Abstract
Resistance to targeted EGFR inhibitors is likely to develop in EGFR mutant lung cancers. Early identification of innate or acquired resistance mechanisms to these agents is essential to direct development of future therapies. We describe the detection of heterogeneous mechanisms of resistance within populations of EGFR mutant cells (PC9 and/or NCI-H1975) with acquired resistance to current and newly developed EGFR TKIs including AZD9291. We report the detection of NRAS mutations, including a novel E63K mutation, and a gain of copy number of WT NRAS or WT KRAS in cell populations resistant to gefitinib, afatinib, WZ4002 or AZD9291. Compared to parental cells, a number of resistant cell populations were more sensitive to inhibition by the MEK inhibitor selumetinib (AZD6244; ARRY-142886) when treated in combination with the originating EGFR inhibitor. In vitro, a combination of AZD9291 with selumetinib prevented emergence of resistance in PC9 cells and delayed resistance in NCI-H1975 cells. In vivo, concomitant dosing of AZD9291 with selumetinib caused regression of AZD9291-resistant tumours in an EGFRm/T790M transgenic model. Our data support the use of a combination of AZD9291 with a MEK inhibitor to delay or prevent resistance to AZD9291 in EGFRm and/or EGFRm/T790M tumours. Further, these findings suggest that NRAS modifications in tumour samples from patients who have progressed on current or EGFR inhibitors in development may support subsequent treatment with a combination of EGFR and MEK inhibition.
Keywords: EGFR, AZD9291, T790M, RAS-MAPK signaling
Introduction
Tumours containing activating epidermal growth factor receptor mutations (EGFRm) (e.g. deletion in exon 19 or an L858R point mutation) account for about 20% of advanced non small cell lung cancer (NSCLC) (1). Although these mutations also sensitise EGFR to inhibition by the established tyrosine kinase inhibitor (TKI) therapies erlotinib and gefitinib (2), almost all tumours will develop acquired resistance to these TKIs within 9–15 months (3, 4). In aproximately 60% of cases, this acquired resistance is associated with an additional T790M mutation in EGFR (5–7). As there are currently no treatments approved for patients with these tumours (8, 9) new EGFR TKIs such as AZD9291, WZ4002 and CO-1686 are being developed which inhibit both EGFRm and T790M mutations in preclinical models (10–12). AZD9291 and CO-1686 have also shown promising Phase 1 activity in patients with T790M advanced NSCLC who have progressed while on prior therapy with an EGFR-TKI (10, 11). These new TKIs may also provide treatment options in the TKI-naive setting for patients with advanced EGFRm NSCLC. However, despite the potential improvements from therapy with these TKIs, experience with targeted agents suggest that resistance to these drugs may also emerge and potentially limit their effectiveness. Therefore identification of resistance mechanisms is important to drive new therapeutic strategies for treating drug resistance in patients.
In vitro, EGFRm cells chronically exposed to escalating doses of gefitinib or erlotinib acquire clinically relevant resistance mechanisms (13, 14), and subsequent studies have identified a range of further potential resistance mechanisms (15–20). Although the clinical importance of many of these mechanisms remains to be determined, trying to predict acquired resistance, especially to new emerging agents such as AZD9291, is a critical area of research. To date, resistance mechanisms have typically been determined from single clonal lines selected from resistant populations of cancer cells and therefore may represent only a small percentage of the original cancer cell population. Since human NSCLC samples are heterogenous (21–23) and tumours are likely to derive acquired resistance through multiple mechanisms, we postulated that it may be better to take a population approach to understanding the diversity and interplay of resistance mechanisms. We studied multiple cell populations resistant to gefitinib, afatinib, WZ4002 or AZD9291 to identify predominant mechanisms of resistance and to investigate signaling pathways activated by various resistance mechanisms.
Materials and Methods
Cell lines, cell culture and compound reagents
All AstraZeneca cell lines were authenticated by short-tandem repeat analysis (STR). PC9 cells (obtained 2011, STR tested May 2013) were from Akiko Hiraide, PreclinicalSciences R&D AZ Japan. NCI-H1975 (CRL-5908, obtained 2004, STR tested Nov 2012), NCI-H820 (HTB-181, obtained 2011, STR tested Jan 2013) and HCC827 (CRL-2868, obtained 2012, STR tested Oct 2012) cells were obtained from ATCC. HCC-2279 (K72279, obtained 2013, STR tested Mar 2013) cells were obtained from KCLB. Cells were cultured in RPMI containing 10% FCS with 2 mmol/L glutamine, supplemented with EGFR inhibitor for resistant cell populations. Selumetinib, gefitinib, afatinib, WZ4002, BMS-536924, AZD5363, AZD2014, AZD8055, GDC-0941, AZD4547, AZD1152-HQPA and AZD9291 were synthesised according to published methods. AZ_6592, AZ_0012, AZ_1902 and AZ_9424 in house compounds (AstraZeneca).
In vitro cell assays
Phenotypic cell assays, immunoblotting and RAS activation assays were carried out as previously described (10, 24) and detailed in Supplementary Methods. Cells were transfected using Lipofectamine RNAiMAX reagent, Invitrogen (Paisley, UK), FuGENE 6 Transfection Reagent, Promega (Madison, USA) or by electroporation, MaxCyte. siRNA and DNA constructs are detailed in Supplementary Methods.
Genetic analysis
SnaPshot mutation analysis was carried as previously described (25). Targeted and whole exome sequencing (WES) were performed on MiSeq and HiSeq platforms, Illumina; Ion Torrent PGM platform, Life Technologies and by Sanger di-deoxy sequencing. Comparative genomic hybridization was performed using SurePrint G3 Human CGH microarrays, Agilent Technologies (Santa Clara, USA). Sequence data processing, mutation detection and gene copy number assessment were carried out as described in Supplementary Material. Data is accessible in NCBI’s Sequence Read Archive accession number SRP044079 and NCBI’s Gene Expression Omnibus (GEO) accession number GSE59239.
Transgenic mouse studies and MRI
In vivo experiments were carried out as previously described using both EGFRL858R+T790M and EGFRL858R transgenic models (10). Details are included in Supplementary Methods.
Results
Generation of EGFR mutant cell populations resistant to AZD9291 and other EGFR TKIs
To carry out a broad investigation into acquired resistance to EGFR inhibitors, we generated in parallel multiple EGFRm (PC9; Ex19del. chosen as a validated cell line for modeling EGFR inhibitor resistance (26)) and EGFRm/T790M (PC9 derivatives and NCI-H1975; L858R/ T790M) cell populations with induced resistance to gefitinib, afatinib, WZ4002 or AZD9291, using either a dose escalation method or by culturing the cells in a single dose of AZD9291 (Supplementary Table S1).
Resistance to AZD9291 and other EGFR TKIs is often associated with increased sensitivity to MEK inhibition
To investigate whether the survival of resistant populations was through activation of alternative signaling pathways that circumvent EGFR dependency, we used a diverse panel of small molecule inhibitors representing key signaling pathways or emerging resistance mechanisms (Supplementary Table S2). The ability of each agent, in the presence of originating EGFR TKI, to inhibit cell growth was measured using an in vitro phenotypic assay, and IC50 values determined following 72 hours treatment (Table 1; Supplementary Table S3). It was striking that 13 of 28 PC9 resistant populations and 2 of 4 NCI-H1975 resistant populations were greater than 5 times more sensitive to the MEK inhibitor selumetinib in combination with the originating EGFR inhibitor, when compared to the corresponding parental cells treated with selumetinib. We therefore focused subsequent studies on understanding mechanisms of selumetinib sensitivity in these populations.
Table 1.
IC50 (μM) values from cell growth inhibition assays comparing compound sensitivity between parental and resistant cell populations.
Cells were treated with dose titrations of indicated inhibitors alone for parental cells and in the presence of original EGFR inhibitor for resistant populations. IC50 values represent an average of at least 2 independent experiments. Errors are standard deviation. Additional compound data is shown in Supplementary Table S3.
| Cell population | Genetic alterations detected within resistant populations | (MEK1/2) | (EGFR) |
|---|---|---|---|
| PC9 | 6.95 (±2.5) | 0.008 (±0.002) | |
| PC9 GR_1 | EGFR T790M / KRAS gain (5.43 fold) | 7.24 (±3.2) | 1.12 (±0.5) |
| PC9 GR_2 | NRAS E63K | 0.62 (±0.3) | 2.8 (±0.4) |
| PC9 GR_3 | EGFR T790M | 6.2 (±3.6) | 0.18 (±0.2) |
| PC9 GR_4 | EGFR T790M | 7.32 (±2.3) | 0.02 (±0.01) |
| PC9 GR_5 | EGFR T790M | 8.77 (±1.5) | 0.14 (±0.06) |
| PC9 GR_6 | EGFR T790M | 7.44 (±2.6) | 0.005 (±0.001) |
| PC9 GR_7 | EGFR T790M | 3.7 (±0.99) | 0.002 (±0.002) |
| PC9 GR_8 | EGFR T790M / KRAS gain (7.06 fold) | 2.48 (±1.4) | 2.40 (±0.97) |
| PC9 AR_1 | KRAS gain (24.6 fold) | 2.7 (±0.23) | 2.41 (±0.5) |
| PC9 AR_4 | EGFR T790M | 1.63 (±1.1) | 0.73 (±0.3) |
| PC9 AR_6 | NRAS gain (4.23 fold) | 0.89 (±0.6) | 2.4 (±0.5) |
| PC9 WZR_1 | NRAS Q61K | 0.23 (±0.04) | 1.99 (±0.03) |
| PC9 WZR_3 | KRAS gain (2.64 fold) | 0.22 (±0.1) | 1.65 (±0.5) |
| PC9 AZDR_1 | NRAS gain (2.5 fold) / MAPK1 gain / CRKL gain | 0.25 (±0.06) | 2.3 (±0.9) |
| PC9 AZDR_2 | NRAS G12V | 1.4 (±0.9) | 3.69 (±1.2) |
| PC9 AZDR_3 | MAPK1 gain / CRKL gain | 2.38 (±0.9) | 1.94 (±0.5) |
| PC9 AZDR_4 | ND | 0.19 (±0.1) | 2.48 (±1.1) |
| PC9 AZDR_5 | NRAS E63K | 0.17 (±0.05) | 2.14 (±0.06) |
| PC9 AZDR_6 | NRAS E63K | 0.11 (±0.03) | 1.6 (±0.02) |
| PC9 AZDR_7 | NRAS G12R | 0.14 (±0.03) | 2.63 (±0.3) |
| PC9 GR_1_AZDR_1 | EGFR T790M / KRAS gain (6.23 fold) | 3.6 (±0.7) | 2.4 (±0.95) |
| PC9 GR_1_AZDR_2 | KRAS gain (5.66 fold) | 6.7 (±1.4) | 2.7 (±1.2) |
| PC9 GR_1_AZDR_3 | EGFR T790M / KRAS gain (4.44 fold) | 3.4 (±0.5) | 2.4 (±0.7) |
| PC9 GR_1_AZDR_4 | EGFR T790M / KRAS gain (5.46 fold) | 3.6 (±2.6) | 2.6 (±0.9) |
| PC9 GR_6_AZDR_1 | ND | 0.28 (±0.2) | 1.35 (±0.05) |
| PC9 GR_6_AZDR_2 | NRAS gain (2.4 fold) | 0.54 (±0.3) | 2.24 (±0.6) |
| PC9 GR_6_AZDR_3 | NRAS gain (3.68 fold) | 0.13 (±0.06) | 1.48 (±0.3) |
| PC9 GR_6_AZDR_4 | ND | 0.73 (±0.5) | 1.74 (±0.8) |
| NCI-H1975 | EGFR T790M | 4.94 (±3) | 0.016 (±0.01) |
| NCI-H1975 AZDR_1 | EGFR T790M | 0.024 (±0.003) | 2.52 (±0.4) |
| NCI-H1975 AZDR_2 | EGFR T790M | 0.15 (±0.1) | 2.21 (±0.2) |
| NCI-H1975 AZDR_3 | EGFR T790M | >10 | 3.04 (±0.4) |
| NCI-H1975 AZDR_4 | EGFR T790M / NRAS Q61K | 5.46 (±3.7) | 2.67 (±0.7) |

DNA from resistant populations was analysed for gene mutation and/or gene copy number across a panel of cancer associated genes. Data represents genetic alterations detected within the resistant populations. Fold gain, relative to parental cells, indicated in brackets for NRAS and KRAS. Non detected (ND)
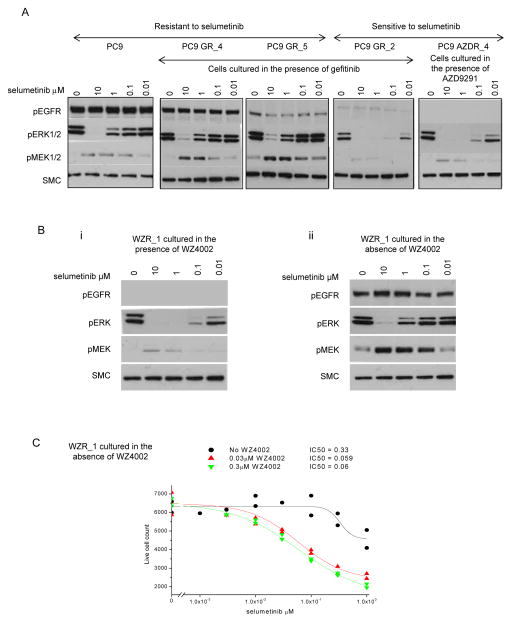
To confirm that increased selumetinib sensitivity was related to RAS-MAPK pathway inhibition, phosphorylation levels of ERK1/2 and MEK1/2 were analysed by immunoblotting PC9 parental and resistant populations grown in the presence of EGFR inhibitor and treated with increasing concentrations of selumetinib. Resistance to selumetinib in the PC9, PC9 GR_4 and PC9 GR_5 cells was associated with a strong induction of phosphorylated MEK and weaker inhibition of ERK phosphorylation when compared to the effects of selumetinib in the sensitive cell populations, PC9 GR_2 and PC9 AZDR_4 (Fig 1A). The dependency of EGFR resistant cell populations on RAS-MAPK activity was further analysed using PC9 WZR_1 cells which showed > 5 fold increased sensitivity to selumetinib (Table 1). Consistent with PC9 GR_2 and AZDR_4 populations, WZR_1 cells maintained in presence of WZ4002 demonstrated expected inhibition of phosphorylated EGFR, and phosphorylated ERK (Fig. 1Bi) and growth inhibition (Fig. 1C) were highly sensitive to selumetinib treatment. In contrast, WZR_1 cells that had been cultured in the absence of WZ4002 displayed an EGFR and selumetinib ERK sensitivity profile similar to that seen in PC9 parental cells namely weak inhibition of ERK phosphorylation and strong induction of pMEK (Fig. 1A, Fig. 1Bii), with associated growth inhibition refractory to selumetinib (Fig. 1C). The strong increase in levels of phosphorylated MEK1/2 in response to selumetinib treatment in the resistant compared to the sensitive populations (Fig. 1A, Fig 1Bi) suggests a difference in pathway activity upstream of MEK between sensitive and resistant populations in response to relief of negative feedback loops upon MEK inhibition.
Figure 1. RAS-MAPK signaling inhibition by selumetinib in EGFR inhibitor resistant cell lines.
(A, B) Cells cultured in the presence or absence of originating EGFR inhibitor as indicated were dosed with titrations of selumetinib for 6 hours. Lysates were analysed by immunoblotting. Data is representative of 2 separate experiments. (C) WZR_1 cells cultured in the absence of WZ4002 prior to the experiment were treated with titrations of selumetinib over 96 hours with no added WZ4002, 0.03 μM or 0.3 μM WZ4002. Data is representative of two separate experiments.
Comparison of genetic alterations across multiple populations of EGFRm or EGFRm/T790M cells resistant to AZD9291 and other EGFR TKIs
In order to investigate the molecular drivers of EGFR TKI resistance we analysed DNA samples prepared from parental and a selection of 32 resistant populations (Supplementary Table S1) for the presence of gene mutations and/or copy number changes across a panel of cancer associated genes using multiple assay platforms. The genetic modifications detected and associated allele frequencies for PC9 and NCI-H1975 populations are summarised in Table 1 and Supplementary Fig. S1. Each mutation detected was confirmed across at least 2 different assay platforms (Supplementary Table S4).
Across the PC9 populations, 7/8 gefitinib and 2/3 afatinib resistant populations had detectable T790M mutations, whereas none of the populations resistant to the WZ4002 or AZD9291 had acquired a detectable T790M mutation (Table 1). The T790M gefitinib resistant populations mostly showed sensitivity to AZD9291, with dose response curves indicating almost all cells in populations PC9 GR_4, 6 and 7 were sensitive to AZD9291 (Supplementary Fig. S2Ai). However, less than 50% of cells in populations PC9 GR_1 (T790M, KRAS gain 5.43 fold), PC9 GR_3 (T790M) and PC9 GR_5 (T790M) were sensitive to growth inhibition by AZD9291 (Supplementary Fig. S2Aii), suggesting these populations contained heterogenous resistant mechanisms. The IC50 values across all AZD9291 sensitive cells were similar (Supplementary Fig S2B). Although T790M was detected within the PC9 GR_8 (T790M, KRAS gain 7.06 fold) population, these cells showed no sensitivity to AZD9291 (Supplementary Fig. S2Aiii), suggesting that the entire population contained resistant clones. This observation of heterogenenous mechanisms of resistance to gefitinib within populations is consistent with clinical setting, supporting use of this population approach for understanding resistance dynamics.
Notably PC9 resistant cell populations lacking detectable T790M frequently displayed increased sensitivity to selumetinib in combination with EGFR inhibition. In selumetinib-sensitive EGFRm/T790M populations with induced resistance to EGFR inhibitors, no additional EGFR mutations were detected. This suggests that RAS-MAPK signaling was important for driving resistance in the absence of EGFR signaling (Table 1).
Interestingly a number of different NRAS alterations were observed in PC9 populations resistant to AZD9291, gefitinib and WZ4002, and NCI-H1975 cells resistant to AZD9291 (Table 1). Notably, we observed a novel non-canonical E63K mutation in NRAS in the only gefitinib-resistant T790M-negative PC9 population and in two AZD9291-resistant PC9 populations (Table 1; Supplementary Fig. S3). This novel NRAS mutation occurs within the functional domain at a sequence position parallel to somatic mutations observed in both KRAS (27) and HRAS (28). We also identified functionally activating NRAS G12V and G12R mutations in 2 different AZD9291-resistant PC9 populations (Table 1). This is the first identification of G12V NRAS in the context of NSCLC. In addition to gene mutations, copy number gains of MAPK1, CRKL, NRAS and KRAS were detected across the resistant populations (Table 1), with the gain of NRAS and KRAS resulting in increased protein levels (Supplementary Fig. S4). Of particular interest KRAS gain was observed in the T790M PC9_GR_1 population that was partially sensitive to AZD9291 (Table 1), suggesting KRAS contributes to the heterogeneous mechanisms of resistance to gefitinib within this population. Indeed 4 separate AZD9291-resistant populations of PC9 GR_1 cells were subsequently generated and KRAS gain was retained within each resistant population (Table 1). Interestingly, although T790M was still present in populations PC9 GR1_AZDR_1, 3 and 4 T790M was no longer detectable in PC9 GR1_AZDR_2 cells.
Modifications of RAS genes can drive resistance to EGFR inhibition
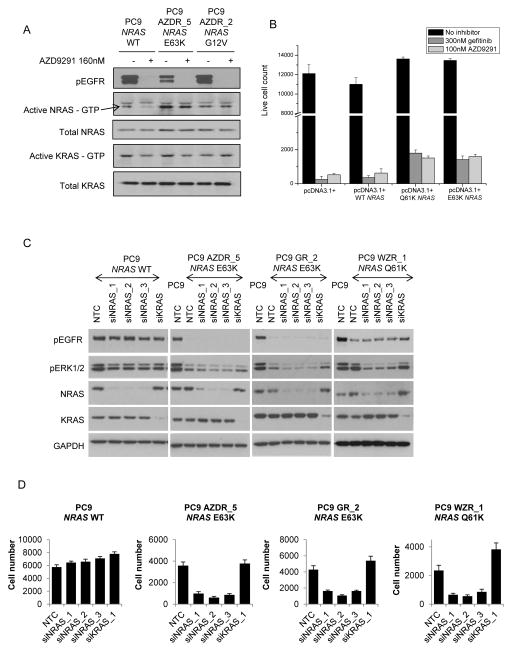
As NRAS mutations were detected in 7 of the resistant populations, and frequently associated with selumetinib sensitivity, it’s role in resistance was further investigated. Basal levels of active NRAS were lower in parental PC9 cells compared to resistant PC9 populations in which E63K, G12V, (Fig. 2A) E63K or Q61K (Supplementary Fig. S4) NRAS mutation had been detected. Treatment of parental PC9 cells with 160nM AZD9291 decreased levels of phosphorylated EGFR and active NRAS. In contrast, in mutant NRAS cells, a decrease in phosphorylated EGFR was not associated with corresponding decrease in active NRAS, suggesting constitutive activation of NRAS disconnected from EGFR in these cells (Fig. 2A; Supplementary Fig. S5A). In transient exogenous expression in PC9 cells, WT and mutant NRAS variants were activating (Supplementary S5Bi,ii), and prevented cell growth inhibition by either AZD9291 or gefitinib compared to control transfected cells (Fig. 2B). Increased resistance to growth inhibition by AZD9291 was also observed in additional parental EGFRm cell lines similarly trasfected with WT and activating mutant NRAS variants (Supplementary Fig. S5C). Knockdown of NRAS in cell populations with 3 separate siRNAs, but not KRAS, for 72 hours resulted in a significant decrease in phosphorylated ERK in the resistant populations harboring NRAS mutations, but to a lesser extent in the PC9 parental cells (Fig. 2C). Moreover, knockdown of NRAS but not KRAS was associated with inhibition of proliferation only in the NRAS mutant populations (Fig. 2D). These data indicate that activating NRAS mutations including the novel E63K NRAS are sufficient to drive resistance to EGFR inhibition. Similarly knockdown of NRAS in the presence of AZD9291 caused a significant decrease in cell growth of PC9 GR_6 AZDR_2 (NRAS gain 2.4 fold) and PC9 GR_6 AZDR 3 (NRAS gain 3.68 fold) populations (data not shown).
Figure 2. Determining the functional role of NRAS modifications in acquired resistance to EGFR inhibitors.
(A) Resistant populations were cultured in media without EGFR inhibitor for 5 days prior to carrying out the assay. Lysates were prepared from parental and resistant cells serum starved overnight and treated for 6 hours +/− 160nM AZD9291. RAS activity was measured using RAS GTPase-specific pulldown assays. (B) PC9 cells transfected with NRAS and control pcDNA 3.1+ constructs for 48 hours were treated with 100nM AZD9291 or 300nM gefitinib for a further 96 hours. Live cell number was determined by nuclei count. The data is representative of three separate experiments. Error bars are standard deviation. (C) Resistant populations were cultured in media supplemented with EGFR inhibitor for all siRNA experiments. Lysates from cells treated with 20nM NTC, NRAS or KRAS siRNA for 48 hours were anlaysed by immunoblotting. (D) Cells treated for 72 hours with 20nM NTC, NRAS or KRAS siRNA were fixed and cell number determined by nuclei count. Data is representative of 3 replicate experiments. Error bars are standard deviation.
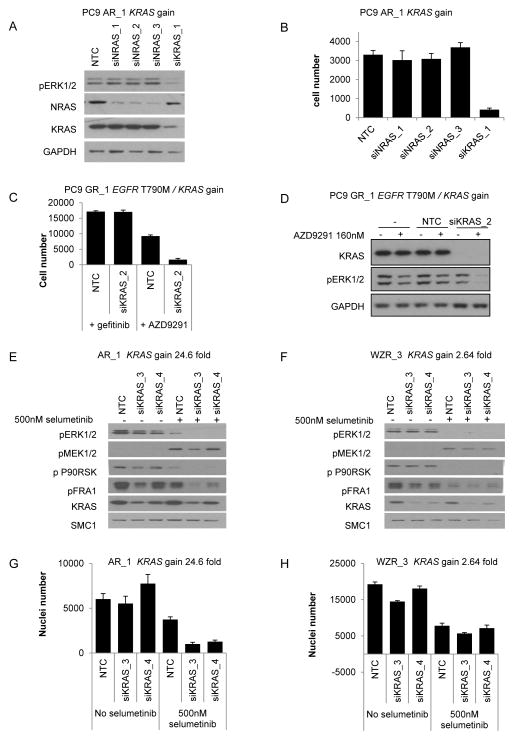
As KRAS gain was detected within 8 resistant populations (Table 1), we determined whether this could similarly drive resistance. Knockdown of KRAS in PC9 parental cells had no effect on cell growth or levels of phosphorylated ERK (Fig. 2C), whereas KRAS knockdown in PC9 AR_1 (KRAS gain 24.6 fold), in the presence of afatinib, caused a significant decrease in both phosphorylated ERK levels after 48 hours (Fig. 3A) and proliferation over 72 hours (Fig. 3B). Interestingly, knockdown of KRAS in the PC9 GR_1 (T790M and KRAS gain 5.43 fold) population, had no effect on cell proliferation alone or when treated in combination with gefitinib (Fig. 3C). However a significant decrease in cell growth was observed when KRAS knockdown was combined with AZD9291 treatment (Fig. 3C). Consistent with this, knockdown of KRAS in the presence of AZD9291 resulted in complete inhibition of phosphorylated ERK, but not in case of gefitinib (Fig. 3D). These observations suggest that KRAS and T790M are both important for driving resistance in the PC9 GR_1 population.
Figure 3. Determining the functional role of KRAS gain in acquired resistance to EGFR inhibitors.
(A) Immunoblotting of PC9 AR_1 (KRAS gain) cells grown in 1.5μM afatinib transfected with 20nM of NRAS, KRAS or control siRNA for 48 hours. (B) PC9 AR_1 (KRAS gain) cells grown in 1.5μM afatinib treated for 96 hours with 20nM of NRAS, KRAS or control siRNA. Cell number was determined by nuclei count. (C) PC9 GR_1 (EGFR T790M / KRAS gain) cells grown in 1.5μM gefitinib were transfected with 20nM of KRAS or control siRNA −/+ 160nM AZD9291. After 4 days cell number was determined by nuclei count. Data shown is representative data. Error bars are standard deviation. (D) Immunoblotting of PC9 GR_1 cells, grown in media containing gefitinib, transfected with 20nM of KRAS or NTC siRNA for 5 days and then treated with 160nM of AZD9291 for 2 hours. (E) Immunoblotting of PC9 AR_1 (KRAS gain) cells grown in 1.5μM afatinib and (F) WZR_3 (KRAS gain) cells grown in 1.5μM WZ4002 transfected with 20nM of KRAS or control siRNA for 48 hours and then treated for 4 hours +/− 500nM selumetinib. (G) PC9 AR_1 (KRAS gain) cells grown in 1.5μM afatinib and (H) WZR_3 (KRAS gain) cells grown in 1.5μM WZ4002 treated for 96 hours with 20nM of KRAS or control siRNA +/− 500 nM selumetinib. Cell number was determined by nuclei count.
Interestingly, we noted that a 2.64 fold gain of KRAS, as detected in WZR_3 cells, was associated with increased sensitivity to selumetinib, but cell populations with KRAS gains of between 4.44 and 24.6 fold were insensitive to selumetinib (Table 1), suggesting a threshold effect of KRAS expression. Indeed, partial knockdown of KRAS for 48 hours in AR_1 cells (KRAS gain 24.6 fold) with afatinib resensitised them to selumetinib inhibition as revealed by decreased phosphorylated ERK, FRA1 and p90RSK levels compared to cells similarly treated with control siRNA (Fig 3E). Moreover partial knockdown of KRAS followed by treatment with selumetinib resulted in significantly greater inhibition of cell growth compared to cells treated with control siRNA (Fig. 3G). In contrast, no significant increase in inhibition of MAPK pathway or cell growth was observed with partial knockdown of KRAS followed by selumetinib treatment in WZR_3 cells (KRAS gain 2.64 fold) cultured in the presence of WZ4002 (Fig. 3F, 3H). Interestingly selumetinib inhibition of MEK1/2 in AR_1 cells resulted in enhanced pMEK1/2 levels compared to that observed in selumetinib-sensitive WZR_3 cells (Figs. 3E, 3F). Collectively, this data is consistent with previous reports in which KRAS amplification in colon cells drives high levels of pathway output and ERK dependent negative feedback, relief of which upon MEK inhibition drives relative insensitivity to MEK inhibitors (29). Similarly, we observed that enhanced exogenous expression of wild-type NRAS in PC9 AR_6 cells (NRAS 4.23 fold gain) reduced the effectiveness of selumetinib treatment on phosphorylated ERK and growth inhibition compared to PC9 AR_6 cells treated with control DNA (data not shown).
In vitro a combination of AZD9291 with selumetinib delays or prevents resistance emerging in EGFRm and EGFRm/T790M cells
Since the data had indicated that RAS-MAPK activation was a frequent mechanism of AZD9291 and other EGFR TKI resistance, we tested whether treatment with a combination of AZD9291 and selumetinib could delay or prevent the emergence of resistance in these settings.
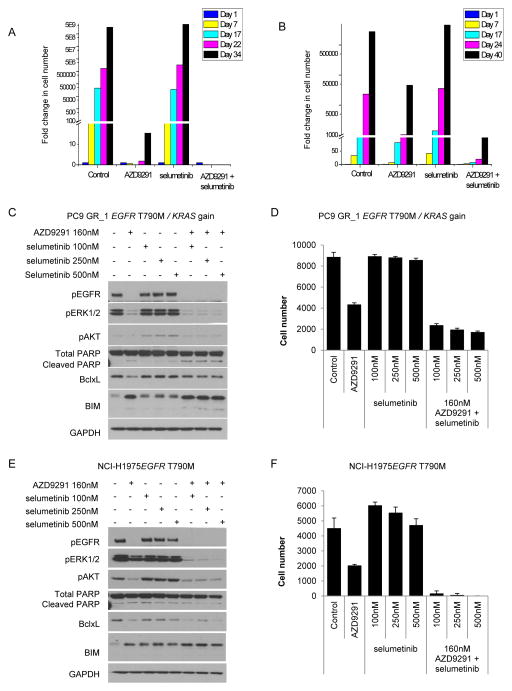
PC9 (EGFRm) cells were treated with 160nM AZD9291 or 100nM selumetinib alone or in combination. Selumetinib alone did not inhibit proliferation of PC9 cells (Fig. 4A). Whereas resistant PC9 cells began to emerge after 34 days treatment with AZD9291, no resistant cells were observed over a similar time with a combination of AZD9291 and selumetinib (Fig. 4A). To investigate the combination in the EGFRm/T790M setting, NCI-H1975 cells were treated with 160nM AZD9291 or 100nM selumetinib alone or in combination. Treatment with 100nM selumetinib alone did not inhibit proliferation of NCI-H1975 cells (Fig. 4B). Treatment with AZD9291 initially slowed the rate of proliferation, however a small resistant population had emerged following 17 days of treatment (Fig. 4B). Treatment with a combination of AZD9291 and selumetinib significantly delayed outgrowth of resistant cells compared to AZD9291 alone (Fig. 4B). Similarly a combination of AZD9291 with selumetinib prevented emergence of resistance in 2 other cell lines, HCC827 (EGFR Ex19del) and NCI-H820 (EGFR Ex19del/T790M+/METamp) (Supplementary Fig. S6A, B).
Figure 4. In vitro combination of AZD9291 with selumetinib induces more profound phenotype inhibition.
(A) PC9 and (B) NCI-H1975 cells were chronically treated for 34 days with DMSO, AZD9291, selumetinib or a combination of AZD9291 with selumetinib. Fold increase in cell number was monitored over time. Lysates from PC9 GR_1 (C) or NCI-H1975 (E) cells treated with AZD9291 and selumetinib alone or in combination for 48 hours were analysed by immunoblotting. PC9 GR_1 (D) or NCI-H1975 (F) cells were treated over 12 days with AZD9291 and selumetinib alone or in combination. Cells were fixed and cell number determined from nuclei count. Error bars represent standard deviation.
To further explore the EGFRm/T790M setting, PC9 GR_1 cells (T790M and KRAS gain) were treated with a combination of AZD9291 and selumetinib. Following treatment an increase in pro-apoptotic markers, cleaved PARP and BIM, and a decrease in anti-apoptotic BclxL was observed together with a more profound effect on phosphorylated ERK levels than either agent alone (Fig. 4C). Moreover the combined effect of inhibition of ERK signaling and apoptotic pathway was associated with a greater decrease in cell number over 12 days compared to either inhibitor alone (Fig. 4D). Although combination of selumetinib and AZD9291 did not increase apoptotic markers in NCI-H1975 (Fig. 4E), a profound anti-proliferative effect was observed following 12 days treatment with the combination compared to each agent alone (Fig. 4F). Overall these in vitro studies indicated that combining AZD9291 with selumetinib lead to more profound mechanistic and phenotypic inhibition of cells.
In vivo a combination of AZD9291 with selumetinib caused regression of AZD9291 resistant tumours in transgenic models
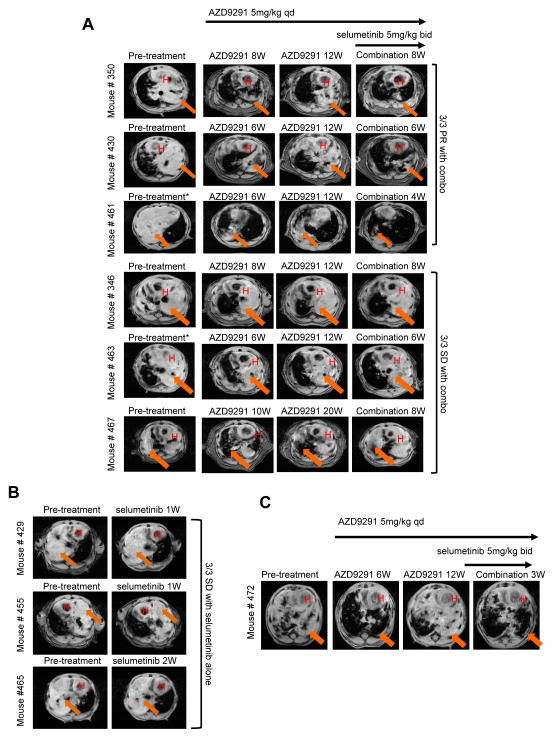
Finally, we tested the concept that MEK pathway inhibition could circumvent resistance to AZD9291 using in vivo mouse tumour models that develop lung adenocarcinomas driven by EGFRL858R + T790M or EGFRL858R (29) and are highly sensitive to inhibition by AZD9291 (10). Three animals with EGFRL858R + T790M transgenic tumours were chronically treated with 5 mg/kg/day AZD9291 and showed initial tumour regression followed by progressive disease after 3 months treatment (Fig. 5A). Animals were subsequently treated with AZD9291 in combination with 5 mg/kg twice daily of selumetinib. Remarkably, resistant tumours in 3/6 animals showed a profound response to the combination, showing strong regression after 1–2 months of combination treatment (Fig. 5A). Tumour regression was not observed when tumour-bearing EGFRL858R + T790M mice were treated with selumetinib alone for 1–2 weeks (Fig. 5B). Similarly, an animal bearing an EGFRL858R tumour showed progression after 3 months of AZD9291 treatment, which regressed following combination of AZD9291 with selumetinib (Fig. 5C). These data provide compelling in vivo evidence to support RAS-MAPK signaling dependency as an important resistance mechanism to AZD9291.
Figure 5. In vivo combination of AZD9291 and selumetinib can overcome acquired resistance to AZD9291 in mutant EGFR trangenic models of lung cancer.
(A) MR images of lungs from tumour-bearing EGFRL858R + T790M transgenic mice pretreatment, after treatment with AZD9291 for 6–20 weeks (W) until progressive disease, and subsequently with the combination of AZD9291/selumetinib for 4–8 weeks. (B) MR images of lung from tumour-bearing EGFRL858R + T790M mice pre- and post- treatment with selumetinib for 1–2 weeks (1W/2W). Combo, AZD9291/selumetinib; H, heart; arrow denotes tumour. (C) MR images of lung from a tumour-bearing EGFRL858R transgenic mouse pretreatment, after treatment with AZD9291 for 6–12 weeks (W) until disease progression, and subsequently with the combination of AZD9291/selumetinib for 3 weeks (3W). *Pretreatment images for mouse #461 and #463 were obtained after these mice received 4 weeks of low dose (1–2.5mg/kg) AZD9291 with no response, before being switched to 5mg/kg dosing.
Basal levels of RAS-MAPK pathway components do not predict MEK inhibitor sensitivity across resistant populations
We also explored whether the activity status of RAS-MAPK pathway could identify tumours that would benefit from combination of EGFR TKI with selumetinib. Unexpectedly, immunoblotting from parental and resistant populations revealed little correlation between basal ERK phosphorylation levels and selumetinib sensitivity (Supplementary Fig. S4). Consistent with this data, analysis of phosphorylated and total protein levels using a Reverse Phase Protein Array (RPPA), showed basal levels of phosphorylated ERK were not indicative of selumetinib sensitivity (Supplementary Table S1; Supplementary Table S5). In conjunction with this, sensitivity to MEK inhibition was not consistently correlated with levels of other phosphorylated or total proteins known to be involved in RAS-MAPK signaling (Supplementary Table S5).
Discussion
Significant advances in our understanding of acquired resistance to EGFR targeted drugs in EGFRm NSCLC, including but not limited to identification of T790M, MET or HER2 amplification and PIK3CA mutants (6), is helping towards the development of new rational treatment strategies to potentially prolong patient benefit such as AZD9291 and CO-1686 which target T790M. However, a large proportion of EGFR inhibitor acquired resistance remains unexplained, and it is anticipated that cells will also find alternative mechanisms to circumvent inhibition by new agents such as AZD9291 and CO-1686.
We have used a novel approach by directly comparing resistance mechanisms across 32 populations with acquired resistance to different EGFR TKIs, and provide the first pre-clinical in-depth analysis of AZD9291 acquired resistance. We took a population approach to try to better emulate the heterogeneity of resistance that occurs in advanced tumours due to competing pressures on both selection of existing clones and gain of new alterations.
A key finding is identification of certain NRAS mutations or NRAS gain as the most frequently detected genetic modifications able to drive resistance to AZD9291. Although previous in vitro data has similarly identified an NRAS Q61K mutation in acquired resistance to gefitinib or erlotinib (24, 30), this is the first report of an NRAS activating mutation conferring acquired resistance to other EGFR inhibitors such as AZD9291. Furthermore this is the first report of the novel NRAS E63K mutation, and together with NRAS G12V, the first report of these NRAS mutations associated with EGFRm NSCLC. The high incidence of NRAS modifications was somewhat surprising in light of recent clinical data in which common NRAS mutations were not detected in lung cancers with acquired resistance to gefitinib or erlotinib (7, 24). However, genetic alterations in NRAS have been associated with resistance to EGFR agents in other disease settings such as colorectal cancer (31, 32), raising the hypothesis that NRAS aberrations may become important in lung cancer too. Copy number changes were not analysed in the previous studies, therefore the clinical relevance of NRAS copy number gain in lung cancer remains unknown. Using more extensive DNA analysis a role for NRAS activation in EGFR TKI resistant EGFRm NSCLC may eventually emerge, and furthermore may only become more apparent as newer agents become established in clinical practice.
Despite the clinical prevalence of specific NRAS molecular alterations being unclear, it was notable how activation of RAS-MAPK signaling independent of EGFR activity was a common biological theme, although the precise molecular nature driving resistance remains unclear for a number of populations. Others have reported alternative mechanisms of resistance to EGFR TKIs associated with increased dependency on RAS-MAPK signaling including loss of NF1, CRKL amplification and EMT (30, 33–35). In addition, amplification of MAPK1 was reported as a resistance mechanism to WZ4002 (15) and has been observed in PC9 AZD9291 resistant populations in the current study. Taken together, these studies suggest that activation of RAS-MAPK signaling independent of EGFR could be a frequent resistance mechanism for the TKIs currently in development, with multiple different aberrations converging on RAS-MAPK signaling. This mirrors other disease areas, where resistance mechanisms to EGFR targeting result in RAS-MAPK pathway activation by various mechanisms e.g. mutations in KRAS, NRAS and BRAF in colorectal cancer (31, 32) or over expression of RAS family in head and neck cancer (36) are associated with cetuximab resistance. Moreover, data presented here and by others (15, 34) support use of MEK inhibitors such as selumetinib in combination with new EGFR TKIs to overcome such acquired resistance mechanisms or potentially in earlier treatment as part of prevention strategies. Interestingly, our data support that this combination may provide benefit in both T790M and EGFRm TKI-naïve settings.
In addition to increased sensitivity to selumetinib across a number of the resistant populations we also observed increased sensitivity to the Aurora kinase B inhibitor AZD1152-HQPA, in combination with AZD9291, in all of the AZD9291 resistant NCI-H1975 populations compared to the parental cells (Supplementary Table S3). This is consistent with recent reports (37) and is worthy of further investigation.
Overall, the emerging pre-clinical evidence presented here supports a picture whereby during chronic exposure to AZD9291, competing selection pressures are likely to influence which clones within a heterogeneous population ultimately become dominant. This could also involve T790M clones becoming less prevalent within a tumour as other resistance clones become more dominant. Moreover, data from ourselves and others provide a compelling rationale for combining inhibitors of the RAS-MAPK signaling such as selumetinib with AZD9291 across EGFRm settings in NSCLC, to tackle RAS-MAPK as a potentially important escape mechanism within such clones. A key challenge will be to develop effective patient selection strategies to identify those patients who may benefit from such a combination. Emerging data suggest multiple genetic and non-genetic alterations, including certain NRAS modifications reported here, could occur that converge on activating the RAS-MAPK pathway, and therefore it is possible that a broad biomarker platform will need to be established. It is important that measuring basal phosphorylation levels of ERK is unlikely to be sufficient to determine dependence on RAS-MAPK signaling or sensitivity to MEK inhibitors (38), thus more sophisticated predictive biomarker strategies will need to be developed. Future studies will determine how clinically prevalent these pre-clinical mechanisms will be, but these pre-clinical observations provide important insights to focus clinical translation efforts.
Supplementary Material
Acknowledgments
Financial Support
This work was supported by AstraZeneca. Some of this work was supported in part by grants from the V Foundation (to W. Pao) and the NCI (R01-CA121210, P01-CA129243, and U54-CA143798 to W. Pao). W. Pao received additional support from Vanderbilt’s Specialized Program of Research Excellence in Lung Cancer grant(CA90949) and the VICC Cancer Center Core grant (P30-CA68485). Imaging support to W. Pao was provided by NCI P30 CA68485.
We would like to thank Carl Barrett for helping support delivery of sequencing studies and Hedley Carr for technical advice and assistance. We would also like to thank Gayle Marshall for support in generating resistance models. At Vanderbilt, we thank Helen Pan for assistance with molecular genotyping assays and Daniel Colvin and Fuxue Xin for magnetic resonance imaging assistance.
Footnotes
Disclosure of Potential Conflicts of Interest
A patent relating to EGFR T790M mutation testing was licensed on behalf of William Pao and others by Memorial Sloan-Kettering Cancer Center to MolecularMD. William Pao received research funding from AstraZeneca. C.A. Eberlein, D Stetson, A.A Markovets, Z Lai, K.J Al-Kadhimi, S.J Ross, M.J Ahdesmaki, A Ahmed, C.H Barnes, H Brown, P.D Smith, J.R Dry, G Beran, K.S Thress, B Dougherty, D.A.E. Cross are employees of AstraZeneca.
References
- 1.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28 (Suppl 1):S24–31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10(11):760–74. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 4.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for european patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–46. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–7. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31(27):3327–34. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 9.Keating GM. Afatinib: A review of its use in the treatment of advanced non-small cell lung cancer. Drugs. 2014;74(2):207–21. doi: 10.1007/s40265-013-0170-8. [DOI] [PubMed] [Google Scholar]
- 10.Cross DA, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter AO, Sjin RT, Haringsma HJ, et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov. 2013;3(12):1404–15. doi: 10.1158/2159-8290.CD-13-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462(7276):1070–4. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y, Ko J, Cui Z, et al. The EGFR T790M mutation in acquired resistance to an irreversible second-generation EGFR inhibitor. Mol Cancer Ther. 2012;11(3):784–91. doi: 10.1158/1535-7163.MCT-11-0750. [DOI] [PubMed] [Google Scholar]
- 14.Nakagawa T, Takeuchi S, Yamada T, et al. Combined therapy with mutant-selective EGFR inhibitor and met kinase inhibitor for overcoming erlotinib resistance in EGFR-mutant lung cancer. Mol Cancer Ther. 2012;11(10):2149–57. doi: 10.1158/1535-7163.MCT-12-0195. [DOI] [PubMed] [Google Scholar]
- 15.Ercan D, Xu C, Yanagita M, et al. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov. 2012;2(10):934–47. doi: 10.1158/2159-8290.CD-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SM, Yun MR, Hong YK, et al. Glycolysis inhibition sensitizes non-small cell lung cancer with T790M mutation to irreversible EGFR inhibitors via translational suppression of mcl-1 by AMPK activation. Mol Cancer Ther. 2013 doi: 10.1158/1535-7163.MCT-12-1188. [DOI] [PubMed] [Google Scholar]
- 17.Shien K, Toyooka S, Yamamoto H, et al. Acquired resistance to EGFR inhibitors is associated with a manifestation of stem cell-like properties in cancer cells. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-12-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vazquez-Martin A, Cufi S, Oliveras-Ferraros C, et al. IGF-1R/epithelial-to-mesenchymal transition (EMT) crosstalk suppresses the erlotinib-sensitizing effect of EGFR exon 19 deletion mutations. Sci Rep. 2013;3:2560. doi: 10.1038/srep02560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ware KE, Hinz TK, Kleczko E, et al. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis. 2013;2:e39. doi: 10.1038/oncsis.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamauchi M, Yoshino I, Yamaguchi R, et al. N-cadherin expression is a potential survival mechanism of gefitinib-resistant lung cancer cells. Am J Cancer Res. 2011;1(7):823–33. [PMC free article] [PubMed] [Google Scholar]
- 21.Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501(7467):355–64. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morabito A, Costanzo R, Rachiglio AM, et al. Activity of gefitinib in a non-small-cell lung cancer patient with both activating and resistance EGFR mutations. J Thorac Oncol. 2013;8(7):e59–60. doi: 10.1097/JTO.0b013e318286cc26. [DOI] [PubMed] [Google Scholar]
- 23.Tan DS, Camilleri-Broet S, Tan EH, et al. Intertumor heterogeneity of non-small-cell lung carcinomas revealed by multiplexed mutation profiling and integrative genomics. Int J Cancer. 2014 doi: 10.1002/ijc.28750. [DOI] [PubMed] [Google Scholar]
- 24.Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109(31):E2127–33. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Z, Dias-Santagata D, Duke M, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13(1):74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chmielecki J, Foo J, Oxnard GR, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3(90):90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukushima S, Otsuka A, Suzuki T, et al. Mutually exclusive mutations of KIT and RAS are associated with KIT mRNA expression and chromosomal instability in primary intracranial pure germinomas. Acta Neuropathol. 2014;127(6):911–25. doi: 10.1007/s00401-014-1247-5. [DOI] [PubMed] [Google Scholar]
- 28.van der Burgt I, Kupsky W, Stassou S, et al. Myopathy caused by HRAS germline mutations: Implications for disturbed myogenic differentiation in the presence of constitutive HRas activation. J Med Genet. 2007;44(7):459–62. doi: 10.1136/jmg.2007.049270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regales L, Balak MN, Gong Y, et al. Development of new mouse lung tumor models expressing EGFR T790M mutants associated with clinical resistance to kinase inhibitors. PLoS One. 2007;2(8):e810. doi: 10.1371/journal.pone.0000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang MH, Lee JH, Chang YJ, et al. MEK inhibitors reverse resistance in epidermal growth factor receptor mutation lung cancer cells with acquired resistance to gefitinib. Mol Oncol. 2012 doi: 10.1016/j.molonc.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meriggi F, Vermi W, Bertocchi P, Zaniboni A. The emerging role of NRAS mutations in colorectal cancer patients selected for anti-EGFR therapies. Rev Recent Clin Trials. 2014 doi: 10.2174/1568026614666140423121525. [DOI] [PubMed] [Google Scholar]
- 32.Misale S, Arena S, Lamba S, et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci Transl Med. 2014;6(224):224ra26. doi: 10.1126/scitranslmed.3007947. [DOI] [PubMed] [Google Scholar]
- 33.Buonato JM, Lazzara MJ. ERK1/2 blockade prevents epithelial-mesenchymal transition in lung cancer cells and promotes their sensitivity to EGFR inhibition. Cancer Res. 2014;74(1):309–19. doi: 10.1158/0008-5472.CAN-12-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Bruin EC, Cowell C, Warne PH, et al. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-13-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung HW, Du J, Boehm JS, et al. Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers. Cancer Discov. 2011;1(7):608–25. doi: 10.1158/2159-8290.CD-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saki M, Toulany M, Rodemann HP. Acquired resistance to cetuximab is associated with the overexpression of ras family members and the loss of radiosensitization in head and neck cancer cells. Radiother Oncol. 2013 doi: 10.1016/j.radonc.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 37.Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346(6216):1480–6. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niepel M, Hafner M, Pace EA, et al. Profiles of basal and stimulated receptor signaling networks predict drug response in breast cancer lines. Sci Signal. 2013;6(294):ra84. doi: 10.1126/scisignal.2004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.