Abstract
The generalist parasite Trypanosoma cruzi has two phylogenetic lineages associated almost exclusively with bats—Trypanosoma cruzi Tcbat and the subspecies T. c. marinkellei. We present new information on the genetic variation, geographic distribution, host associations, and potential vectors of these lineages. We conducted field surveys of bats and triatomines in southern Ecuador, a country endemic for Chagas disease, and screened for trypanosomes by microscopy and PCR. We identified parasites at species and genotype levels through phylogenetic approaches based on 18S ribosomal RNA (18S rRNA) and cytochrome b (cytb) genes and conducted a comparison of nucleotide diversity of the cytb gene. We document for the first time T. cruzi Tcbat and T. c. marinkellei in Ecuador, expanding their distribution in South America to the western side of the Andes. In addition, we found the triatomines Cavernicola pilosa and Triatoma dispar sharing shelters with bats. The comparisons of nucleotide diversity revealed a higher diversity for T. c. marinkellei than any of the T. c. cruzi genotypes associated with Chagas disease. Findings from this study increased both the number of host species and known geographical ranges of both parasites and suggest potential vectors for these two trypanosomes associated with bats in rural areas of southern Ecuador. The higher nucleotide diversity of T. c. marinkellei supports a long evolutionary relationship between T. cruzi and bats, implying that bats are the original hosts of this important parasite.
Introduction
The protozoan parasite Trypanosoma cruzi (order Kinetoplastida) causes Chagas disease, one of the main tropical diseases in the Americas [1]. The intraspecific genetic variation of this species is complex, and several attempts have been made to establish main intraspecific lineages. Currently, T. cruzi is divided in two subspecies: T. c. cruzi and T. c. marinkellei [2], and even further, T. c. cruzi is divided in seven discrete typing units (DTUs) (i.e., TcI-TcVI and Tcbat) [3–6]. Moreover, new investigations have unveiled pronounced structuring within some DTUs e.g., [7–10]. However, among these subdivisions of T. cruzi, those lineages associated almost exclusively with bats (i.e., Tcbat and T. c. marinkellei) are arguably the most enigmatic for their biology and scant documentation.
T. c. marinkellei is restricted to bats, does not infect laboratory mice, and is thought to be exclusively transmitted by triatomines of the genus Cavernicola [11,12]. Nevertheless, the high prevalence and wide distribution of this trypanosome subspecies suggest the participation of other vectors [13]. In contrast, Tcbat like all T. c. cruzi DTUs, proved to be infective to mice [3]. In addition, Tcbat was recently found in a Colombian child and in pre-Columbian mummies of the Cabuza and Camarones cultures in Chile [14,15]. Tcbat is unable to develop in T. infestans and R. prolixus [3], and although its vectors remain to be discovered, the occurrence of this genotype in Brazil, Panamá, and Colombia suggests that it can be transmitted by diverse vector species.
Phylogenetic studies have demonstrated that Tcbat is sister to TcI, and it has been long established that T. c. marinkellei is sister to the monophyletic group formed for all DTUs [3,5,6,16]. Given that T. cruzi sensu lato belongs to the T. cruzi clade, a group of ~18 species that mostly parasitize bats [17–19], here we hypothesized that bats are the ancestral hosts of T. cruzi. Thus, it would be expected that the bat lineages of T. cruzi—specially the most basal lineage that is T. c. marinkellei—would have a greater genetic diversity than the other subdivisions of T. cruzi.
Here we provide information on the genetic variation, geographic distribution, host associations, and potential vectors of bat lineages of T. cruzi. We analyze the nucleotide diversity of the T. cruzi lineages to test the hypothesis that bats are the ancestral hosts of this parasite, and discuss the implications of our results to understand the origins of T. cruzi and Chagas disease.
Methods
Ethics statement
Permits for field research, allowing handling and euthanizing bats, were granted by the Ministerio del Ambiente de Ecuador (002–07 IC-FAU-DNBAPVS/MA, and 008-IC-INSEC-DPL-MA). Bats were euthanized in the field by thoracic compression in 2007 and by inhalation of carbon dioxide in 2012. The methods we used for manipulation and euthanizing of bats are in agreement with the guidelines of the American Society of Mammalogists for the use of wild mammals in research [20]. No protocol of an institutional animal care and use committee (IACUC) was required for this research because Pontificia Universidad Católica del Ecuador does not have an IACUC.
Field surveys of bats and triatomines in Ecuador
A total of 74 bats were caught during January 2007 and July 2012 using a butterfly net at roosts inside human constructions or by mist netting forest patches in the following rural communities: Bellamaría Chica, Bellamaría, Chaquizhca and Chinguilamaca in Loja province, and Rancho Alegre in Zamora Chinchipe province. Additionally, at the Chinguilamaca and Rancho Alegre locations, opportunistic triatomine insect searches within the bat roosts were performed manually. Taxonomic identification of bats and triatomines was conducted by morphological examination following [21] and [22], respectively.
Captured bats were euthanized; samples of blood, liver, heart, and muscle tissues were collected, and from the animals captured in 2012, blood smears and trypanosome cultures were also prepared. A blood aliquot of 150μl was inoculated in biphasic Novy-Nicolle-MacNeal (NNN) culture and the growth of parasites was tracked weekly by microscopy during the first month, and then again at three and six months after initial inoculation. Positive samples containing Trypanosoma-like parasites were grown until counts reached more than 100 parasites per field and were then transferred to Liver Infusion Tryptose (LIT) medium for further growth.
Detection of trypanosomes
Direct microscopic detection of trypanosomes was performed on the animals captured in 2012. DNA was extracted from samples of blood, liver, muscle, or positive cultures with the DNeasy kit (Qiagen, Valencia, CA) following manufacturer´s protocol. Samples were PCR amplified with the primer sets S35/S36 and 121/122 to detect infections with T. cruzi and T. rangeli, respectively [23–26]. Products were visualized on 2% agarose gels. No triatomines were analyzed because of problems with extracting DNA from improperly preserved specimens.
Phylogenetic identification of trypanosome lineages
We sequenced fragments of the 18S rRNA and cytb genes of Trypanosoma from a subset of eight positive animals from three localities that yielded positive results (S1 Table). Not all the positive samples were sequenced because animals were collected in groups and most likely share the same parasite genotypes. To amplify the 18S rRNA, we followed a nested PCR protocol [27] with the following modifications: the initial PCR amplification was conducted with the newly designed primers SSU4_F (GTGCCAGCACCCGCGGTAAT) and 18Sq1R (CCACCGACCAAAAGCGGCCA); both nested PCR amplifications were run with a touchdown PCR profile [28]. The cytb gene was amplified following a published protocol [16]. After cleaning the PCR products with ExoSAP-IT (Affymetrix, Santa Clara, CA), we did sequencing reactions in both directions with the ABI BigDye chemistry (Applied Biosystems, Inc., Foster City, CA), and sequenced the fragments on an ABI 3730xl DNA Analyzer automatic sequencer (Applied Biosystems, Inc., Foster City, CA).
We built a matrix for each gene with sequences of previous studies of lineage diversity of T. cruzi e.g., [3,5,29,30] and also used sequences of Trypanosoma dionisii and Trypanosoma erneyi as outgroups [31] (S1 Dataset). We assembled each gene fragment with the Geneious Alignment tool in Geneious v. 6.1.8 [32], and the alignments were checked and corrected manually. The 18S rRNA and cytb alignments were cropped at 863 bp (99 sequences) (S2 Dataset) and 490 bp (362 sequences) (S3 Dataset) respectively. We built a network genealogy for the 18S rRNA gene with the program SplitsTree v. 4.11.3 using the NeighborNet method [33]. Internode supports were estimated by performing 100 bootstrap replicates using the same parameters optimized for network inferences. For the alignment of the cytb gene we started a maximum likelihood run in RAxML v. 8 [34] and interrupted it after obtaining the reduced matrix that contains only one sequence per unique haplotype (S4 Dataset). Following this, we ran to completion the analysis with the GTR-CAT approximation on the reduced matrix. The GTR-CAT approximation is a rapid algorithm for ML analyses that resembles the GTR-G model, but it is optimized for faster performance [35].
Also, to test the combinability of the 18S rRNA and cytb genes for concatenated phylogenetic analysis we used the software MLSTest [36] to run the analysis ILD-BIONJ [37] that is an efficient variant of the incongruence length difference test [38]. We ran ILD-BIONJ with reduced alignments that only contained strains represented for both genes. This analysis determined that our loci had significantly different branching patterns (p = 0.0099), so no further concatenated analyses were performed.
Comparisons of nucleotide diversity among T. cruzi lineages
We calculated the nucleotide diversity (π) of each T. cruzi subdivision on the cytb tree (i.e., TcI, TcII, TcIII-TcVI, Tcbat, and the subspecies T. c. marinkellei). In Mega v. 6 [39] we used the option “compute mean diversity in entire population”, which calculates a nucleotide diversity index that is independent of sample size (equation 12.73 in [40]). Standard errors were estimated by 1,000 bootstrap replicates. The 18S rRNA fragment was not used for these calculations because of indels within the alignment, which are removed from the calculations producing severe underestimations of nucleotide diversity.
Results
Detection of trypanosomes
Of the 74 bats examined, 27 (36.5%) were positive for T. cruzi, and only one was positive for T. rangeli. The species with the highest infection rate was Artibeus fraterculus (90.1%), while Myotis sp. and Glossophaga soricina were positive in one locality each (Table 1). All positive bats were negative by direct microscopy, and trypanosomes were detected only by PCR.
Table 1. Results of PCR screenings for Trypanosoma cruzi and Trypanosoma rangeli.
| Bat species | Locality | Bats examined | T. cruzi | T. rangeli |
|---|---|---|---|---|
| Artibeus fraterculus | Bella Maria Chica, Loja | 22 | 20 (90.1%) | 0 |
| Desmodus rotundus | Bella Maria, Loja | 7 | 3 (42.9%) | 1 (14.3%) |
| Glossophaga soricina | Bella Maria, Loja | 5 | 0 | 0 |
| Glossophaga soricina | Chaquizhca, Loja | 12 | 3 (25%) | 0 |
| Molossus molossus | Chinguilamaca, Loja | 3 | 0 | 0 |
| Myotis sp. | Chinguilamaca, Loja | 18 | 1 (5.6%) | 0 |
| Myotis sp. | Rancho Alegre, Zamora | 7 | 0 | 0 |
| 5 species | 5 localities | 74 bats | 27 (36.5%) | 1 (1.6%) |
Phylogenetic identification of trypanosome lineages
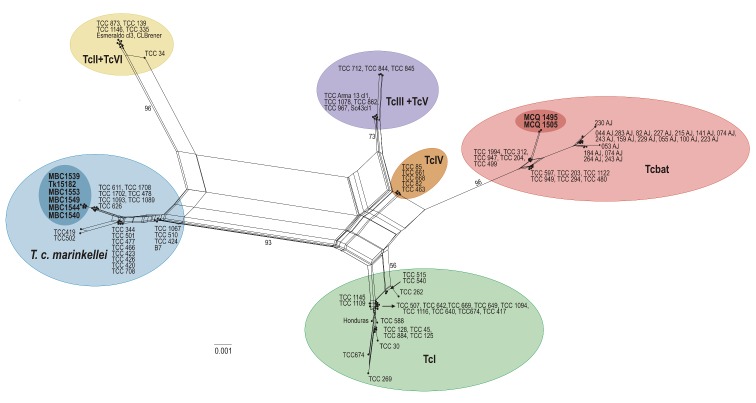
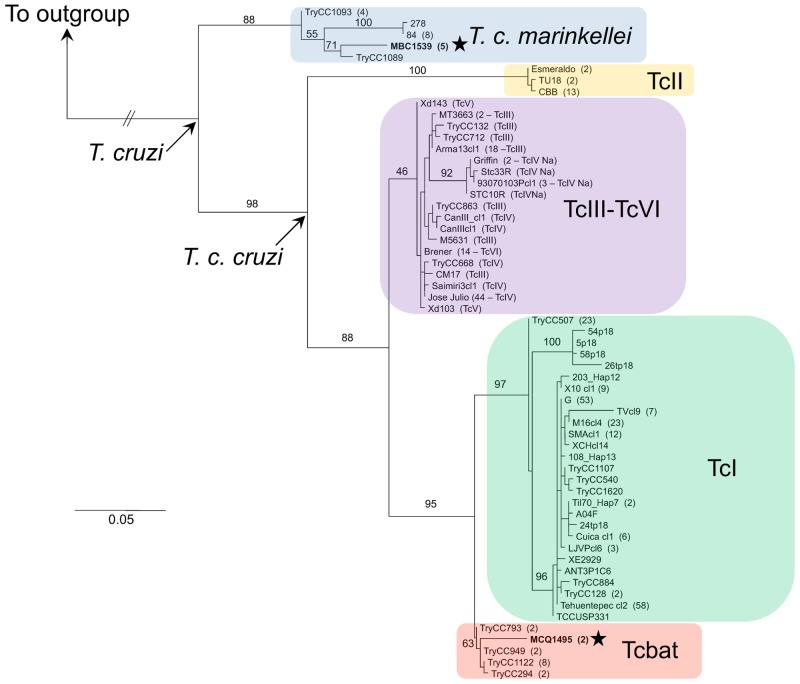
Sequences of the 18S rRNA and cytb genes were obtained from seven out of the eight positive samples; for one sample (TK 151852), only the 18S rRNA fragment was successfully acquired. Six out the eight samples were found infected with T. cruzi marinkellei, and two were positive for Tcbat (S1 Table). The network genealogy of the 18S rRNA gene resulted in distinctive clusters for each genetic lineage (Fig 1). The phylogenetic analysis of the cytb gene revealed five mitochondrial lineages within T. cruzi sensu lato, differing from the traditional DTU classification by clustering in a single clade the DTUs TcIII, TcIV, TcV, and TcVI (Fig 2).
Fig 1. Network genealogy using partial 18S rRNA gene sequences.
from eight new trypanosomes characterised in this study (in bold) plus 70 other sequences from all DTUs (TcI-TcVI and Tcbat) of T. cruzi and 22 sequences from T. c. marinkellei. Network constructed with the NeighborNet algorithm excluding all conserved sites and with uncorrected p-distance. Numbers in nodes correspond to bootstrap support values using the same parameter optimized for network inferences.
Fig 2. Mitochondrial phylogeny of Trypanosoma cruzi.
Maximum likelihood tree of a fragment of the cytb gene of Trypanosoma cruzi and T. dionisii as outgroup, representing 60 haplotypes from 362 sequences. The parentheses at the tip labels contain the number of identical sequences per each haplotype, and in the TcIII-TcVI group the DTU identity for each haplotype is indicated. Numbers on branches correspond to bootstrap support values. Stars indicate the haplotypes found in Ecuador.
Comparisons of nucleotide diversity among T. cruzi lineages
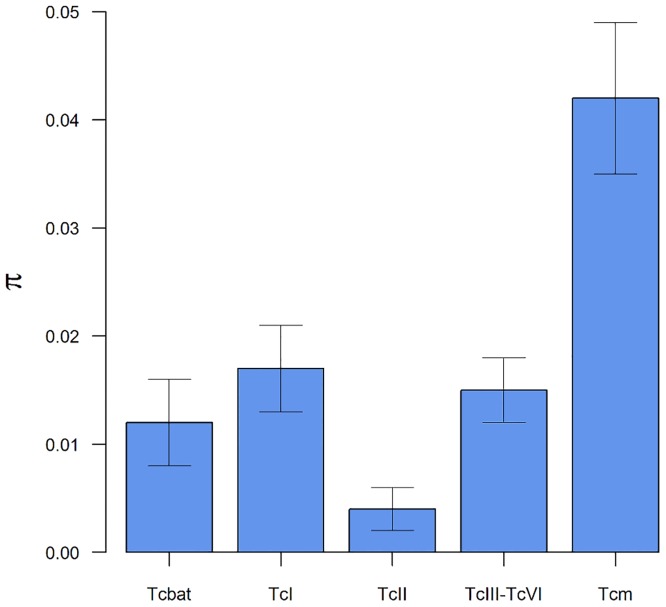
Among the mitochondrial lineages, T. c. marinkellei shows the largest nucleotide diversity in the cytb gene, followed in descending order by TcI, TcIII-TcVI, Tcbat, and TcII. The nucleotide diversity within the T. c. marinkellei is 2.6 times larger than in TcI (0.042 vs. 0.017), and Tcbat nucleotide diversity (0.012) is comparable with the diversity of the pooled DTUs TcIII-TcVI (0.015) (Fig 3).
Fig 3. Mitochondrial diversity of Trypanosoma cruzi.
Nucleotide diversity (π) of mitochondrial lineages of Trypanosoma cruzi calculated for the haplotypes of the cytb gene. The subspecies T. c. marinkellei shows larger nucleotide diversity than the other examined lineages. Whiskers in each bar indicate the standard error.
Triatomine vectors associated with bats
Triatomine bugs were found in two bat roosting sites. One live adult Cavernicola pilosa associated with Myotis sp. was found inside the walls of a two-story cinderblock house in Barrio Rancho Alegre, close to the town of Zamora, Zamora Chinchipe Province (WGS84, 78.86497°W, 03.98589°S; 885 m) (Fig 4). One live adult of Triatoma dispar was found associated with Molossus molossus and Myotis sp. in the window crevice of an adobe barn that is used to raise chickens. One of the Myotis sp. was positive for T. c. marinkellei. This barn is close to a two-story house located in Chinguilamaca, Loja Province (WGS84, 79.32769°W, 04.18938°S; 1358 m) (Fig 4).
Fig 4. Constructions in Ecuador where Cavernicola pilosa and Triatoma dispar were found in association with bats.
A. House with cinderblock walls at Rancho Alegre, Zamora Chinchipe where C. pilosa (inset) was inhabiting a roost of Myotis sp. B. Adobe barn in Chinguilamaca, Loja where T. dispar (inset) was found associated with Molossus molossus and Myotis sp. Arrows indicate the entrances to the bat roosts where the insects were collected.
Discussion
The large genetic diversity for the bat lineages of T. cruzi sensu lato (i.e., Tcbat and T. c. marinkellei), together with the phylogenetic pattern of bat-exclusive species basal to T. cruzi (e.g., T. dionisii, T. erneyii), strongly suggest a long evolutionary history between these parasites and bats; a history that is longer and more complex than with any other group of T. cruzi reservoir hosts (e.g., didelphid marsupials). This may indicate that bats are the original reservoir hosts of T. cruzi, as it has been suggested in the bat-seeding hypothesis [17,18]. Similar rationale has been implemented in deciphering the origins of the malaria parasite Plasmodium falciparum in non-human African apes; P. falciparum has higher nucleotide diversity in chimpanzees than in humans [41], and human parasites nest within gorilla lineages [42].
Trypanosoma cruzi seems to have switched from bats to non-volant mammals more than three million years ago [43], with humans among the most recent hosts of this parasite, since the peopling of the Americas occurred less than 15 thousand years ago [44]. Thus, the origins of Chagas disease can be explained in two series of host switching events. First, a single host switch likely occurred from bats to non-volant mammals; this should have been facilitated by generalist triatomines able to interact with bats and non-volant mammals, such as some Panstrongylus and Triatoma species [22]. Second, several host switches of T. cruzi from bats and wild, non-volant mammals to humans are required to explain the diversity of lineages circulating in human populations, since all DTUs have diverged earlier than the arrival of humans to the Americas [43]. These host switches to humans could have occurred directly from wild triatomine populations during the human colonization of the Americas, since the domestication process of triatomines occurred several times and likely over extensive periods of time [45].
The findings of Tcbat and T. c. marinkellei in southern Ecuador greatly extend the known distribution of these lineages. Tcbat has been previously known from few localities in Brazil, Colombia and Panamá [3,5,6,15]; T. c. marinkellei has been reported in Bolivia, Brazil, Colombia, Panamá, and Venezuela e.g., [2,13,19,29,30,46]. Most of these records are from South America, east of the Andes, thus these Ecuadorian records from west of the Andes, could prove useful to track the biogeographic history and dispersal patterns of these trypanosomes. Nonetheless, it is surprising that in surveys of bat trypanosomes in Venezuela and Bolivia, Tcbat has not been detected [30,47], suggesting that this lineage may have a wide but patchy distribution, with locally high prevalences.
Previously, Tcbat has been reported in association with nine bat species that are either frugivorous or insectivorous and belong to the families Emballonuridae, Noctilionidae, Phyllostomidae, and Vespertilionidae [3,5,6,48]. Our findings of Tcbat in the phyllostomid bat Glossophaga soricina, a mostly nectar feeding species, increase the breadth of dietary guilds of hosts and reaffirms the host-generalist condition among bats of this T. cruzi lineage. On the other hand, T. c. marinkellei has been detected in at least 11 species of only phyllostomid bats [2,13,19,29,30,46]. The positive animals reported here add new host species—A. fraterculus and Myotis sp.—to T. c. marinkellei.
In addition, Triatoma dispar could be a second potential vector of T. c. marinkellei, since the trapped individual was found in the roosting site were Myotis sp. was captured. Previously, only C. pilosa was thought to be a vector of T. c. marinkellei following the observations of Marinkelle [12]. The paucity in triatomine searches in bat roosts may be responsible for the lack of information on vectors of the bat lineages of T. cruzi. There are several sylvatic species of triatomines, some already reported in association with bats (e.g., Eratyrus cuspidatus, E. mucronatus, Triatoma rubida, Panstrongylus geniculatus) [22], but new surveys are required to try to isolate trypanosomes from these triatomines and their bat hosts.
Not surprisingly, PCR detection was more sensitive than direct microscopy [49]. PCR detection is highly encouraged to detect trypanosomes from preserved tissues and in degraded or difficult samples e.g., [50–52]. Although the assays with primers S35/S36 and 121/122 work generally well, we recommend amplification and sequencing of a fragment of the 18S rRNA gene (see materials and methods), because the amplification bands are bright and easy to visualize, and other trypanosomes species could be detected (e.g., [19,27]).
The T. cruzi prevalence of 36.5% in bats with 90.1% prevalence in Artibeus fraterculus found in this study is unusually high. Previous T. cruzi surveys in similar environments in Ecuador detected lower prevalences in non-volant mammals [53,54], indicating that transmission cycles involving bats could be more active than those in rodents and marsupials in this region. Some life-history traits make bats ideal long-term reservoir hosts of pathogens: bats are long-lived animals, are able to use different shelters, and can live in large aggregations that might attract vectors and facilitate transmission [55]. In particular, A. fraterculus in Loja province are resilient to human disruption of the environment, live in large aggregations, and use man-made structures as roosts [56,57]. Further studies are required to determine the ecological variables (i.e., triatomine vectors) associated with the high prevalence of T. cruzi in bats.
The triatomines associated with bats reported herein increase the number of the species previously considered to be restricted to sylvatic environments but which now are also known to occur in human environments. Previously, Cavernicola pilosa and Triatoma dispar had been found only in sylvatic conditions and associated with wild mammals [22,58], with exception of one report of a C. pilosa on a house roof [59], and a recent report of individuals of T. dispar in domestic areas [60]. C. pilosa has been reported in roost sites of at least nine bat species within five families [22,58]. Triatoma dispar has been reported associated with the sloth Choloepus hoffmanni [22,61]. Our findings are the first records of C. pilosa and T. dispar associated with vespertilionid bats (Myotis sp.), and T. dispar associated to a molossid bat (Molossus molossus).
The findings of C. pilosa and T. dispar within human constructions may be attributed as accidental colonization, because of our failure to find established colonies with eggs and nymphs. Nonetheless, bats roost in deep cracks in the walls and roofs, making the collecting process difficult, and therefore bat roosts may be systematically overlooked in triatomine surveys inside houses. Despite recent intensive triatomine surveys in Ecuador, this is the first report of T. dispar in Loja province [62–64]. The association of C. pilosa and T. dispar with bats roosting in human constructions is remarkable as it may have human health implications. Tcbat has been reported in bats, and recently in association with humans [14,15]. Potentially, triatomines associated with bats might opportunistically feed on and be able to transmit trypanosomes to humans. The triatomines reported in this study were collected in constructions with characteristics associated with risk factors for T. cruzi infections (Fig 4); living in houses with adobe walls in Loja province or houses with open and mixed walls in the Ecuadorian Amazon have been identified as risk factors for anti-T. cruzi seropositivity in human populations [65,66].
The high nucleotide diversity of T. c. marinkellei supports a long evolutionary relationship between T. cruzi and bats, implying that bats are the original hosts of this important parasite. Tcbat and T. c. marinkellei are for the first time recorded in Ecuador, expanding their distribution in South America to the western side of the Andes, demonstrating that both lineages are widely distributed in South America. Also, we suggest potential vectors for these two genetic lineages of trypanosomes associated with bats in rural areas of southern Ecuador.
Supporting Information
Trypanosome-positive samples selected for DNA sequencing with respective GenBank accession numbers, locality, bat species, and whether trypanosome sequences were obtained from LIT cultures or directly from liver tissue.
(DOCX)
18S rRNA and cytb sequences used in this study with corresponding GenBank accession numbers within brackets. Codes in bold indicate the sequences newly generated for this research.
(DOCX)
Alignment of an 806 bp fragment of the 18S rRNA gene from 100 sequences of Trypanosoma cruzi sensu lato. This file was used for the network genealogy (Fig 2).
(TXT)
Alignment of a 490 bp fragment of the cytb gene from 361 sequences of Trypanosoma cruzi and one T. dionisii.
(TXT)
A subset of the alignment of Additional file 3 containing one sequence per each unique haplotype. This file was used for the cytb maximum likelihood tree and estimations of nucleotide diversity (Fig 3).
(TXT)
Acknowledgments
CMP thanks V. Crespo-Pérez for help provided during the preparation of this manuscript. We thank G. Acosta, S. Alemán, M. Granda, and N. Muñoz, for their help with the fieldwork. We thank L. Messenger and an anonymous referee for reviewing this manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. DNA sequences are deposited in GenBank under accession numbers KT829450-KT829472.
Funding Statement
Fieldwork support was provided by Proyecto PUCE I13048 to SO-M and MJG, NIH Academic Research Enhancement Award 1R15AI077896-01 to MJG, and a Latin American Student Field Research Award from the American Society of Mammalogists to CMP. Laboratory work was supported by a NSF-DDIG grant DEB-1311582 to SLP and CMP, and Global Infectious Disease Training Grant 1D43TW008261-01A1 of Fogarty International Center to MJG. Data analyses was supported by a Graduate Student Fellowship of the American Museum of Natural History to CMP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rassi A Jr, Rassi A, Marin-Neto JA. Chagas disease. The Lancet. 2010;375: 1388–1402. 10.1016/S0140-6736(10)60061-X [DOI] [PubMed] [Google Scholar]
- 2. Baker JR, Miles MA, Godfrey DG, Barrett TV. Biochemical characterization of some species of Trypanosoma (Schyzotrypanum) from bats (Microchiroptera). Am J Trop Med Hyg. 1978;27: 483–491. [DOI] [PubMed] [Google Scholar]
- 3. Marcili A, Lima L, Cavazzana M, Junqueira ACV, Veludo HH, Maia Da Silva F, et al. A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology. 2009;136: 641–655. 10.1017/S0031182009005861 [DOI] [PubMed] [Google Scholar]
- 4. Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MMG, et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect Genet Evol. 2012;12: 240–253. 10.1016/j.meegid.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 5. Pinto CM, Kalko EKV, Cottontail I, Wellinghausen N, Cottontail VM. TcBat a bat-exclusive lineage of Trypanosoma cruzi in the Panama Canal Zone, with comments on its classification and the use of the 18S rRNA gene for lineage identification. Infect Genet Evol. 2012;12: 1328–1332. 10.1016/j.meegid.2012.04.013 [DOI] [PubMed] [Google Scholar]
- 6. Lima L, Espinosa-Álvarez O, Ortiz PA, Trejo-Varón JA, Carranza JC, Pinto CM, et al. Genetic diversity of Trypanosoma cruzi in bats, and multilocus phylogenetic and phylogeographical analyses supporting Tcbat as an independent DTU (discrete typing unit). Acta Trop. 10.1016/j.actatropica.2015.07.015 [DOI] [PubMed] [Google Scholar]
- 7. Llewellyn MS, Miles MA, Carrasco HJ, Lewis MD, Yeo M, Vargas J, et al. Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit I reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathog. 2009;5: e1000410 10.1371/journal.ppat.1000410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marcili A, Lima L, Valente VC, Valente SA, Batista JS, Junqueira ACV, et al. Comparative phylogeography of Trypanosoma cruzi TCIIc: New hosts, association with terrestrial ecotopes, and spatial clustering. Infect Genet Evol. 2009;9: 1265–1274. 10.1016/j.meegid.2009.07.003 [DOI] [PubMed] [Google Scholar]
- 9. Guhl F, Ramírez JD. Trypanosoma cruzi I diversity: Towards the need of genetic subdivision? Acta Trop. 2011;119: 1–4. 10.1016/j.actatropica.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 10. Ramírez JD, Duque MC, Guhl F. Phylogenetic reconstruction based on Cytochrome b (Cytb) gene sequences reveals distinct genotypes within Colombian Trypanosoma cruzi I populations. Acta Trop. 2011;119: 61–65. 10.1016/j.actatropica.2011.04.009 [DOI] [PubMed] [Google Scholar]
- 11. Marinkelle CJ. Biology of the trypanosomes of bats In: Lumsden WHR, Evans DA, editors. Biology of the Kinetoplastida. Academic Press; 1976. pp. 175–216. [Google Scholar]
- 12. Marinkelle CJ. Developmental stages of Trypanosoma cruzi-like flagellates in Cavernicola pilosa . Rev Biol Trop. 1982;30: 107–111. [PubMed] [Google Scholar]
- 13. Cavazzana M, Marcili A, Lima L, da Silva FM, Junqueira ÂCV, Veludo HH, et al. Phylogeographical, ecological and biological patterns shown by nuclear (ssrRNA and gGAPDH) and mitochondrial (Cyt b) genes of trypanosomes of the subgenus Schizotrypanum parasitic in Brazilian bats. Int J Parasitol. 2010;40: 345–355. 10.1016/j.ijpara.2009.08.015 [DOI] [PubMed] [Google Scholar]
- 14. Guhl F, Auderheide A, Ramírez JD. From ancient to contemporary molecular eco-epidemiology of Chagas disease in the Americas. Int J Parasitol. 2014;44: 605–612. 10.1016/j.ijpara.2014.02.005 [DOI] [PubMed] [Google Scholar]
- 15. Ramírez JD, Hernández C, Montilla M, Zambrano P, Flórez AC, Parra E, et al. First report of human Trypanosoma cruzi infection attributed to TcBat genotype. Zoonoses Public Health. 2014;61: 477–479. 10.1111/zph.12094 [DOI] [PubMed] [Google Scholar]
- 16. Barnabe C, Brisse S, Tibayrenc M. Phylogenetic diversity of bat trypanosomes of subgenus Schizotrypanum based on multilocus enzyme electrophoresis, random amplified polymorphic DNA, and cytochrome b nucleotide sequence analyses. Infect Genet Evol. 2003;2: 201–208. 10.1016/S1567-1348(02)00130-2 [DOI] [PubMed] [Google Scholar]
- 17. Lima L, Espinosa-Álvarez O, Hamilton PB, Neves L, Takata CS, Campaner M, et al. Trypanosoma livingstonei: a new species from African bats supports the bat seeding hypothesis for the Trypanosoma cruzi clade. Parasit Vectors. 2013;6: 1–17. 10.1186/1756-3305-6-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamilton PB, Teixeira MMG, Stevens JR. The evolution of Trypanosoma cruzi: the “bat seeding” hypothesis. Trends Parasitol. 2012;28: 136–141. 10.1016/j.pt.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 19. Cottontail VM, Kalko EKV, Cottontail I, Wellinghausen N, Tschapka M, Perkins SL, et al. High local diversity of Trypanosoma in a common bat species, and implications for the biogeography and taxonomy of the T. cruzi clade. PLoS ONE. 2014;9: e108603 10.1371/journal.pone.0108603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sikes RS, Gannon WL. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J Mammal. 2011;92: 235–253. 10.1644/10-MAMM-F-355.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Albuja L. Murciélagos del Ecuador. 2nd ed Quito: Cicetrónic Cía Ltda; Offset; 1999. [Google Scholar]
- 22. Lent H, Wygodzinsky P. Revision of the Triatominae (Hemiptera, Reduviidae), and their significance as vectors of Chagas’ disease. Bull Am Mus Nat Hist. 1979;163: 123–520. [Google Scholar]
- 23. Sturm NR, Degrave W, Morel C, Simpson L. Sensitive detection and schizodeme classification of Trypanosoma cruzi cells by amplification of kinetoplast minicircle DNA sequences: use in diagnosis of Chagas’ disease. Mol Biochem Parasitol. 1989;33: 205–214. 10.1016/0166-6851(89)90082-0 [DOI] [PubMed] [Google Scholar]
- 24. Wincker P, Britto C, Pereira JB, Cardoso MA, Oelemann W, Morel CM. Use of a simplified polymerase chain reaction procedure to detect Trypanosoma cruzi in blood samples from chronic chagasic patients in a rural endemic area. Am J Trop Med Hyg. 1994;51: 771–777. [DOI] [PubMed] [Google Scholar]
- 25. Vallejo GA, Guhl F, Chiari E, Macedo AM. Species specific detection of Trypanosoma cruzi and Trypanosoma rangeli in vector and mammalian hosts by polymerase chain reaction amplification of kinetoplast minicircle DNA. Acta Trop. 1999;72: 203–212. 10.1016/S0001-706X(98)00085-0 [DOI] [PubMed] [Google Scholar]
- 26. Sousa de MA. On opportunist infections by Trypanosoma lewisi in humans and its differential diagnosis from T. cruzi and T. rangeli . Parasitol Res. 2014;113: 4471–4475. 10.1007/s00436-014-4132-1 [DOI] [PubMed] [Google Scholar]
- 27. Noyes HA, Stevens JR, Teixeira M, Phelan J, Holz P. A nested PCR for the ssrRNA gene detects Trypanosoma binneyi in the platypus and Trypanosoma sp. in wombats and kangaroos in Australia. Int J Parasitol. 1999;29: 331–339. [DOI] [PubMed] [Google Scholar]
- 28. Murphy WJ, O’Brien SJ. Designing and optimizing comparative anchor primers for comparative gene mapping and phylogenetic inference. Nat Protoc. 2007;2: 3022–3030. 10.1038/nprot.2007.429 [DOI] [PubMed] [Google Scholar]
- 29. Marcili A, da Costa AP, Soares HS, Acosta I da CL, de Lima JTR, Minervino AHH, et al. Isolation and phylogenetic relationships of bat trypanosomes from different biomes in Mato Grosso, Brazil. J Parasitol. 2013;99: 1071–1076. 10.1645/12-156.1 [DOI] [PubMed] [Google Scholar]
- 30. García L, Ortiz S, Osorio G, Torrico MC, Torrico F, Solari A. Phylogenetic analysis of Bolivian bat trypanosomes of the subgenus Schizotrypanum based on cytochrome b sequence and minicircle analyses. PLoS ONE. 2012;7: e36578 10.1371/journal.pone.0036578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lima L, da Silva FM, Neves L, Attias M, Takata CSA, Campaner M, et al. Evolutionary insights from bat trypanosomes: morphological, developmental and phylogenetic evidence of a new species, Trypanosoma (Schizotrypanum) erneyi sp. nov., in African bats closely related to Trypanosoma (Schizotrypanum) cruzi and allied species. Protist. 2012;163: 856–872. 10.1016/j.protis.2011.12.003 [DOI] [PubMed] [Google Scholar]
- 32.Biomatters. Geneious v. 6.1.8 [Internet]. 2014. Available: http://www.geneious.com/
- 33. Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23: 254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 34. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 36. Tomasini N, Lauthier JJ, Llewellyn MS, Diosque P. MLSTest: Novel software for multi-locus sequence data analysis in eukaryotic organisms. Infect Genet Evol. 2013;20: 188–196. 10.1016/j.meegid.2013.08.029 [DOI] [PubMed] [Google Scholar]
- 37. Zelwer M, Daubin V. Detecting phylogenetic incongruence using BIONJ: an improvement of the ILD test. Mol Phylogenet Evol. 2004;33: 687–693. 10.1016/j.ympev.2004.08.013 [DOI] [PubMed] [Google Scholar]
- 38. Farris JS, Källersjö M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1994;10: 315–319. 10.1111/j.1096-0031.1994.tb00181.x [DOI] [Google Scholar]
- 39. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford and New York: Oxford University Press; 2000. [Google Scholar]
- 41. Rich SM, Leendertz FH, Xu G, LeBreton M, Djoko CF, Aminake MN, et al. The origin of malignant malaria. Proc Natl Acad Sci. 2009;106: 14902–14907. 10.1073/pnas.0907740106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu W, Li Y, Learn GH, Rudicell RS, Robertson JD, Keele BF, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467: 420–425. 10.1038/nature09442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Flores-López CA, Machado CA. Analyses of 32 loci clarify phylogenetic relationships among Trypanosoma cruzi lineages and support a single hybridization prior to human contact. PLoS Negl Trop Dis. 2011;5: e1272 10.1371/journal.pntd.0001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goebel T, Waters MR, O’Rourke DH. The late Pleistocene dispersal of modern humans in the Americas. Science. 2008;319: 1497–1502. 10.1126/science.1153569 [DOI] [PubMed] [Google Scholar]
- 45. Schofield CJ, Diotaiuti L, Dujardin JP. The process of domestication in triatominae. Mem Inst Oswaldo Cruz. 1999;94: 375–378. 10.1590/S0074-02761999000700073 [DOI] [PubMed] [Google Scholar]
- 46. Cottontail VM, Wellinghausen N, Kalko EKV. Habitat fragmentation and haemoparasites in the common fruit bat, Artibeus jamaicensis (Phyllostomidae) in a tropical lowland forest in Panamá. Parasitology. 2009;136: 1133–1145. 10.1017/S0031182009990485 [DOI] [PubMed] [Google Scholar]
- 47. Silva-Iturriza A, Nassar JM, García-Rawlins AM, Rosales R, Mijares A. Trypanosoma evansi kDNA minicircle found in the Venezuelan nectar-feeding bat Leptonycteris curasoae (Glossophaginae), supports the hypothesis of multiple origins of that parasite in South America. Parasitol Int. 2013;62: 95–99. 10.1016/j.parint.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 48. Ramírez JD, Tapia-Calle G, Muñoz-Cruz G, Poveda C, Rendón LM, Hincapié E, et al. Trypanosome species in neo-tropical bats: Biological, evolutionary and epidemiological implications. Infect Genet Evol. 2014;22: 250–256. 10.1016/j.meegid.2013.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kirchhoff LV, Votava JR, Ochs DE, Moser DR. Comparison of PCR and microscopic methods for detecting Trypanosoma cruzi . J Clin Microbiol. 1996;34: 1171–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guhl F, Jaramillo C, Yockteng R, Vallejo GA, Caárdenas-Arroyo F. Trypanosoma cruzi DNA in human mummies. The Lancet. 1997;349: 1370 10.1016/S0140-6736(05)63207-2 [DOI] [PubMed] [Google Scholar]
- 51. Wyatt KB, Campos PF, Gilbert MTP, Kolokotronis S- O, Hynes WH, DeSalle R, et al. Historical mammal extinction on Christmas Island (Indian Ocean) correlates with introduced infectious disease. PLoS ONE. 2008;3: e3602 10.1371/journal.pone.0003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pinto CM, Baxter BD, Hanson JD, Mendez-Harclerode FM, Suchecki JR, Grijalva MJ, et al. Using museum collections to detect pathogens. Emerg Infect Dis. 2010;16: 356–357. 10.3201/eid1602.090998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pinto CM, Grijalva MJ, Costales JA. Prevalencia de Trypanosoma cruzi en roedores y marsupiales en dos localidades de Manabí, Ecuador. Rev Pontif Univ Católica Ecuad. 2003;71: 225–233. [Google Scholar]
- 54. Pinto CM, Ocaña-Mayorga S, Lascano MS, Grijalva MJ. Infection by trypanosomes in marsupials and rodents associated with human dwellings in Ecuador. J Parasitol. 2006;92: 1251–1255. [DOI] [PubMed] [Google Scholar]
- 55. Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19: 531–545. 10.1128/CMR.00017-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pinto CM, Marchán-Rivadeneira MR, Tapia EE, Carrera JP, Baker RJ. Distribution, abundance and roosts of the fruit bat Artibeus fraterculus (Chiroptera: Phyllostomidae). Acta Chiropterologica. 2013;15: 85–94. 10.3161/150811013X667885 [DOI] [Google Scholar]
- 57.Pinto CM, Boada C. Artibeus fraterculus [Internet]. Pinto CM, editor. MammaliaWebEcuador. Museo de Zoología, Pontificia Universidad Católica del Ecuador; 2015. Available: http://zoologia.puce.edu.ec/vertebrados/mamiferos/FichaEspecie.aspx?Id=1930
- 58. Oliveira RLF, Carneiro MA, Diotaiuti L. Ecology of Cavernicola pilosa Barber, 1937, Hemiptera: Reduviidae: Triatominae in the Boa Esperanca cave, Tocantins, Brazil. Ecotropica. 2008;14: 63–68. [Google Scholar]
- 59. Gomes A de C, Pereira JLA. On the finding of Cavernicola pilosa Barber, 1937, in the State of Paraná, Brazil. Rev Saúde Pública. 1977;11: 427–428. [PubMed] [Google Scholar]
- 60. Guevara Á, Moreira J, Criollo H, Vivero S, Racines M, Cevallos V, et al. First description of Trypanosoma cruzi human infection in Esmeraldas province, Ecuador. Parasit Vectors. 2014;7: 1–3. 10.1186/1756-3305-7-358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Barreto M, Barreto P. Triatoma dispar (Hemiptera: Reduviidae), a new record for Colombia. J Med Entomol. 1984;21: 750–750. [Google Scholar]
- 62. Abad-Franch F, Paucar C, Carpio C, Cuba Cuba CA, Aguilar V, Marcelo H, et al. Biogeography of Triatominae (Hemiptera: Reduviidae) in Ecuador: implications for the design of control strategies. Mem Inst Oswaldo Cruz. 2001;96: 611–620. [DOI] [PubMed] [Google Scholar]
- 63. Grijalva MJ, Palomeque-Rodriguez FS, Costales JA, Davila S, Arcos-Teran L. High household infestation rates by synanthropic vectors of Chagas disease in southern Ecuador. J Med Entomol. 2005;42: 68–74. [DOI] [PubMed] [Google Scholar]
- 64. Grijalva MJ, Suarez-Davalos V, Villacis AG, Ocaña-Mayorga S, Dangles O. Ecological factors related to the widespread distribution of sylvatic Rhodnius ecuadoriensis populations in southern Ecuador. Parasit Vectors. 2012;5: 17 10.1186/1756-3305-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grijalva MJ, Escalante L, Paredes RA, Costales JA, Padilla A, Rowland EC, et al. Seroprevalence and risk factors for Trypanosoma cruzi infection in the Amazon region of Ecuador. Am J Trop Med Hyg. 2003;69: 380–385. [PubMed] [Google Scholar]
- 66. Black CL, Ocaña S, Riner D, Costales JA, Lascano MS, Davila S, et al. Household risk factors for Trypanosoma cruzi seropositivity in two geographic regions of Ecuador. J Parasitol. 2007;93: 12–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trypanosome-positive samples selected for DNA sequencing with respective GenBank accession numbers, locality, bat species, and whether trypanosome sequences were obtained from LIT cultures or directly from liver tissue.
(DOCX)
18S rRNA and cytb sequences used in this study with corresponding GenBank accession numbers within brackets. Codes in bold indicate the sequences newly generated for this research.
(DOCX)
Alignment of an 806 bp fragment of the 18S rRNA gene from 100 sequences of Trypanosoma cruzi sensu lato. This file was used for the network genealogy (Fig 2).
(TXT)
Alignment of a 490 bp fragment of the cytb gene from 361 sequences of Trypanosoma cruzi and one T. dionisii.
(TXT)
A subset of the alignment of Additional file 3 containing one sequence per each unique haplotype. This file was used for the cytb maximum likelihood tree and estimations of nucleotide diversity (Fig 3).
(TXT)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. DNA sequences are deposited in GenBank under accession numbers KT829450-KT829472.