Abstract
During eukaryotic translation initiation, the small ribosomal subunit, assisted by initiation factors, locates the messenger RNA start codon by scanning from the 5′ cap. This process is powered by the eukaryotic initiation factor 4A (eIF4A), a DEAD-box helicase. eIF4A has been thought to unwind structures formed in the untranslated 5′ region via a nonprocessive mechanism. Using a single-molecule assay, we found that eIF4A functions instead as an adenosine triphosphate–dependent processive helicase when complexed with two accessory proteins, eIF4G and eIF4B. Translocation occurred in discrete steps of 11 ± 2 base pairs, irrespective of the accessory factor combination. Our findings support a memory-less stepwise mechanism for translation initiation and suggest that similar factor-dependent processivity may be shared by other members of the DEAD-box helicase family.
DEAD-box proteins are ubiquitous enzymes responsible for RNA remodeling. Found in eukaryotes as well as eubacteria and archaea, DEAD-box proteins participate in different stages of the mRNA life cycle, including translation initiation (1–3), ribosome biogenesis (4), pre-mRNA splicing (5), and RNA chaperoning (6). A key initiation factor that unwinds secondary structure during mRNA recruitment and scanning is eukaryotic initiation factor 4A (eIF4A) (1). This protein is a member of helicase superfamily 2 (SF2) and comprises the minimal helicase module within the DEAD-box family (7). As such, eIF4A possesses two RecA-like domains that contain the core conserved motifs necessary for adenosine triphosphate (ATP) binding and hydrolysis, as well as RNA binding and melting (8). Like most DEAD-box proteins, eIF4A is generally associated with a number of accessory proteins, including eIF4G, eIF4B, and eIF4H (1). These proteins help recruit eIF4A to the 5′ end of the mRNA, where it unwinds duplex regions to promote mRNA recruitment to the ribosome (9). eIF4G, eIF4B, and eIF4H synergistically activate the duplex unwinding activity of eIF4A, enhancing its RNA binding affinity and accelerating the cycling of its RecA-like domains between open and closed conformations (10–14). Nevertheless, it is still not understood whether duplex unwinding by eIF4A occurs via a distributive or processive mechanism. In fact, both mechanisms have been previously suggested (2, 13, 15–17).
To determine the mechanism used by eIF4A during translation initiation, we developed a high-resolution, single-molecule, optical trapping assay to study the helicase activity of eIF4A in conjunction with known accessory factors eIF4B, eIF4H, and eIF4G. The assay used purified human eIF4A, eIF4B, and eIF4H, as well as a previously characterized truncation mutant of eIF4G (eIF4G682-1105) that retains the evolutionarily conserved eIF4A binding domain (18–20). The experimental geometry consisted of a “dumbbell” arrangement formed by two optically trapped beads, with a nascent RNA transcript stretched between them. This transcript was attached to one bead via hybridization to a DNA “handle” and to the other bead via RNA polymerase (RNAP) arrested at a road-block (Fig. 1). The RNA transcript comprised a reporter hairpin [with a 72–base pair (bp) stem and a 4-nucleotide (nt) loop, and either 25 or 50% GC content] placed adjacent to a 20-nt, single-stranded RNA region at the 5′ end of the hairpin stem, which formed a target for the loading of a single eIF4A helicase (based on helicase footprint size) (Fig. 1 and fig. S1) (21, 22). In this assay, loading of the helicase onto the RNA, followed by subsequent unwinding of the reporter hairpin during directional translocation (5′-to-3′, based on assay geometry), increases the bead-to-bead distance, which can be measured with nanometer-level accuracy (Fig. 1B). Any reannealing of a previously opened hairpin, arising from either enzyme dissociation or reverse translocation along the RNA, decreases this distance.
Fig. 1. Experimental geometry of the single-molecule eIF4A helicase assay (not to scale).
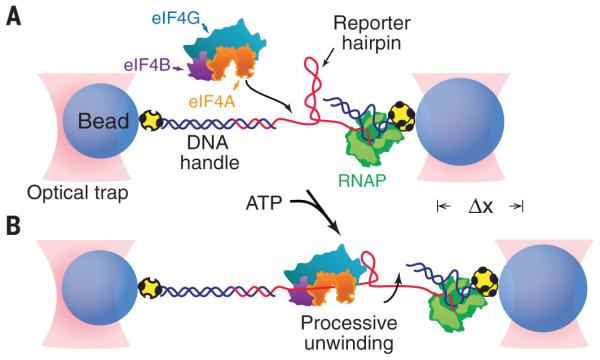
(A) A single 72-bp RNA reporter hairpin (red) is tethered between two microscopic avidincoated beads (blue) held in optical traps (pink) by a DNA handle and a biotinylated RNA polymerase (green) transcriptionally stalled at a biotin-avidin roadblock (yellow). The tether contains a short single-stranded RNA flanking sequence adjacent to the 5′ side of the hairpin for loading eIF4A helicase, shown here complexed with eIF4B and eIF4G. (B) As eIF4A, alone or bound to combinations of eIF4B, eIF4H, and eIF4G, translocates along the RNA, its helicase activity unwinds the hairpin, leading to an increase in distance (Dx) between the trapped beads. In all figures, “eIF4G” corresponds to the truncation mutant eIF4G682-1105.
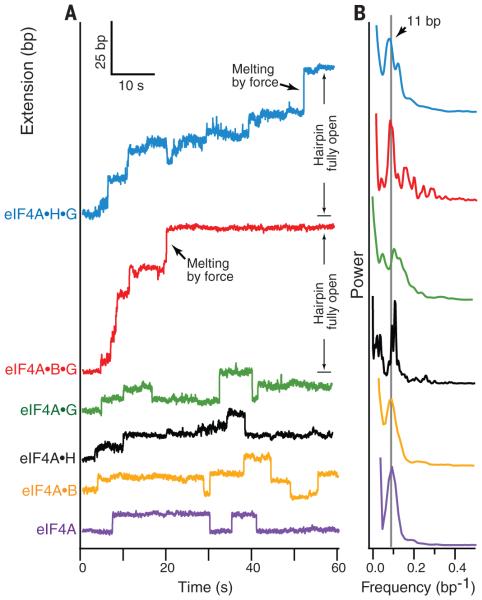
When eIF4A helicase was studied alone in the presence of ATP, only a very few stepwise unwinding events were observed, consistent with previous reports of nonprocessivity in bulk assays (23). Any RNA unwinding by eIF4A was typically characterized by a single step forward, followed either by a single step backward or by enzyme dissociation (Fig. 2A and fig. S3). The introduction of a single additional factor, whether eIF4B, eIF4H, or eIF4G682-1105, slightly enhanced eIF4A’s ability to unwind RNA in a forward or backward direction. The addition of factors eIF4G682-1105 and eIF4B together increased the 5′-to-3′ processivity of eIF4A synergistically, permitting the ternary complex to efficiently melt the entire 72-bp reporter hairpin. This finding suggests that the minimal unit for ribosomal scanning may include a minimum of three initiation factors: eIF4A, eIF4B, and eIF4G. Although eIF4B and eIF4H are homologous proteins, and although each conferred an increase in processivity when complexed with eIF4A and eIF4G682-1105, the eIF4A•B•G682-1105 complex melted hairpins more efficiently than eIF4A•H•G682-1105 (Fig. 2A and fig. S3). Previous experiments have shown that at least three translation initiation factors are required for efficient strand separation (10, 19, 24–26). The present observations argue that strand separation by eIF4A•B/H•G682-1105 can be a direct consequence of processive translocation by a helicase complex based on eIF4A, not of a distributive mechanism requiring multiple binding events (supplementary text).
Fig. 2. Factordependent processivity of eIF4A.
(A) Representative singlemolecule records of helicase activity of eIF4A (purple), eIF4A•B (yellow), eIF4A•H (black), eIF4A•G (green), eIF4A•B•G (red), and eIF4A•H•G (blue) over a 60-s interval under constant load; traces are offset vertically for clarity. Note instances of forward motion corresponding to hairpin unwinding (extension increase) and rearward motion corresponding to hairpin reannealing (extension decrease). The final extension increase leading to full opening of the hairpin was facilitated by force, once the ever-shortening duplex region remaining became unstable under the constant load (black arrows; red and blue traces). (B) Normalized power spectra of the pairwise distances derived from multiple records of helicase and cofactor activities, color-coded as in (A), showing prominent peaks at a spatial frequency (0.09 bp−1, gray line) corresponding to an ~11-bp step. Each power spectrum represents an average of at least 50 different single-molecule records.
We next identified the specific changes in eIF4A behavior responsible for the factor-mediated increase in processivity. Because thermal noise in displacement often masks individual steps, the step size for eIF4A was determined by computing the pairwise distribution of distances moved in single records: Power spectra derived from such distributions display peaks at the corresponding spatial frequencies of any underlying steps (27). Whether eIF4A was scored alone or in complex with other factors, the power spectra from records of its activity all displayed peaks near 11 ± 2 bp (mean ± SEM), a spacing comparable to the helicase footprint and which corresponds closely to a single turn of the RNA duplex (Fig. 2B) (8). The measured step size did not differ among assays conducted under conditions that produced a comparatively high proportion of back-steps (eIF4A, eIF4A•B, eIF4A•H, and eIF4A•G682-1105) and those that did not (eIF4A•B•G682-1105 and eIF4A•H•G682-1105).
When the noise levels in individual records are sufficiently low, it is possible to search for the presence of substeps within the ~11-bp steps. An analysis was therefore carried out by fitting selected segments of individual records using a step-finding algorithm whose robustness has been previously established (28). Substeps were identified, ranging in size from 2 to 4 bp (3 ± 1 bp, mean ± SEM; figs. S8 and S9). In this respect, the behavior of eIF4A is similar to that of the NS3 DEAD-box helicase from hepatitis C virus (supplementary text) (29).
To examine whether different cofactors might affect the directionality of eIF4A, we computed the probability ratio of forward to backward stepping, a measure of directional persistence (23). For eIF4A alone, this ratio was slightly higher than unity (~1.1), consistent with nonprocessive motion (Fig. 3A and fig. S11). For binary complexes eIF4A•B, eIF4A•H, and eIF4A•G682-1105, the ratio increased only moderately (to ~1.7) and was, within error, the same for each complex. In contrast, ternary complexes eIF4A•B•G682-1105 and eIF4A•H•G682-1105 displayed a dramatic increase in persistence ratio, with values of ~7 and ~4, respectively. These results indicate that one role of cofactors eIF4B, eIF4H, and eIF4G682-1105 is to enhance the directionality of eIF4A translocation. The topology of eIF4A•H•G has been studied (10), but unlike in the case of NS3, it is unclear which eIF4A residues mediate changes in processivity (supplementary text) (30).
Fig. 3. Translocation properties of eIF4A helicase and accessory factor combinations.
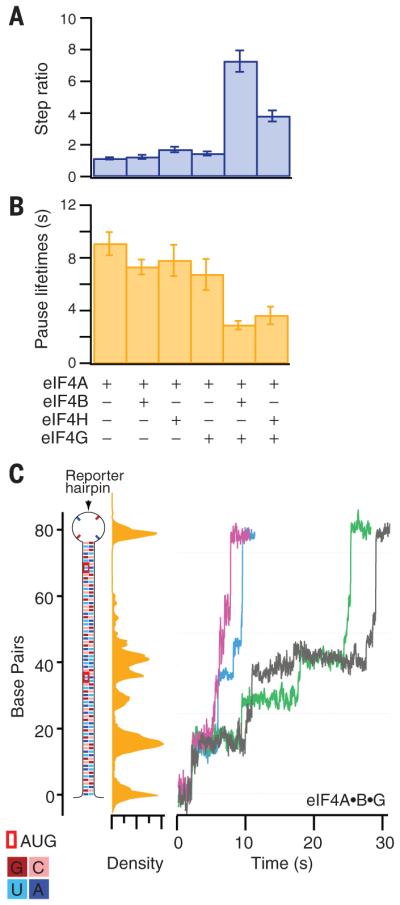
(A) Forward-to-backward stepping ratio for the indicated complexes (legend, bottom), with errors (SEM). (B) Mean pause lifetime with errors (SEM) for the indicated complexes (legend, bottom). (C) eIF4A•B•G melting of a reporter hairpin with the sequence shown (left), color-coded according to the legend (bottom). Five representative single-molecule records, expressed as base pairs unwound, are shown over a 30-s interval (right). The positions of these records were histogrammed in a single density plot (middle, yellow); strong peaks in this plot indicate locations where the complex pauses, which were not correlated with the location of the AUGs.
In single-molecule records, mechanical steps were occasionally separated by pauses, corresponding to comparatively longer periods of invariant tether extension. Pause lifetimes were exponentially distributed for eIF4A, both alone and in combination with accessory factors, and were well fitted by single exponentials (Fig. 3B and fig. S12). We found that eIF4A displayed the longest measured pause lifetime, 9.0 ± 0.4 s (mean ± SEM). The complexes eIF4A•B, eIF4A•H, and eIF4A•G682-1105 exhibited similar lifetimes within experimental error (~7 s), but these were not statistically different from that of eIF4A alone. However, ternary complexes eIF4A•B•G682-1105 and eIF4A•H•G682-1105 had pause lifetimes three times shorter than that of eIF4A, with durations of 2.9 ± 0.1 s and 3.6 ± 0.2 s (mean ± SEM), respectively. These findings suggest that, in addition to enhancing the directionality of eIF4A, the accessory factors eIF4B, eIF4H, and eIF4G682-1105 also increase the mRNA unwinding efficiency by decreasing pausing. Pause locations were distributed along the reporter hairpin (Fig. 3C and fig. S3). The two reporter hairpins used in this study were based on randomized nucleotide sequences, and we could identify no obvious relationship between the locations and durations of pauses and the local base composition. However, many additional sequences would need to be scored to identify any possible sequence dependence of helicase activity.
Finally, analysis of unwinding activity indicates that eIF4A, alone or in complexes, has no memory associated with a given series of steps and that the translocation mechanism is governed by a single rate-limiting step (supplementary text).
Although DEAD-box helicases have sometimes been regarded as nonprocessive enzymes (29), we find here that eIF4A displays a factormediated processivity requiring at least two cofactors (eIF4B/H and eIF4G682-1105). This acquired processivity is characterized by a preferential directionality (presumably 5'-to-3'), with burst openings of 11 ± 2 bp (mean ± SEM) and dramatically reduced pausing. In eukaryotic translation initiation, the eIF4AβBβG682-1105 complex comprises a minimal processive unit that is thought to facilitate sequential, step-by-step ribosomal scanning. In light of our findings, we anticipate that the processivity and dynamics of other DEAD-box family members may be influenced by accessory cofactors, so that enzymatic activities scored in their absence may not fully reflect the in vivo function of the proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Landick for providing RNAP and J. Hershey, E. Abrash, and members of the Fraser and Block labs for helpful comments. This work was supported by National Institute of General Medical Sciences grants R37GM057035 (S.M.B.) and R01GM092927 (C.S.F.), an Alejandro and Lisa Zaffaroni Graduate Fellowship (C.G.G.), a Stanford Graduate Fellowship (K.L.F.), and training grant T32 GM-007377 (K.F.).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/348/6242/1486/suppl/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S15
References (31-51)
REFERENCES AND NOTES
- 1.Parsyan A, et al. Nat. Rev. Mol. Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 2.Linder P, Jankowsky E. Nat. Rev. Mol. Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 3.Jarmoskaite I, Russell R. Annu. Rev. Biochem. 2014;83:697–725. doi: 10.1146/annurev-biochem-060713-035546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocak S, Linder P. Nat. Rev. Mol. Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 5.Jarmoskaite I, Russell R. Wiley Interdiscip. Rev.: RNA. 2011;2:135–152. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodson SA. RNA Biol. 2010;7:677–686. doi: 10.4161/rna.7.6.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fairman-Williams ME, Guenther U-P, Jankowsky E. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caruthers JM, Johnson ER, McKay DB. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aitken CE, Lorsch JR. Nat. Struct. Mol. Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 10.Marintchev A, et al. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen KH, et al. Nucleic Acids Res. 2011;39:2678–2689. doi: 10.1093/nar/gkq1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, et al. Nucleic Acids Res. 2012;40:6199–6207. doi: 10.1093/nar/gks278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abramson RD, Dever TE, Merrick WC. J. Biol. Chem. 1988;263:6016–6019. [PubMed] [Google Scholar]
- 14.Sun Y, et al. Structure. 2014;22:941–948. doi: 10.1016/j.str.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Liu F, Putnam A, Jankowsky E. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreou AZ, Klostermeier D. RNA Biol. 2013;10:19–32. doi: 10.4161/rna.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Gregorio E, Preiss T, Hentze MW. EMBO J. 1999;18:4865–4874. doi: 10.1093/emboj/18.17.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13339–13344. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korneeva NL, First EA, Benoit CA, Rhoads RE. J. Biol. Chem. 2005;280:1872–1881. doi: 10.1074/jbc.M406168200. [DOI] [PubMed] [Google Scholar]
- 21.Lorsch JR, Herschlag D. Biochemistry. 1998;37:2194–2206. doi: 10.1021/bi9724319. [DOI] [PubMed] [Google Scholar]
- 22.Peck ML, Herschlag D. RNA. 1999;5:1210–1221. doi: 10.1017/s1355838299990817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozen F, et al. Mol. Cell. Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers GW, Jr., Richter NJ, Lima WF, Merrick WC. J. Biol. Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 25.Rogers GW, Jr., Richter NJ, Merrick WC. J. Biol. Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 26.Andreou AZ, Klostermeier D. J. Mol. Biol. 2014;426:51–61. doi: 10.1016/j.jmb.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 27.Schnitzer MJ, Block SM. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 28.Kerssemakers JWJ, et al. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 29.Pyle AM. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 30.Myong S, Bruno MM, Pyle AM, Ha T. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.