levenson et al., 2004; Chwang et al., 2006; Federman et al., 2009; for a review, see Barrett and Wood, 2008). Mice with mutations that decrease HAT activity and histone acetylation show impairments in synaptic plasticity and long-term memory formation (Bourtchouladze et al., 2003; Alarcón et al., 2004; Korzus et al., 2004; Wood et al., 2005, 2006). For example, focal homozygous deletion of CBP in the adult dorsal hippocampus can disrupt histone acetylation and gene expression during memory consolidation, which correlates with impaired hippocampusdependent long-term memory and hippocampal longterm potentiation (Barrett et al., 2011).
The first studies on the role of HDACs in learning and memory utilized general HDAC inhibitors that blocked HDAC function by chelating the zinc ion at the active site (Finnin et al., 1999). For example sodium butyrate, valproic acid, and suberoylanilide hydroxamic acid (SAHA) all primarily target class I HDACs with little effect on class IIa (Kilgore et al., 2010), indicating a critical role for class I HDACs in memory formation and plasticity. However, the HDAC deacetylases assays performed by Kilgore et al. (2010) measured the deacetylase activity of purified, recombinant human HDACs on acetylated tripeptide substrates and it is currently unclear how well these results translate to the inhibition of specific HDACs within the brain an animal. Class I and II HDACs also have varying enzymatic activity on different acetylated peptide substrates (i.e., acetylated tripeptides vs. trifluoroacetyllysine tripeptides), potentially confounding the half maximal inhibitory concentration (IC50) readings if a non-optimal substrate is chosen for that HDAC (Lahm et al., 2007; Bradner et al., 2010). Furthermore, current techno logy allows for only an approximation of HDAC inhibitor specificity in vivo by comparing the IC50 for each HDAC on purified recombinant proteins to the maximum drug concentration (Cmax ) within the targeted brain region. For example, McQuown et al. (2011) found that RGFP136 has an IC50 of 5.2 μm for HDAC1, 3.0 μm for HDAC2 and 0.4 μm for HDAC3 on purified recombinant HDACs. Following a systemic injection of RGFP136, the Cmax in brain lysates was found to be ~1.7 μm, suggesting that systemic administration resulted in a brain concentration of RGFP136 sufficient to specifically inhibit only HDAC3.
The first experiments to examine the role of HDACs as negative regulators of transcriptional processes underlying synaptic plasticity were conducted by Guan and colleagues (2002) in Aplysia. In this system induction of long-term facilitation (five pulses of serotonin, 5HT) leads to increased histone acetylation, HAT binding and gene expression, while induction of long-term depression (a pulse of a neuropeptide related to enkephalins, FMRFa) recruited HDAC5 binding, and decreased histone acetylation and gene ex pression. As further evidence for a functional role of HDACs as negative regulators, blocking HDAC function by treatment with the HDAC inhibitor TSA transformed short-term facilitation (1×5HT) into longterm facilitation and blocked the induction of long-term depression (Guan et al., 2002). It was previously demonstrated by this group that a single pulse of serotonin (1×5HT) produced facilitation that was both short-term and transcription independent while five pulses produced long-term facilitation that required both transcription and translation (Bartsch et al., 1995). Collectively, these findings indicate that HDAC inhibition could transform a transcription-independent form of short-term facilitation into a transcription-dependent long-term form. This idea was further explored in rodents, where HDAC inhibitors have been shown to enhance long-term potentiation (LTP) within the lateral amygdala (TSA; Yeh et al., 2004) and hippocampal slices [suberoylanilide hydroxamic acid (SAHA); TSA/sodium butyrate; TSA; Alarcón et al., 2004; Levenson et al., 2004; Vecsey et al., 2007]. Similar to the findings in invertebrates, HDAC inhibition (TSA) can transform a transcription-independent, transient form of long-term potentiation into a transcriptionally-dependent and long lasting form of LTP (Vecsey et al., 2007). Furthermore, HDAC inhibition (TSA) enhancement of LTP required activation of NMDA receptors, ERK signaling and transcription (Levenson et al., 2004) as well as the interaction between CREB and CBP in the hippocampus (TSA, sodium butyrate; Vecsey et al., 2007; Barrett et al., 2011).
HDAC inhibition has also been shown to increase histone acetylation and enhance memory in a variety of tasks including long-term contextual fear memories (sodium butyrate, TSA; Levenson et al., 2004; Vecsey et al., 2007), novel object recognition (sodium butyrate; Stefanko et al., 2009; Haettig et al., 2011) and fear extinction (sodium butyrate, TSA, valproic acid; Bredy et al., 2007; Lattal et al., 2007). Similar to the transformation of a transcription-independent form of LTP into a longterm transcription-dependent form, HDAC inhibition can change a subthreshold learning event that does not result in short- or long-term memory into one that does (sodium butyrate, RGFP136; Stefanko et al., 2009; Haettig et al., 2011; McQuown et al., 2011). Stefanko and colleagues demonstrated that a subthreshold object recognition training paradigm (3 min training) does not produced short-term (90-min) or long-term (24-h) memory for a novel object. Post training systemic deli very of the HDAC inhibitor sodium butyrate produced long-term memory and this memory was persistent for at least 7 days, a time at which normal object recognition memory fails. There were no effects on short-term memory, indicating that the observed enhancement was dependent on transcriptional activation (Stefanko et al., 2009). Recently, inhibition of HDAC3 (see section below) was shown to transform a subthreshold learning event into a long-term memory that persisted for at least 7 days (McQuown et al., 2011). In crabs, weak context-signal training can be enhanced with post training HDAC inhibition (sodium butyrate, TSA) to produce longterm memory (Federman et al., 2009). Together these findings indicate that HDACs serve as critical negative regulators of long-term memory formation and that removing the molecular ‘brake pads’ can transform subthreshold learning events into robust long-term memories.
The role of individual HDACs in learning and memory
Since the finding that sodium butyrate, valproic acid, and SAHA ( Table 1 ) all predominately target class I HDACs (Kilgore et al., 2010), recent work has focused on examining the role of individual class I HDACs in the brain. A number of studies have utilized both pharmacological and genetic approaches to examine the function of individual HDACs in the brain ( Table 1 ), including HDAC1 (Bahari-Javan et al., 2012), HDAC2 (Guan et al., 2009), HDAC3 (McQuown et al., 2011), HDAC5 (Renthal et al., 2007), HDAC6 (Fukada et al., 2012) and SIRT1 (Gao et al., 2010). The following sections are not intended to provide a comprehensive review of the individual HDACs, but instead to highlight several current studies examining the role of specific HDACs in memory formation and the regulation of emotional behaviors.
Table 1.
The role of individual HDACs in learning and memory.
| Paper | HDAC | Manipulation | Brain region | Histone acetylation | Gene expression | Behavior | LTP |
|---|---|---|---|---|---|---|---|
| Bahari-Javan et al. (2012) | 1 | AAV over-expression | Dorsal Hc | Decreased H3K9ac at c-fos promoter | Decreased c-fos mRNA | Enhanced extinction of contextual fear conditioning, normal STM and working memory; normal LTM for novel object recognition; spatial water maze and contextual fear conditioning | – |
| Pharmacological inhibition (MS-275) | Dorsal Hc | Increased H3K9ac at c-fos promoter | Increased c-fos mRNA | Impaired extinction of contextual fear conditioning | – | ||
| siRNA knockdown | Dorsal Hc | Increased H3K9ac at c-fos promoter | Increased c-fos mRNA | Impaired extinction of contextual fear conditioning | – | ||
| Guan et al. (2009) | 1 | Germline over-expression | Neuronal | Decreased Kac global | Decreased NR2A protein, other plasticity genes normal | Normal cued and contextual fear LTM; normal acquisition and LTM for spatial MWM; normal spatial working memory, locomotion and pain sensitivity | Normal LTP (CA1 Schaffer collateral HFS) |
| 2 | Germline over-expression | Neuronal | Decreased H4K12ac global; decreased acH3 in hippocampal Bdnf-pII, Fos, GluR1 promoters | Decreased hippocampal CaMKIIA, CREB, NR2B, NR2A ERG1, c-FOS protein | Impaired cued and contextual fear LTM; normal STM,; impaired acquisition of spatial MWM; impaired spatial working memory; normal locomotion and pain sensitivity | Impaired LTP maintenance (CA1 Schaffer collateral HFS) | |
| Homozygous germline knockout | Neuronal | Increased H4K5ac, H4K12ac, H2Bac global; increased hippocampal acH3 & acH4 in Bdnf-pII, Egr1, Fos, GluR1 promoters | Increased hippocampal CaMKIIA, CREB, SVP, NR2B, NR2A, ERG1, c-FOS protein | Enhanced cued and contextual fear LTM; enhanced contextual (but not cued) fear STM; enhanced spatial working memory; normal locomotion and pain sensitivity | enhanced LTP maintenance (subthreshold CA1 Schaffer collateral HFS) | ||
| McQuown et al. (2011) | 3 | HDAC3-FLOX mice and AAV-CRE infusions | Dorsal Hc | Increased H4K8ac in hippocampal CA1 | Increased/extended expression of c-fos and Nr4a2 mRNA | Transform subthreshold object training into persistent object location LTM | – |
| Homozygous knockin single-point mutation in the NcoR DAD domain (Y478A) disrupts the HDAC3/NCoR | All cells with NCoR | – | – | Transform subthreshold object training into persistent LTM for object location and recognition | – | ||
| Pharmacological inhibition (RGFP136) | Dorsal Hc | Increased H4K8ac in hippocampal CA1 | – | Transform subthreshold object training into persistent LTM for object location | – | ||
| Renthal et al. (2007) | 5 | Homozygous germline knockout | All cells with HDAC5 | Increased H3ac at promoters of gene targets identified in microarray | Altered expression in NAc of gene families associated with dopamine transmission, excitability and cocaine responses | Increased social avoidance following chronic (but not acute) social defeat stress; anhedonic to sucrose reward; sensitization to cocaine; normal anxiety and locomotion | – |
| HSV over-expression | NAc | – | – | Impaired conditioned place preference to cocaine | – | ||
| 9 | Homozygous germline knockout | All cells with HDAC9 | – | – | Normal cocaine sensitivity | – | |
| HSV over-expression | NAc | – | – | Normal conditioned place preference to cocaine | – | ||
| Fukada et al. (2012) | 6 | Homozygous germline knockout | All cells with HDAC6 | increased Ac-α-tubulin | – | Elevated exploration in open field; normal home cage activity; decreased anxiety (elevated plus maze); reduced immobility on tail suspension test | – |
| Pharmacological inhibition (NCT-14b) | Systemic | Increased ac-α-tubulin | – | Reduced immobility on tail suspension test | – | ||
| Gao et al. (2010) | SIRT1 | Over-expression of SIRT1 with a deletion of the catalytic domain | Neuronal | – | Release miR-134 repression; decrease in CREB protein, decrease in Bdnf mRNA | Impaired LTM for contextual and cued fear novel object recognition and spatial Morris water maze; normal locomotion, shock sensitivity and visual platform water maze | Impaired LTP maintenance in CA1 (2×TBS of Schaffer collaterals) |
Abbreviations: AAV, adeno-associated virus; CRE, cre-recombinase; HSV, Herpes Simplex Virus; Hc, hippocampus; NAc, Nucleus accumbens; ac, acetylation; STM, short-term memory; LTM, long-term memory; LTP, long-term potentiation; HFS, high frequency stimulation; TBS, theta burst stimulation; BDNF, brain-derived neurotrophic factor.
HDAC1
HDAC1 has recently been implicated in several neurological disease states (Gräff et al., 2011); however, mouse models with germline over-expression or homozygous knockout of HDAC1 display normal memory formation (Guan et al., 2009). Bahari-Javen and colleagues (2012) used acute manipulations of HDAC1 to reveal a specific role for HDAC1 in the formation of extinction memory for contextual fear conditioning. Over-expression of HDAC1 in the dorsal hippocampus did produce a modest enhancement in pre-pulse inhibition of the startle response, but did not alter exploration, motor function, working memory or short-term memory. There was also no impact on long-term memory for novel object recognition, spatial Morris water maze or contextual fear conditioning. Over-expression of HDAC1 in the dorsal hippocampus did, however, facilitate extinction of contextual fear conditioning. Conversely, an HDAC1-specific inhibitor delivered into the dorsal hippocampus following each extinction trial impaired contextual fear extinction but had no effect when delivered following the initial training, indicating a unique role for HDAC1 in the extinction of fear memories (Bahari-Javan et al., 2012).
Repeated extinction trials led to increased levels of HDAC1 at the c-fos promoter and a corresponding decrease in c-fos expression, indicating that HDAC1 may repress c-fos expression during extinction training. Consistent with this idea, the authors found a decrease in H3K9 acetylation (H3K9Ac), a histone modification linked to active transcription, and HDAC1 activity at the c-fos promoter following extinction training. To directly test the role of HDAC1 in regulating c-fos expression during extinction training, the authors examined the impact of manipulating HDAC1 expression of H3K9Ac levels at the c-fos promoter and c-fos expression. Over-expression of HDAC1 in the dorsal hippocampus using adeno-associated virus decreased H3K9Ac levels at the c-fos promoter, decreased c-fos expression and facilitated extinction. Conversely, blocking HDAC1 pharmacologically or knocking down expression with siRNA increased H3K9Ac levels, increased c-fos expression and blocked extinction (Bahari-Javan et al., 2012).
These findings suggest a unique role for HDAC1 in repressing gene expression underlying the formation of extinction memory. In contrast, the non-selective HDAC inhibitors sodium butyrate (systemic) and TSA (intrahippocampal infusion) facilitate fear extinction when delivered following extinction training (Lattal et al., 2007). However, these inhibitors may target other HDAC family members that serve as negative regulators of gene expression required for fear extinction memory, such as HDAC3 (McQuown et al., 2011).
HDAC2
Guan and colleagues (2009) showed that HDAC2 binds the promoter of a variety of activity-regulated genes and is found within the coREST, NURD and mSin3a co-repressor complexes (see above). Over-expression of HDAC2 (but not HDAC1) decreased histone acetylation and impaired long-term but not short-term contextual and cued fear conditioning. These animals were also impaired on acquisition and probe test for the spatial version of the Morris water maze task and on a T-maze non-matching-to-place working memory task. In contrast, homozygous germline HDAC2 knockout mice have increased levels of histone acetylation, increased contextual (long- and short-term) and cued fear conditioning (long-term) and enhanced spatial working memory. Similarly, HDAC2 over-expressing mice have impaired LTP while HDAC2 knockout mice have enhanced LTP. HDAC2 also appears to regulate the spine density and synapse number, with HDAC2 overexpression decreasing and HDAC2 knockout increasing spine density. Furthermore, both the behavioral deficits and spine loss in the HDAC2 over-expressing mice could be rescued by chronic (10-day) treatment with the HDAC inhibitor SAHA, indicating that HDAC2 is a critical target for SAHA (Guan et al., 2009). Overall, these findings indicate that HDAC2 serves as a critical negative regulator of both synaptic plasticity and memory processes.
HDAC3
HDAC3 has also recently been shown to be a negative regulator of long-term memory formation. McQuown and colleagues (2011) used genetically-modified HDAC3-FLOX mice in combination with surgical infusions of adenoassociated virus expressing Cre recombinase to selectively delete HDAC3 within the dorsal hippocampus of adult mice. This genetic deletion approach avoids any potential developmental confounds often observed in traditional homozygous knockout animals. Deletion of hippocampal HDAC3 increased H4K8 acetylation and the expression of both c-fos and Nr4a2 were maintained beyond their normal expression window ( Figure 1 ). In addition, hippocampal HDAC3 deletions transformed a subthreshold training event into long-term object location memory, similar to general HDAC inhibitors (Stefanko et al., 2009; Haettig et al., 2011). Hippocampal HDAC3 deletions also transform a subthreshold training experience into a longterm memory that persists beyond a point at which normal memory fails in wild-type mice (7 days). The transformation of a training event that does not normally form a persistent long-term memory is reminiscent of the modulatory role of stress and arousal in enhancing memory for emotionally salient events (see sections below).
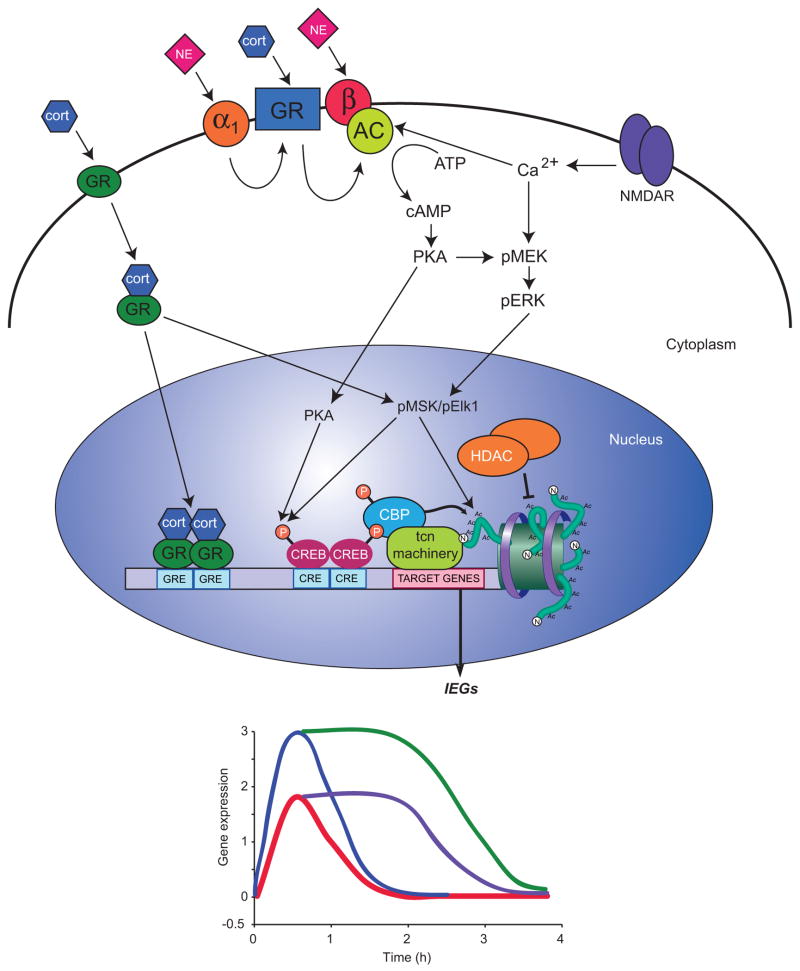
Figure 1. Regulation of gene expression by emotional memory.
In the basal state, the dynamic exchange between HATs and HDACs at the promoter and regulatory regions favor HDACs, which serve as molecular ‘brake pads’ by repressing gene expression. Emotionally salient stimuli activate the release of HDACs and activate the expression of immediate early genes (IEGs) through the activation of multiple neurochemical signaling pathways. The best-characterized pathways and components in mediating epigenetic regulation of gene expression for emotional memory are illustrated here. Emotionally salient events trigger the release of cortisol (cort) and norepinephrine (NE) that bind their respective receptors on post-synaptic neurons. Cortisol binds to both membrane-bound and nuclear hormone glucocorticoid receptors (GRs). Once activated, nuclear hormone GRs and the bound cortisol translocate to the nucleus where GRs dimerize and bind glucocorticoid-response elements (GREs) within regulatory and promoter regions. GRs can also regulate transcription through other protein - protein interactions, such as pMSK/pElk1. NE binds to post synaptic β- and α 1 -adrenoceptors, leading to the activation of adenylate cyclase (AC), cAMP formation, cAMP-dependent protein kinase (PKA) activation and phosphorylation of cAMP response binding protein (CREB). Neuronal activity also activates NMDA receptors (NMDAR) that increase intracellular calcium (Ca 2+ ) concentration, leading to the activation of AC and the ERK/MAPK signaling cascade (pMEK/perk/pMSK/pElk1). pMSK and PKA can both phosphorylate CREB bound to cAMP response elements (CREs) within the promoter of actively transcribed genes. Once bound to pCREB, CREB binding protein (CBP) interacts with the basal transcription machinery and acetylates histones. During this process, the molecular ‘brake pads’ (HDACs and associated co-repressors) are released from the promoter, allowing histone acetylation. The combined action of these three unique but interacting signaling cascades is to regulate transcription required for long-term memory formation. As shown in the gene expression time course shown at the bottom of the figure, IEGs are rapidly induced following a learning event and return to baseline within approximately two hours (red line). The remaining lines represent hypothesized changes in gene expression with HDAC inhibition. With HDAC inhibition, IEG expression is predicted to be extended beyond the time at which it would normally return to baseline (purple line; see Vecsey et al., 2007; McQuown et al., 2011). The blue line represents the predicted rapid increase in IEG expression following a learning event and HDAC inhibition at ‘primed’ promoters (see text). The expression of these gene targets are then predicted to be extended beyond their normal expression window (green line). Genes that have been shown to have extended expression following HDAC inhibition are critical for HDAC inhibition-dependent effects on enhanced/persistent long-term memory (e.g., nr4a2; McQuown et al., 2011).
Long-term memory for both object location and object recognition were impaired in mice with a single point mutation in the DAD domain (Y478A) of NCoR that disrupts the HDAC3/NCoR interaction, indicating that HDAC3 plays a critical role in memory for the object’s location as well as the object itself. Furthermore, post-training administration of an HDAC3 selective inhibitor produced similar effects on acetylation and long-term memory formation, indicating that the memory enhancements were due to the role of HDAC3’s regulation of transcription during memory consolidation. This was further evidenced by the required interaction between CBP and pCREB for HDAC3 to enhance memory. The HDAC3 inhibitor failed to enhance memory in transgenic mice in which interaction between CBP and CREB is disrupted, indicating that HDAC3 enhances memory through the regulation of CBP/CREB target genes. Specifically, blocking the CREB target gene Nr4a2, a known target of general HDAC inhibitors (Vecsey et al., 2007), blocked memory enhancement in mice with hippocampal deletions of HDAC3. These findings demonstrate the first evidence for the role of a specific gene target in the mechanism underlying HDAC regulation of memory formation. Furthermore, this is the first evidence that HDAC3 negatively regulates the gene expression required for long-term memory and that removing or blocking HDAC3 function can pull the ‘brakes’ off of the system, allowing the formation of long-term, persistent memories.
HDAC5
Renthal and colleagues (2007) recently examined the role of HDAC5 as a negative regulator of behavioral adaptations to chronic emotional stimuli. They utilized a social defeat stress paradigm that models some of the behavioral components of human depression. To examine the impact of chronic defeat stress on HDAC5, mice were placed in the home cage of an aggressive CD1 mouse for 10 min/day for 10 days. Between defeat sessions, a porous barrier allowing non-physical aggressive interactions separated the experimental mouse and aggressive mouse. Twenty-four hours after the last defeat session, social interactions between the experimental mouse and a novel mouse were measured. HDAC5 knockout mice displayed increased social avoidance following chronic (but not an acute single session) stress, indicating a role for HDAC5 in regulating behavioral adaptations to repeated stressful stimuli. In wild-type mice chronic, but not acute, stress down-regulated hdac5 expression in the nucleus accumbens (NAc), suggesting a potential mechanism for removing the HDAC5 ‘brake pads’ and allowing the expression of a subset of specific target genes in response to repeated stressful stimuli.
In comparison, chronic, but not acute, cocaine treatment does not alter hdac5 expression, but does increase the phosphorylation of HDAC5 (Ser259) and its export from the nucleus in the NAc (Renthal et al., 2007). This export mechanism is consistent with the activity-dependent phosphorylation and nuclear export of HDAC5 in hippocampal cultures (Chawla et al., 2003; but see Taniguchi et al., 2012) and together with the findings from the social defeat paradigm suggest that there are at least two independent mechanisms for regulating HDAC5 function ( hdac5 expression and HDAC5 nuclear export). In further support of the role of HDAC5 in regulating long-term behavioral adaptations to cocaine, over-expression of HDAC5 in the NAc decreases the rewarding effects of cocaine, while knockout of HDAC5 results in sensitization. Chronic cocaine treatment in wildtype mice also alters gene expression and histone H3 acetylation patterns in the NAc. These results led the authors to propose a model by which HDAC5 negatively regulates the gene expression required for responses to chronically stressful stimuli. In this model, chronic stress can reduce HDAC5 function by decreasing mRNA expression or increasing nuclear export. The subsequent decrease in HDAC5 function removes the ‘brakes’ on the gene expression underlying behavioral adaptations to chronic stress.
HDAC6
In the adult mouse, HDAC6 is expressed within the medial and dorsal raphe nuclei and expression co-localizes with a marker of serotonergic neurons (tryptophan hydroxylase 2; Fukada et al., 2012). Given the role of serotonin in regulating emotional behaviors, Fukada and colleagues used HDAC6 germline knockout mice to evaluate the role of HDAC6 in regulating a series of emotional behaviors. HDAC6 knockout mice are hyperactive in a novel environment (increased distance travelled in open field testing) but show normal locomotion in the home cage. HDAC6 knockout mice also demonstrate decreased levels of anxiety (increased entries into the open arm of an elevated plus maze) and lower levels of immobility on a tail suspension test, indicating that a lack of HDAC6 may lead to an anti-depressant-like phenotype. Similarly, acute treatment with an HDAC6 inhibitor (NCT-14b) produced similar anti-depressant-like effects on immobility, indicating that results with the germline knockout animals were not developmental. The HDAC6 knockout mice also have normal serum corticosterone levels at baseline and in response to stress, normal serotonin levels, and normal behavioral responses to both serotonin and noradrenaline reuptake inhibitors (fluoxetine and desipramine). Both HDAC6 knockout mice and wild-type mice treated with NCT-14b show normal levels of histone acetylation but increased levels of acetylated α-tubulin, a protein implicated in cytoskeletal dynamics and cell motility (reviewed in Valenzuela-Fernández et al., 2008). Presently, the mechanism for how HDAC6 may regulate emotional behavior is unknown and may involve the deacetylation of multiple cytoplasmic protein targets including α-tubulin, cortactin (actin cytoskeleton dynamics) and HSP90 (chaperone) (reviewed in Valenzuela-Fernández et al., 2008). Interestingly, not all emotional behaviors appear to be regulated by HDAC6, as chronic HDAC6 inhibition (10 days) does not alter contextual fear memory (Guan et al., 2009). Chronic HDAC6 inhibitor administration may, however, result in either molecular or cellular compensation, hence masking any potential enhancements of a single, acute dose.
SIRT1
The majority of work on the role of HDACs in learning and memory has focused on class I and II HDACs; however the class III HDAC SIRT1 has recently been demonstrated to play a role in regulating memory formation. Gao and colleagues (2010) used transgenic mice with brain-specific deletion mutations of the catalytic domain of SIRT1 to examine the role of neuronal SIRT1 in memory. SIRT1 mutant mice had impaired long-term memory for contextual and cued fear, novel object recognition and spatial Morris water maze as well as impaired maintenance of LTP. These effects were mediated by post-translational down-regulation of CREB by miR-134. Expression of miR-134 is normally repressed by SIRT1. In SIRT1 mutant mice this repression is released, resulting in increased miR-134 expression, decreased CREB protein levels, and subsequently a decrease in BDNF levels. Knockdown of miR-134 in the CA1 region of the dorsal hippocampus of SIRT1 mutant mice restored CREB and BDNF levels and rescued deficits in contextual fear memory and LTP maintenance. While these results demonstrate the role of SIRT1 as a negative regulator of microRNA expression, it is unclear what the impact of developmental abnormalities in the SIRT1 mutant mice may be. The genetically modified SIRT1 mutant mice used by Gao et al. (2010) were previously shown to have metabolic abnormalities including disrupted somatotropic signaling and dwarfism (Cohen et al., 2009). While these mutant mice did have normal shock sensitivity, locomotor activity, cued water maze and basal synaptic transmission (Gao et al., 2010), future studies would benefit from utilizing inducible models that bypass the potential confound of the known role of SIRT1 in development (Chen et al., 2003; McBurney et al., 2003; Cohen et al., 2009).
Collectively, the highlighted papers above provide a flavor for the type of pharmacological, genetic and behavioral approaches currently being used to explore the role of individual HDACs in learning and memory. Both the findings with non-selective HDAC inhibitors and more HDAC-specific manipulations strongly support a role for HDACs as negative regulators of gene expression required for long-term memory formation.
Molecular brake pad hypothesis: forming enduring emotional memories
We recently proposed the molecular brake path hypothesis (McQuown et al., 2011), which postulates that HDACs and their associated co-repressors form complexes (molecular brake pads) that normally keep a specific set of genes required for long-term memory processes in a silent state. As discussed in the preceding section, studies using both general HDAC inhibitors as well as manipulations of specific HDAC enzymes provide evidence that HDACs serve as critical negative regulators of long-term memory formation. This is consistent with the in vitro findings discussed above indicating a dynamic interaction between HATs and HDACs at active promoters and the belief that HDACs may prevent gene expression at genes ‘primed’ for activation (Wang et al., 2009). In this model a sufficiently strong activity-dependent stimulus is required to remove these ‘brake pads’ in order to initiate the gene expression required for longterm memory formation. Emotionally arousing experiences tend to be better remembered and these memories can last for the lifetime of the organism (reviewed in Cahill and McGaugh, 1996; McGaugh, 2000, 2004). We propose here that the neuromodulatory signaling activated by emotionally arousing events may serve as powerful regulators for HDAC removal and consequently the activation of gene expression required for the formation of persistent long-term memories.
Emotionally salient events lead to enduring long-term memories
This next section of this review will focus on the role of several neuromodulators [corticotropin-releasing hormone (CRH); epinephrine and glucocorticoids] in forming enduring memories following an acute, emotionally salient event. These types of long-lasting memories of stressful or arousing events may confer an evolutionary advantage by allowing the organism to adjust its behavioral response in similar future situations. The ability of emotional arousal to modulate memory formation to produce a robust and enduring memory has been extensively explored and the basic signaling cascades are highlighted below. This section is not intended to serve as a comprehensive review of research on emotional memory, but instead to provide a context for the proposed role of molecular ‘brake pads’ in modulating long-term memory formation for emotionally salient events.
Emotional arousal triggers a diverse set of neurochemical signals within both the peripheral and central nervous system that can vary depending on the type and duration of the arousing event (for a review, see Joëls et al., 2011; McClelland et al., 2011). For example, following an emotionally arousing training event, such as fear conditioning in rodents, CRH is released from cells within the hypothalamus (Vale et al., 1981), hippocampus (Chen et al., 2001) and amygdala (Roozendaal et al., 2002). CRH within the hypothalamus induces the release of corticotropin from the pituitary that subsequently promotes the release of glucocorticoids (cortisol in humans and corticosterone in rodents; for a review, see McClelland et al., 2011). In parallel, epinephrine is released from the adrenal glands and activates the release of norepinephrine in the basal lateral amygdala (BLA). As shown in Figure 1, norepinephrine binds to post-synaptic β- and α 1 -adrenoceptors in the BLA, leading to the activation of adenylate cyclase, cAMP formation, cAMP-dependent protein kinase (PKA) activation and phosphorylation of CREB (Roozendaal and Quirarte, 2002; Roozendaal et al., 2006).
In contrast, glucocorticoids can diffuse directly across the blood-brain barrier and bind glucocorticoid receptors (GRs) within the brain. GRs are nuclear hormone receptors that translocate to the nucleus upon glucocorticoid binding and bind as homodimers to glucocorticoid-responsive DNA elements at the promoters and enhancers of a subset of target genes. Once bound to DNA, GRs interact with the basal transcription machinery, regulatory transcription factors and chromatin remodeling enzymes, including HATs and HDACs, leading to transcriptional activation or repression (Beato and Sánchez-Pacheco, 1996; reviewed in Revollo and Cidlowski, 2009). GRs can also regulate transcription and other non-genomic signaling cascades through protein-protein interactions independent of DNA binding (reviewed in Revollo and Cidlowski, 2009). In addition, within the BLA the interaction between GRs and the membrane bound β-adrenoceptor–cAMP system indicates that glucocorticoids may also signal through membrane-bound GRs to modulate cAMP production by adrenoceptor activation ( Figure 1; reviewed in Roozendaal et al., 2007).
CRH, norepinephrine and glucocorticoids are all involved in modulating memory consolidation for emotionally salient events (reviewed in McGaugh, 2004; Roozendaal et al., 2007; Joëls et al., 2011). For example, inhibitory avoidance training activates CRH release in the amygdala and blocking this release impairs memory consolidation (Roozendaal and Quirarte, 2002). Similarly, extensive evidence has implicated a role for norepinephrine and glucocorticoid release in the amygdala in modulating memory formation (reviewed in McGaugh, 2004; Roozendaal et al., 2007). Inhibitory avoidance training induces the release of norepinephrine in the amygdala (McIntyre et al., 2002), post-training infusions of norepinephrine or adrenoreceptor agonists enhance memory formation (Liang et al., 1986; Ferry and McGaugh, 1999) and blocking of adrenoreceptors impairs epineph-rine’s enhancing effect on memory consolidation (Liang et al., 1986). Systemic delivery of corticosterone (Kovács et al., 1977) or direct infusions of GR agonists into the amygdala enhance inhibitory avoidance memory formation (Roozendaal and McGaugh, 1997; reviewed in Roozendaal, 2000). Blocking corticosterone synthesis prevents epinephrine’s enhancing effects on memory consolidation (Roozendaal et al., 1996) and, conversely, blocking β-adrenergic receptor activation blocks the memory-enhancing effects of a post-training GR agonist (Roozendaal et al., 1999), indicating an interaction between glucocorticoid- and adrenergic-mediated enhancement of memory consolidation. This hypothesis was further supported by evidence that activation of GRs on the post-synaptic BLA neurons facilitates norepinephrine signaling through an interaction with α 1 -adrenoceptors and the β-adrenoceptor–cAMP–PKA–pCREB signaling cascade (see Figure 1; Roozendaal and Quirarte, 2002; Roozendaal et al., 2006). Furthermore, CRH also appears to facilitate the formation of emotional memories by interacting with GRs and the β-adrenoceptor–cAMP signaling pathway (Roozendaal et al., 2008).
Epigenetic regulation of emotional memory formation
While the role of CRH and adrenal hormones in enhancing memory formation for emotionally salient events has been extensively explored (reviewed in Roozendaal et al. 2007; Joëls et al., 2011), how activation of the subsequent downstream signaling cascades leads to stronger and more persistent memory remains an area of active research. Recent work has begun to explore the role of epigenetic mechanisms in regulating the gene expression thought to underlie stress-related learning and memory (reviewed in Trollope et al., 2012). For example, a single stressful event such as exposure to the scent of a predator, forced swimming or exposure to a novel environment increases phosphorylation (serine 10) and acetylation (lysine 14) of histone H3 (H3S10p-K14ac; Bilang Bleuel et al., 2005; Chandramohan et al., 2007, 2008) and activation of gene expression ( erg-1 and c-fos ) in the rodent dentate gyrus (Gutièrrez-Mecinas et al., 2011). Both the increases in H3S10p-K14ac and gene expression require GR and NMDA receptor activity, indicating an interaction between the two signaling cascades (Gutièrrez-Mecinas et al., 2011). Similarly, the long-term memory of a forced swim session (measured by a decreased latency to initiate an immobility response at test (Porsolt et al., 1977; De Pablo et al., 1989; Bilang Bleuel et al., 2005) requires both GR and NMDA receptor activity during training (Bilang Bleuel et al., 2005; Chandramohan et al., 2008; Gutièrrez-Mecinas et al., 2011). Forced swim training results in rapid and transient activation of several signaling molecules downstream of NMDA receptors ( Figure 1 ), including phosphorylation of extracellular-signal-regulated kinase 1/2 (ERK1/2), mitogen-and-stress activated protein kinase 1 (MSK1) and ETS-like transcription factor 1 (Elk1). In addition, phosphorylation of MSK and Elk1 (but not ERK1/2) also requires GR activity, further indicating a synergy between these two signaling cascades, potentially through direct protein - protein inter actions between activated GRs and ERK/MSK/Elk1 (Gutièrrez-Mecinas et al., 2011). Activation of this signaling pathway would lead to phosphorylation of histone 3 at serine 10 (presumably by pMSK1) and acetylation of histone 3 lysine 14 by the recruitment of specific HATs (Chandramohan et al., 2008; Gutièrrez-Mecinas et al., 2011). Collectively, these findings indicate that both NMDA receptor and GR activity initiate signaling cascades critical for the epigenetic regulation of gene expression following stressful experiences, but it is unclear if these findings extend beyond dentate gyrus granule neurons.
We recently investigated the role of glucocorticoid activity in modulating long-term object memory by histone acetylation within the hippocampus and insular cortex (Roozendaal et al., 2010). Systemic post-training corticosterone injections increased H3K14 acetylation in both brain regions and enhanced long-term memory for both object location and recognition (Okuda et al., 2004; Roozendaal et al., 2010). HDAC inhibition post training with infusions of sodium butyrate directly into the hippocampus enhanced object location but not object recognition memory, whereas infusions into the insular cortex showed the opposite effect with enhanced object recognition but not object location memory (Roozendaal et al., 2010). These findings are consistent with the dorsal hippocampus having a critical role in the retrieval of object location but not object recognition memory (Haettig et al., 2011) and implicate glucocorticoid signaling in modulating histone acetylation and object memory.
To further examine the mechanism of how glucocorticoids interact with histone acetylation, we used a membrane impermeable corticosterone (cort:BSA) to activate membrane-bound GRs. Cort:BSA delivered into the insular cortex after training enhanced object recognition memory and increased pCREB levels. GR antagonists or PKA inhibition blocked this enhancement. These findings indicate that in insular cortex corticosterone activates membrane-bound GRs that signal through PKA to increase CREB phosphorylation. However, this does not rule out a role for glucocorticoid hormone receptors in directly regulating gene expression in response to an emotionally arousing experience or in regulating HDAC function. It was recently shown that neurotoxic insults, such as those thought to occur during Alzheimer’s disease progression, up-regulate hdac2 expression through glucocorticoid nuclear hormone receptors (Gräff et al., 2012).
We previously demonstrated that the ability of HDAC inhibitors to increase in histone acetylation and enhance memory required intact interaction between CBP and pCREB in the hippocampus (Vecsey et al., 2007). Given that glucocorticoids increase histone acetylation and pCREB levels, we next asked whether memory enhancements by HDAC inhibition require glucocorticoid signaling. Both a glucocorticoid antagonist and PKA inhibitor blocked the HDAC inhibition-mediated enhancement of object recognition memory, indicating that memory enhancement by HDAC inhibition in the insular cortex requires glucocorticoid and PKA signaling, presumably to activate CBP/pCREB interactions ( Figure 1 ). This is consistent with previous findings that PKA and PKC can activate histone acetylation by MEK/ERK signaling (Levenson et al., 2004; Chwang et al., 2006). To further explore this hypothesis, we tested whether corticosterone could enhance memory in the absence of CBP/ pCREB interactions using Cbp KIX/KIX mutant mice. Cbp KIX/ KIX homozygous knock-in mice express CBP with a triplepoint mutation in the CREB-binding (KIX) domain that disrupts CBP/pCREB interaction (Kasper et al., 2002). CbpK IX/KIX mice have normal short-term but impaired long-term object recognition and location memory (Wood et al., 2006; Haettig et al., 2011). Interestingly, post-training corticosterone rescued memory deficits for object recognition but not for object location, indicating that corticosterone enhances memory for object location (but not object recognition) in a CBP/pCREBdependent manner. Furthermore, these findings implicate a differential role for CBP in the insular cortex and hippocampus, consistent with our previous findings that HDAC inhibition enhances hippocampal synaptic plasticity and object location memory in a CBP-dependent manner (Vecsey et al., 2007; Barrett et al., 2011; Haettig et al., 2011; McQuown et al., 2011) and enhances object recognition memory in a CBP-independent manner (Stefanko et al., 2009; Haettig et al., 2011). The enhancement of object recognition memory in the absence of CBP/pCREB interaction indicates that different HATs may play a critical role in brain regions such as the insular cortex.
The molecular brake pad hypothesis: making emotional memories
The role of glucocorticoid signaling in modulating histone acetylation through HDAC inhibition is an area of active investigation. It is currently unclear how glucocorticoids or their downstream signaling cascades lead to the presumed removal of HDACs from the gene promoters required for long-term memory. In the working model proposed here, HDACs are maintained at ‘primed’ promoters (see the above section on gene regulation) of a subset of genes required for long-term memory. When a sufficiently strong stimulus is engaged, such as that produced during emotional arousal, signaling cascades are engaged (i.e., glucocorticoids) that induce the removal of HDACs allowing gene transcription and subsequent memory formation. Administering corticosterone or an HDAC inhibitor simply mimics the endogenous process by engaging or bypassing the signaling cascades required for HDAC removal. Histone acetylation alone is not sufficient for gene activation, however, and additional signaling cascades that lead to pCREB and HAT (CBP or other as yet unidentified HATs) engagement at the promoter are also required. HDAC inhibition tips the dynamic interaction between HATs and HDACs in favor of histone acetylation, allowing the extended expression of a subset of activity-regulated genes required for long-term memory formation (Figure 1; Vecsey et al., 2007). These changes in gene expression are thought to then underlie the transformation of a subthreshold learning event into a persistent long-term memory (Stefanko et al., 2009; McQuown et al., 2011).
These proposed mechanisms potentially play a powerful role in generating strong and persistent emotional memories by removing the molecular ‘brake pads’ that otherwise prevent memory formation. Misregulation of this process may be critically involved in the development of abnormally strong and intrusive memories, such as those that occur in post-traumatic stress disorder. In contrast, removal of the ‘brake pads’ may be powerfully therapeutic for disorders in which gene expression is misregulated. Recently HDAC inhibitors have been investigated as potential therapeutics in models of a variety of neurodegenerative diseases including Alzheimer’s (Fischer et al., 2007; Kilgore et al., 2010; Gräff et al., 2012), Parkinson’s (Kontopoulos et al., 2006) and Huntington’s disease (Ferrante et al., 2003; Hockly et al., 2003; Gardian et al., 2005). In many of these diseases, HDAC inhibitors were able to ameliorate both the underlying disease pathology and memory impairments (reviewed in Gäff et al., 2011). In addition, HDACs are also targets for treating depression (Tsankova et al., 2006; Schroeder et al., 2007), drug addiction (Malvaez et al., 2010, 2011), stroke (Faraco et al., 2006) and even aging (Peleg et al., 2010). As interest in targeting individual HDACs as potential therapeutics increases (see sections above), it will be of critical importance to gain a better understanding of how these individual enzymes and complexes are regulated and how they in turn regulate gene expression in different disease models. More targeted approaches to regulating HDAC function could serve to reestablish the balance between HDACs and HATs, correcting aberrant gene expression changes associated with a disease state. Overall, the proposed role of HDACs as negative regulators of gene expression required for persistent, emotionally salient memory formation provides an attractive target for future work on therapeutic development.
Biographies
 Dr. Wood received his PhD from the Department of Molecular Biology at Princeton University in molecular cancer biology. He then switched fields to study the neurobiology of learning and memory at the University of Pennsylvania for his postdoctoral fellowship. He is currently an associate professor in the Department of Neurobiology and Behavior at the University of California Irvine. He is also the Director of the Interdepartmental Neuroscience Program. His research program focuses on understanding the molecular mechanisms underlying long-term memory formation and drugseeking behavior.
Dr. Wood received his PhD from the Department of Molecular Biology at Princeton University in molecular cancer biology. He then switched fields to study the neurobiology of learning and memory at the University of Pennsylvania for his postdoctoral fellowship. He is currently an associate professor in the Department of Neurobiology and Behavior at the University of California Irvine. He is also the Director of the Interdepartmental Neuroscience Program. His research program focuses on understanding the molecular mechanisms underlying long-term memory formation and drugseeking behavior.
 Annie Vogel-Ciernia is a graduate student in Dr. Wood’s lab in the Dep artment of Neurobiology & Behavior at the University of California, Irvine. Annie received her undergraduate degree from North Dakota State University with double majors in biotechnology and psychology. Annie’s current research focuses on the role of nucleosome remodeling in long-term memory formation.
Annie Vogel-Ciernia is a graduate student in Dr. Wood’s lab in the Dep artment of Neurobiology & Behavior at the University of California, Irvine. Annie received her undergraduate degree from North Dakota State University with double majors in biotechnology and psychology. Annie’s current research focuses on the role of nucleosome remodeling in long-term memory formation.
Contributor Information
Annie Vogel-Ciernia, Department of Neurobiology and Behavior, Center for the Neurobiology of Learning and Memory, University of California Irvine Irvine, CA 92697–3800, USA.
Marcelo A. Wood, Email: mwood@uci.edu, Department of Neurobiology and Behavior, Center for the Neurobiology of Learning and Memory, University of California Irvine Irvine, CA 92697–3800, USA
References
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;4:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahari-Javan S, Maddalena A, Kerimoglu C, Wittnam J, Held T, Bähr M, Burkhardt S, Delalle I, Kügler S, Fischer A, et al. HDAC1 regulates fear extinction in mice. J Neurosci. 2012;32:5062–5073. doi: 10.1523/JNEUROSCI.0079-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RM, Wood MA. Beyond transcription factors: the role of chromatin modifying enzymes in regulating transcription required for memory. Learn Mem. 2008;15:460–467. doi: 10.1101/lm.917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacol. 2011;36:1545–1556. doi: 10.1038/npp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- Beato M, Sánchez-Pacheco A. Interaction of steroid hormone receptors with the transcription initiation complex. Endocr Rev. 1996;17:587–609. doi: 10.1210/edrv-17-6-587. [DOI] [PubMed] [Google Scholar]
- Bilang Bleuel A, Ulbricht S, Chandramohan Y, De Carli S, Droste SK, Reul JMHM. Psychological stress increases histone H3 phosphorylation in adult dentate gyrus granule neurons: involvement in a glucocorticoid receptor-dependent behavioural response. Eur J Neurosci. 2005;22:1691–1700. doi: 10.1111/j.1460-9568.2005.04358.x. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Bourtchouladze R, Lidge R, Catapano R, Stanley J, Gossweiler S, Romashko D, Scott R, Tully T. A mouse model of Rubinstein-Taybi syndrome: defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc Natl Acad Sci USA. 2003;100:10518–10522. doi: 10.1073/pnas.1834280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1–11 in the rat brain. J Mol Neurosci. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- Cahill L, McGaugh JL. Modulation of memory storage. Curr Opin Neurobiol. 1996;6:237–242. doi: 10.1016/s0959-4388(96)80078-x. [DOI] [PubMed] [Google Scholar]
- Chandramohan Y, Droste SK, Reul JMHM. Novelty stress induces phospho-acetylation of histone H3 in rat dentate gyrus granule neurons through coincident signalling via the N-methyl-D-aspartate receptor and the glucocorticoid receptor: relevance for c-fos induction. J Neurochem. 2007;101:815– 828. doi: 10.1111/j.1471-4159.2006.04396.x. [DOI] [PubMed] [Google Scholar]
- Chandramohan Y, Droste SK, Arthur JSC, Reul JMHM. The forced swimming-induced behavioural immobility response involves histone H3 phospho-acetylation and c-Fos induction in dentate gyrus granule neurons via activation of the N-methyl-d-aspartate/extracellular signal-regulated kinase/mitogen-and stress-activated kinase signalling pathway. Eur J Neurosci. 2008;27:2701–2713. doi: 10.1111/j.1460-9568.2008.06230.x. [DOI] [PubMed] [Google Scholar]
- Chawla S, Vanhoutte P, Arnold FJL, Huang CLH, Bading H. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem. 2003;85:151–159. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bender RA, Frotscher M, Baram TZ. Novel and transient populations of corticotropin-releasing hormone-expressing neurons in developing hippocampus suggest unique functional roles: a quantitative spatiotemporal analysis. J Neurosci. 2001;21:7171–7181. doi: 10.1523/JNEUROSCI.21-18-07171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Yokosuka O, Arai M, Tada M, Fukai K, Imazeki F, Kato M, Seki N, Saisho H. Identification of genes up-regulated by histone deacetylase inhibition with cDNA microarray and exploration of epigenetic alterations on hepatoma cells. J Hepatol. 2004;41:436–445. doi: 10.1016/j.jhep.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Chwang WB, O’Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13:322– 328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pablo JM, Parra A, Segovia S, Guillamón A. Learned immobility explains the behavior of rats in the forced swimming test. Physiol Behav. 1989;46:229–237. doi: 10.1016/0031-9384(89)90261-8. [DOI] [PubMed] [Google Scholar]
- de Ruijter AJM, van Gennip AH, Caron HN, Kemp S, van Kuilenburg ABP. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraco G, Pancani T, Formentini L, Mascagni P, Fossati G, Leoni F, Moroni F, Chiarugi A. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol. 2006;70:1876–1884. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- Fass DM, Butler JEF, Goodman RH. Deacetylase activity is required for cAMP activation of a subset of CREB target genes. J Biol Chem. 2003;278:43014–43019. doi: 10.1074/jbc.M305905200. [DOI] [PubMed] [Google Scholar]
- Federman N, Fustinana MS, Romano A. Histone acetylation is recruited in consolidation as a molecular feature of stronger memories. Learn Mem. 2009;16:600–606. doi: 10.1101/lm.1537009. [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Kubilus JK, Lee J, Ryu H, Beesen A, Zucker B, Smith K, Kowall NW, Ratan RR, Luthi-Carter R, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry B, McGaugh JL. Clenbuterol administration into the basolateral amygdala post-training enhances retention in an inhibitory avoidance task. Neurobiol Learn Mem. 1999;72:8–12. doi: 10.1006/nlme.1998.3904. [DOI] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, Verdin E. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J Biol Chem. 2001;276:35826– 35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45– 57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Fukada M, Hanai A, Nakayama A, Suzuki T, Miyata N, Rodriguiz RM, Wetsel WC, Yao TP, Kawaguchi Y. Loss of deacetylation activity of hdac6 affects emotional behavior in mice. PloS One. 2012;7:e30924. doi: 10.1371/journal.pone.0030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Gräff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardian G, Browne SE, Choi DK, Klivenyi P, Gregorio J, Kubilus JK, Ryu H, Langley B, Ratan RR, Ferrante RJ, et al. Neuroprotective effects of phenylbutyrate in the N171–82Q transgenic mouse model of Huntington’s disease. J Biol Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Therap. 2003;2:151–163. [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Gräff J, Kim D, Dobbin MM, Tsai LH. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiol Rev. 2011;91:603–649. doi: 10.1152/physrev.00012.2010. [DOI] [PubMed] [Google Scholar]
- Gräff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJF, Fass DM, Kao PF, Kahn M, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, Kandel ER. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJF, Zhou Y, Wang X, Mazitschek R, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutièrrez-Mecinas M, Trollope AF, Collins A, Morfett H, Hesketh SA, Kersanté F, Reul JMHM. Long-lasting behavioral responses to stress involve a direct interaction of glucocorticoid receptors with ERK1/2–MSK1–Elk-1 signaling. Proc Natl Acad Sci USA. 2011;108:13806–13811. doi: 10.1073/pnas.1104383108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haettig J, Stefanko DP, Multani ML, Figueroa DX, McQuown SC, Wood MA. HDAC inhibition modulates hippocampus-dependent long-term memory for object location in a CBP-dependent manner. Learn Mem. 2011;18:71–79. doi: 10.1101/lm.1986911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PA, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci USA. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Joëls M, Fernandez G, Roozendaal B. Stress and emotional memory: a matter of timing. Trends Cogn Sci. 2011;15:280–288. doi: 10.1016/j.tics.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Juan LJ, Shia WJ, Chen MH, Yang WM, Seto E, Lin YS, Wu CW. Histone deacetylases specifically down-regulate p53-dependent gene activation. J Biol Chem. 2000;275:20436–20443. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Boussouar F, Ney PA, Jackson CW, Rehg J, van Deursen JM, Brindle PK. A transcriptionfactor- binding surface of coactivator p300 is required for haematopoiesis. Nature. 2002;419:738–743. doi: 10.1038/nature01062. [DOI] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontopoulos E, Parvin JD, Feany MB. Alphasynuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Gen. 2006;15:3012–3023. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kovács GL, Telegdy G, Lissák K. Dose-dependent action of corticosteroids on brain serotonin content and passive avoidance behavior. Horm Behav. 1977;8:155–165. doi: 10.1016/0018-506x(77)90032-0. [DOI] [PubMed] [Google Scholar]
- Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfí A, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattal KM, Barrett RM, Wood MA. Systemic or intrahippocampal delivery of histone deacetylase inhibitors facilitates fear extinction. Behav Neurosci. 2007;121:1125–1131. doi: 10.1037/0735-7044.121.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang KC, Juler RG, McGaugh JL. Modulating effects of posttraining epinephrine on memory: involvement of the amygdala noradrenergic system. Brain Res. 1986;368:125–133. doi: 10.1016/0006-8993(86)91049-8. [DOI] [PubMed] [Google Scholar]
- Malvaez M, Sanchis-Segura C, Vo D, Lattal KM, Wood MA. Modulation of chromatin modification facilitates extinction of cocaine-induced conditioned place preference. Biol Psychiatry. 2010;67:36–43. doi: 10.1016/j.biopsych.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Mhillaj E, Matheos DP, Palmery M, Wood MA. CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaineassociated behaviors. J Neurosci. 2011;31:16941–16948. doi: 10.1523/JNEUROSCI.2747-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S, Korosi A, Cope J, Ivy A, Baram TZ. Emerging roles of epigenetic mechanisms in the enduring effects of early-life stress and experience on learning and memory. Neurobiol Learn Mem. 2011;96:79–88. doi: 10.1016/j.nlm.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh JL. Memory–a century of consolidation. Science. 2000;287:248–251. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- McGaugh JL. The amygdala modulates the consolidation of memories of emotionally arousing experiences. Ann Rev Neurosci. 2004;27:1–28. doi: 10.1146/annurev.neuro.27.070203.144157. [DOI] [PubMed] [Google Scholar]
- McIntyre CK, Hatfield T, McGaugh JL. Amygdala norepinephrine levels after training predict inhibitory avoidance retention performance in rats. Eur J Neurosci. 2002;16:1223– 1226. doi: 10.1046/j.1460-9568.2002.02188.x. [DOI] [PubMed] [Google Scholar]
- McQuown SC, Wood MA. HDAC3 and the molecular brake pad hypothesis. Neurobiol Learn Mem. 2011;96:27–34. doi: 10.1016/j.nlm.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métivier R, Penot G, Hübner MR, Reid G, Brand H, Koš M, Gannon F. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Michán S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle. 2008;7:1539–1544. doi: 10.4161/cc.7.11.6006. [DOI] [PubMed] [Google Scholar]
- Ng HH, Bird A. Histone deacetylases: silencers for hire. Trends Biochem Sci. 2000;25:121–126. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- Okuda S, Roozendaal B, McGaugh JL. Glucocorticoid effects on object recognition memory require trainingassociated emotional arousal. Proc Natl Acad Sci USA. 2004;101:853–858. doi: 10.1073/pnas.0307803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Perissi V, Scafoglio C, Zhang J, Ohgi KA, Rose DW, Glass CK, Rosenfeld MG. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol Cell. 2008;29:755–766. doi: 10.1016/j.molcel.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Porsolt R, Bertin A, Jalfre M. Behavioral despair in mice: a primary screening test for antidepressants. Arch Int Pharmacodyn Ther. 1977;229:327–336. [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Revollo JR, Cidlowski JA. Mechanisms generating diversity in glucocorticoid receptor signaling. Ann NY Acad Sci. 2009;1179:167–178. doi: 10.1111/j.1749-6632.2009.04986.x. [DOI] [PubMed] [Google Scholar]
- Roozendaal B. Glucocorticoids and the regulation of memory consolidation. Psychoneuroendocrinology. 2000;25:213– 238. doi: 10.1016/s0306-4530(99)00058-x. [DOI] [PubMed] [Google Scholar]
- Roozendaal B, McGaugh JL. Glucocorticoid receptor agonist and antagonist administration into the basolateral but not central amygdala modulates memory storage. Neurobiol Learn Mem. 1997;67:176–179. doi: 10.1006/nlme.1996.3765. [DOI] [PubMed] [Google Scholar]
- Roozendaal B, Quirarte G. Glucocorticoids interact with the basolateral amygdala β-adrenoceptor– cAMP/cAMP/ PKA system in influencing memory consolidation. Eur J Neurosci. 2002;15:553–560. doi: 10.1046/j.0953-816x.2001.01876.x. [DOI] [PubMed] [Google Scholar]
- Roozendaal B, Carmi O, McGaugh JL. Adrenocortical suppression blocks the memory-enhancing effects of amphetamine and epinephrine. Proc Natl Acad Sci USA. 1996;93:1429–1433. doi: 10.1073/pnas.93.4.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B, Nguyen BT, Power AE, McGaugh JL. Basolateral amygdala noradrenergic influence enables enhancement of memory consolidation induced by hippocampal glucocorticoid receptor activation. Proc Natl Acad Sci USA. 1999;96:11642–11647. doi: 10.1073/pnas.96.20.11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B, Brunson KL, Holloway BL, McGaugh JL, Baram TZ. Involvement of stress-released corticotropin-releasing hormone in the basolateral amygdala in regulating memory consolidation. Proc Natl Acad Sci USA. 2002;99:13908–13913. doi: 10.1073/pnas.212504599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B, Okuda S, Van der Zee EA, McGaugh JL. Glucocorticoid enhancement of memory requires arousal-induced noradrenergic activation in the basolateral amygdala. Proc Natl Acad Sci USA. 2006;103:6741–6746. doi: 10.1073/pnas.0601874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B, Barsegyan A, Lee S. Adrenal stress hormones, amygdala activation, and memory for emotionally arousing experiences. Prog Brain Res. 2007;167:79–97. doi: 10.1016/S0079-6123(07)67006-X. [DOI] [PubMed] [Google Scholar]
- Roozendaal B, Schelling G, McGaugh JL. Corticotropin-releasing factor in the basolateral amygdala enhances memory consolidation via an interaction with the beta-adrenoceptor-cAMP pathway: dependence on glucocorticoid receptor activation. J Neurosci. 2008;28:6642–6651. doi: 10.1523/JNEUROSCI.1336-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B, Hernandez A, Cabrera SM, Hagewoud R, Malvaez M, Stefanko DP, Haettig J, Wood MA. Membrane-associated glucocorticoid activity is necessary for modulation of long-term memory via chromatin modification. J Neurosci. 2010;30:5037–5046. doi: 10.1523/JNEUROSCI.5717-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, Daigle KR, Fenn JD, Adelman K, Dudek SM. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat Neurosci. 2011;14:848–856. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Schroeder FA, Lin CL, Crusio WE, Akbarian S. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol Psychiatry. 2007;62:55– 64. doi: 10.1016/j.biopsych.2006.06.036. [DOI] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA. Modulation of long-term memory for object recognition via HDAC inhibition. Proc Natl Acad Sci USA. 2009;106:9447–9452. doi: 10.1073/pnas.0903964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swank MW, Sweatt JD. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/RSK cascade in insular cortex during novel taste learning. J Neurosci. 2001;21:3383–3391. doi: 10.1523/JNEUROSCI.21-10-03383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Carreira MB, Smith LN, Zirlin BC, Neve RL, Cowan CW. Histone deacetylase 5 limits cocaine reward through cAMP-induced nuclear import. Neuron. 2012;73:108–120. doi: 10.1016/j.neuron.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trollope AF, Gutièrrez-Mecinas M, Mifsud KR, Collins A, Saunderson EA, Reul JMHM. Stress, epigenetic control of gene expression and memory formation. Exp Neurol. 2012;233:3–11. doi: 10.1016/j.expneurol.2011.03.022. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Vale W, Spiess J, Rivier C, Rivier J. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- Valenzuela-Fernández A, Cabrero JR, Serrador JM, Sánchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 2008;18:291–297. doi: 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB: CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PJ, Fairall L, Santos GM, Schwabe JWR. Structure of HDAC3 bound to co-repressor and inositol tetra phosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Wood M. Introduction to the special issue of Neurobiology of Learning and Memory on epigenetics and memory. Neurobiol Learn Mem. 2011;96:1. doi: 10.1016/j.nlm.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AMM, Lombardi TL, Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Attner MA, Oliveira AMM, Brindle PK, Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn Mem. 2006;13:609–617. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh SH, Lin CH, Gean PW. Acetylation of nuclear factor-κB in rat amygdala improves long-term but not short-term retention of fear memory. Mol Pharmacol. 2004;65:1286–1292. doi: 10.1124/mol.65.5.1286. [DOI] [PubMed] [Google Scholar]
- Yu TT, McIntyre JC, Bose SC, Hardin D, Owen MC, McClintock TS. Differentially expressed transcripts from phenotypically identified olfactory sensory neurons. J Comp Neurol. 2005;483:251–262. doi: 10.1002/cne.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]