Abstract
Background
Entamoeba histolytica, E. dispar and E. moshkovskii are the most frequent species described in human infection where E. histolytica is the only true pathogen. The epidemiology of this infection is complex due to the absence of a routine exam that allows a correct discrimination of the Entamoeba species complex. Therefore, molecular methods appear as the unique epidemiological tool to accomplish the species discrimination. Herein, we conducted a cross-sectional study to determine the frequency of Entamoeba species infections in a group of asymptomatic individuals from a rural area in central Colombia.
Methodology/Principal Findings
A total of 181 fecal samples from asymptomatic children under 16 years old from the hamlet La Vírgen, Cundinamarca (Colombia) that voluntarily accepted to participate in the study were collected. The fecal samples were examined by light microscopy and DNA-extracted, subsequently submitted to molecular discrimination of E. dispar/E. histolytica/E. moshkovskii infection based on a multiplex PCR assay targeting the 18S rRNA fragment. To confirm the species description, twenty samples were randomly submitted to DNA sequencing of the aforementioned fragment. By direct microscopic examination, frequency of the complex E. histolytica/E. dispar/E. moshkovskii was 18.8% (34/181). PCR showed a frequency of 49.1% (89/181), discriminated as 23.2% (42/181) that were positive for E. dispar, 25.4% (46/181) for E. moshkovskii and 0.55% (1/ 181) for E. histolytica. Also, mixed infections were detected between E. dispar and E. moshkovskii at 4.42% (8/181) of the samples. Molecular barcoding confirmed the diagnosis depicted by the multiplex PCR assay.
Conclusions/Significance
This is the first description of E. moshkovskii in Colombia and the second report in South-America to our knowledge. Our results suggest the need to unravel the true epidemiology of Entamoeba infections around the world, including the real pathogenic role that E. moshkovskii may have.
Introduction
Amoebiasis is the parasitism caused by Entamoeba histolytica, the only pathogenic species among the amoebas that inhabit in the human digestive tract and other organs [1]. It is one of the most prevalent parasitic diseases worldwide, particularly in developing countries where sanitation is insufficient [2]. The genus Entamoeba includes six species: E. histolytica, E. dispar, E. moshkovskii, E. bangladeshi, E. poleki, E. coli and E. hartmanni. The first three species are morphologically similar but with different biochemical and genetic features [3]. Previously, it was considered that about 500 million people were infected with E. histolytica worldwide. However, this frequency is overestimated due to the use of light microscopy instead of molecular tools able to differentiate the species [2, 4]. Asymptomatic presentation is the most common clinical form of amoebiasis (around 80–90% of the cases) and might be caused by three independent species: The pathogenic amoeba E. histolytica, E. dispar considered as a commensal and E. moshkovskii in which the pathogenic potential is still under discussion. This premise reflects the need to understand the molecular epidemiology of these species in endemic countries [5].
Recent studies have reported the presence of E. moshkovskii in fecal samples from humans in different countries such as United States, Italy, Iran, Turkey, Bangladesh, India, Australia and Brazil [3, 6, 7, 8, 9, 10]. The role that E. moshkovskii plays in human health is controversial, some authors still consider this species as a commensal but recent studies in India and Bangladesh have identified this species as the only likely pathogen in individuals with gastrointestinal clinical manifestations including dysentery [5, 7, 8, 11]. However, in these patients, no studies of viral or bacterial agents were conducted to rule out other pathogens or potential pathology agents.
At present, the prevalence of infection discriminated by species of Entamoeba is barely known [12, 13, 14, 15]. To address this problem, different molecular methodologies have been used to distinguish the three species (E. histolytica, E. dispar and E. moshkovskii) and have proved to be useful as an epidemiological and surveillance tool [3, 12, 15, 16, 17]. In studies deploying these methodologies, predominance of infection with E. dispar has been reported, while E. histolytica infection looks to be less frequent. Although the distribution of species seems to change according to the region studied [12, 5, 18]. In Latin American countries: Nicaragua, Brazil and Ecuador have reported a higher prevalence of E. dispar compared to E. histolytica [19, 20, 21]. However, countries such as Venezuela and Mexico have reported that E. histolytica is more frequent than E. dispar [22, 23, 24]. In Colombia there is no data (by molecular methods) on the prevalence of these three amoebas in humans and also if there exist the presence of E. moshkovskii. Therefore, the objective of this study was to differentially detect the presence of E. histolytica, E. dispar and E. moshkovskii by PCR in stool samples from school children in a rural community of Cundinamarca, Colombia.
Materials and Methods
Ethical statement
We obtained 181 fecal samples from asymptomatic children under 16 years old from the hamlet La Vírgen, Cundinamarca (Colombia) that voluntarily accepted to participate in the study. The children were physically examined to verify their asymptomatic status. La Virgen hamlet is located in the department of Cundinamarca at 4° 45´ north latitude and 74° 32´west longitude with an altitude of 1050 meters above sea level [25, 26]. The overall percentage of unsatisfied basic needs (UBS) is 58%. A parent or guardian of any child participant of this study provided written informed consent on their behalf. The ethical clearance of this study was followed by the ethics of Helsinki declaration and resolution No. 008430 of 1993 from the Ministry of Health from Colombia and “El Código del Menor”. The study protocol was approved by the ethics committee from the faculty of Medicine of the Universidad Nacional de Colombia under the Number 0045763.
Fecal samples collection, microscopic diagnosis and DNA extraction
The fecal samples were collected in plastic recipients, labeled and conserved in refrigerated boxes. The samples were divided in two parts: one part was fixed in a proportion (1:4) in ethanol 70% and stored at -20°C for DNA extraction. The other part was used for conducting Kato-Katz, modified Richie-Frick method and direct microcopy examination for diagnosis of intestinal parasites [27, 28]. From each sample, 250 mg were submitted to DNA extraction using the QIAmp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to manufacturer´s instructions. Genomic DNA was preserved at -20°C until analysis.
E. histolytica/dispar/moshkovskii PCR discrimination and statistical analyses
Differential identification of E. histolytica/E. dispar/E. moshkovskii was performed by a simple PCR protocol described by Hamzah et al., 2006 that targets the small rRNA subunit. [29]. Positive controls used for carrying out the PCR were reference strains listed as follows: E. histolytica HM1 strain, E. dispar ISS strain and E. moshkoskii SAW760 Laredo strain were kindly provided by Dr. Graham Clark, London School of Hygiene and Tropical Medicine. As negative control was used DNA extracted from a sample of stool from a healthy individual, whom tested negative by light microscopy, Gal / GalNAc lectin determination and PCR [30].
The four oligonucleotides used in PCR were described by Hamzah et al., 2006 [29]. The mixture of the three oligonucleotides allowed the specific amplification of genomic DNA of E. histolytica, E. dispar and E. moshkovskii. The sequence of the sense oligonucleotide (ENTAF) represents the central region of the gene encoding the small subunit ribosomal RNA, conserved in all three species of Entamoeba; antisense oligonucleotides EhR, EmR and EdR are specific for E. histolytica, E. moshkovskii and E. dispar respectively. The primers sequences: ENTAF: 5'-GAG CAC AGC ATG AGC GAA AT-3 '; EhR: 5'-GAT CTA GAA CTC ACA CTT ATG T-3 '; EdR: 5'-CAC CAC TCC CTA CTA TTA DC-3, 'EmR, 5'-TGA GCC CCA GAG GAG ACA T-3'. The combination of oligonucleotides generated specifically products of 166 bp for E. histolytica DNA, 752 bp for E. dispar DNA, and 580-bp for E. moshkovskii DNA [31]. A total of 20 samples were submitted to DNA sequencing of the partial region of the 18S rRNA gene for the confirmation of the diagnosis depicted by PCR. The PCR products were digested with EXOSAP (Affymetrix, USA) and sequenced by the dideoxy-terminal method in an automated capillary sequencer (AB3730, Applied Biosystems) by both strands in Macrogen (Korea). The sequences were submitted to BLASTn for similarity search with Entamoeba sequences deposited on the databases. The resulting sequences were edited in MEGA 5.0 and aligned using ClustalW 1.8 with reference sequences from E. histolytica (FJ888636), E. dispar (KJ719489), E. moshkovskii (KJ719489), E. bangladeshi (JQ412862), E. polecki (FR686399), E. coli (FR686448) and E. hartmanni (FR686382) retrieved from GeneBank [32, 33]. All edited sequences were deposited in GenBank and assigned accession numbers (Under submission). A maximum composite likelihood (MCL) analysis using a Tamura-3 parameter was run in RaxML Phylogeny.fr platform. To evaluate the robustness of the nodes in the resulting phylogenetic tree, 1000 bootstrap replicates were performed.
Bivariate and multivariate analyses by logistic regression model were conducted to determine the factors (age and sex) potentially related to the infections and parasitic co-infections by estimation of odds ratio (OR) and confidence intervals of 95%. In all cases, p value < 0.05 was considered significant. The analysis was done using the Stata version 10.0 (Stata Corporation, College Station, USA). The Kappa (κ) test assessed by the scale [34] was used to measure the correlation between microscopy methods for the complex E. histolytica/ E. dispar/E. moshkovskii and PCR for differential diagnosis of the three species. The relationship between the variables of age, gender, and other co-infections with the prevalence of the species E. histolytica/E. dispar/E. moshkovskii measured by PCR was determined by testing Χ2. Statistical analyses were performed using the EPIDAT v3.1 program (Directorate of Public Saúde Xeral, Xunta de Galicia (Spain) -PAHO).
Results
Prevalence of E. histolytica, E. dispar, E.moshkovskii according to different diagnostic methods
When compared by age and gender, no significant associations were observed in the infection by E. dispar, E. moshkovskii and/or E. histolytica (Table 1). However, infection with E. moshkovskii in the group of 12–15 years is observed as a factor risk compared to the age group of 5–8 years old. By direct microscopic examination, the frequency of the complex E. histolytica/E. dispar/E. moshkovskii was 18.8% (34/181). PCR showed a frequency of 49.2% (89/181), discriminated as 23.2% (42/181) positive for E. dispar, 25.4% (46/181) for E. moshkovskii and 0.55% (1/ 181) for E. histolytica. Also, mixed infections were detected between E. dispar and E. moshkovskii at 4.42% (8/181) of the samples. Finally, the correlation between PCR and light microscopy was moderate on the scale of Altman: Kappa 0.451 (p <0.05; 95% CI: 0342–0560).
Table 1. Bivariate and Multivariate analysis of the E. dispar and E. moshkovskii infections and other co-infections in relation to age and sex.
| Parasite | Associations | Bivariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|---|
| Coefficient | OR (CI 95%) | P | Coefficient | OR (CI 95%) | P | ||
| E. histolytica | Sex | ||||||
| Male | Ref | Ref | Ref | Ref | |||
| Female | -0.49 | 0.95 (0.47–1.91) | 0.891 | -0.01 | 0.98 (0.48–2.01) | 0.971 | |
| Age | |||||||
| 5–8 | Ref | Ref | Ref | Ref | |||
| 9–11 | -1.41 | 0.87 (0.39–1.93) | 0.73 | -0.14 | 0.87 (0.37–1.97) | 0.739 | |
| 12–15 | 0.14 | 1.14 (0.47–2.79) | 0.765 | 0.13 | 1.15 (0.48–2.79) | 0.764 | |
| E. moshkovskii | Sex | ||||||
| Male | Ref | Ref | Ref | Ref | |||
| Female | -0.55 | 0.58 (0.29–1.13) | 0.113 | -0.51 | 0.59 (0.29–1.20) | 0.149 | |
| Age | |||||||
| 5–8 | Ref | Ref | Ref | Ref | |||
| 9–11 | -0.05 | 0.94 (0.42–2.12) | 0.89 | 0.05 | 1.05 (0.46–2.41) | 1.05 | |
| 12–15 | 0.78 | 2.18 (0.93–5.10) | 0.074 | 0.79 | 2.21 (0.95–5.23) | 0.069 | |
| E. dispar | Sex | ||||||
| Male | Ref | Ref | Ref | Ref | |||
| Female | -0.08 | 0.92 (0.42–1.97) | 0.82 | -0.1 | 0.90 (0.41–1.96) | 0.798 | |
| Age | |||||||
| 5–8 | Ref | Ref | Ref | Ref | |||
| 9–11 | -0.11 | 0.89 (0.18–4.41) | 0.891 | -0.01 | 0.98 (0.19–4.98) | 0.988 | |
| 12–15 | 1.27 | 3.57 (0.21–58.62) | 0.372 | 1.14 | 3.15 (0.17–55.41) | 0.432 | |
| Co-infection`A. lumbricoides—T. trichiura | Sex | ||||||
| Male | Ref | Ref | Ref | Ref | |||
| Female | 0.79 | 2.22 (0.56–8.67) | 0.25 | 1.01 | 2.76 (0.68–11.2) | 0.151 | |
| Age | |||||||
| 5–8 | Ref | Ref | Ref | Ref | |||
| 9–11 | -0.77 | 0.46 (0.08–2.61) | 0.383 | -0.96 | 0.38 (0.07–2.19) | 0.28 | |
| 12–15 | 0.77 | 2.15 (0.54–8.53) | 0.275 | 0.76 | 2.14 (0.53–8.62) | 0.282 | |
| Co-infection Hookworms—T. trichiura | Sex | ||||||
| Male | Ref | Ref | Ref | Ref | |||
| Female | 1.07 | 2.92 (0.59–14.42) | 0.189 | 1.25 | 3.52 (0.69–17.93) | 0.13 | |
| Age | |||||||
| 5–8 | Ref | Ref | Ref | Ref | |||
| 9–11 | -0.46 | 0.62 (0.10–3.87) | 0.615 | -0.69 | 0.49 (0.07–3.15) | 0.46 | |
| 12–15 | 0.81 | 2.27 (0.48–10.70) | 0.3 | 0.89 | 2.26 (0.47–10.85) | 0.308 | |
Ref = reference; OR = odds ratio; CI = Confidence Interval
DNA sequence analysis
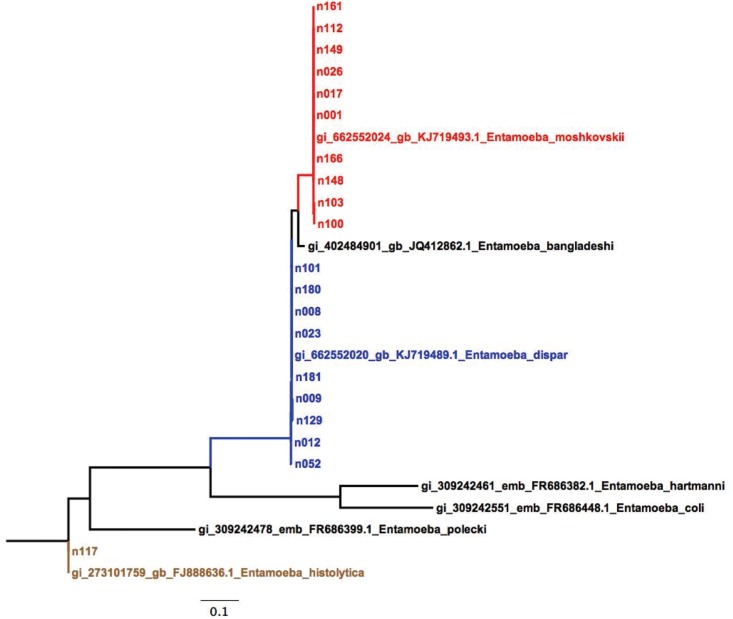
18S rRNA sequences were obtained from 20 samples (one E. histolytica, nine E. dispar and ten E. moshkovskii). Sequences were deposited on GenBank under the accession numbers KT825974-KT825993. The BLAST results showed a 100% similarity for one E. histolytica sample, seven E. dispar and seven E. moshkovskii samples. Two samples from E. dispar (n103 and n100) and three from E. moshkovskii (n009, n129 and n181) showed a similarity of 97%. A ML tree was built according to the 18S sequences to confirm the detection of E. dispar, E. histolytica and E. moshkovskii (Fig 1). The results showed the correct assignment of the Entamoeba species and the congruence with the PCR assay. Albeit, the first and novel description of E. moshkovskii in Colombia.
Fig 1. Maximum Composite Likelihood reconstruction of phylogenetically related 18S rRNA sequences from Amoeba species including 20 DNA sequences from the samples analyzed.
Presence of co-infections
Among the patients included in the study, 85.1% (154/181) were diagnosed with at least one intestinal parasite. Twenty-five percent (45/181) of individuals were infected by one parasite, 26.5% (48/181) with 2 parasites and 30.9% (56/181) with 3 or more parasites. In total, the frequency of intestinal parasites in collected stool samples were: Ascaris lumbricoides (7%), Trichuris trichiura (14%), Hookworm (15%), Hymenolepis nana (1%), Chilomastix mesnili (3%), E. coli (34%), Endolimax nana (45%), Iodamoeba butschlii (7%), Blastocystis (34%), E. hartmanni (3%) and Giardia duodenalis (13%). Associations of co-infections with E. dispar and E. moshkovskii are shown in Table 2. No statistical association between the co-infections with the frequency of the Entamoeba species was observed (p = 0.123).
Table 2. Presence of co-infections (protozoan of helminth parasites).
| Patients infected by E. moshkovskii (n = 46) | Patients not infected by E. moshkovskii (n = 135) | Patients infected by E. dispar (n = 42) | Patients not infected by E. dispar (n = 139) | |
|---|---|---|---|---|
| Protozoans | ||||
| Giardia duodenalis | 5 | 9 | 4 | 10 |
| Blastocystis spp | 15 | 31 | 19 | 23 |
| Entamoeba coli | 1 | 3 | 3 | 1 |
| Entamoeba hartmanni | 1 | 0 | 4 | 0 |
| Endolimax nana | 19 | 44 | 18 | 45 |
| Iodamoeba butschilli | 3 | 10 | 6 | 7 |
| Helminths | ||||
| Ascaris lumbricoides | 2 | 4 | 4 | 2 |
| Trichiuris Trichiura | 0 | 4 | 3 | 1 |
| Hookworms | 6 | 19 | 5 | 19 |
Discussion
Amoebiasis is a diagnostic challenge, clearly due to the existence of three morphologically indistinguishable species leading to the overdiagnosis of the parasitism produced by E. histolytica. In this study, by light microscopy the most commonly method employed for diagnosis, 34 samples were found positive for the complex E. histolytica/E.dispar/E.moshkovskii but only one patient presented infection by E. histolytica, 42 with E. dispar and 46 with E. moshkovskii when PCR was used as a diagnostic method. This test increased the sensitivity and specificity leading to an appropriate treatment for the infected individual. If the diagnosis is conducted only by microscopy medication had to be administered unnecessarily from the clinical standpoint. This reinforces the need for differential diagnosis of E. histolytica with the other species that form the complex because so far this is the only species of the genus Entamoeba that requires one level treatment action. On the other hand, resistance to metronidazole in the clinical context has been reported in cases of amebic liver abscess [35, 36]. This has been more difficult to demonstrate in cases of intestinal amoebiasis [37].
The finding of E. dispar as responsible for most of the infections of the complex E. histolytica/E. dispar/E. moshkovskii has occurred in different studies [21, 38] and in those studies that use molecular methods to split the complex has been the most prevalent species [18, 19, 20, 29, 39, 40]. The reported frequency of E. dispar by PCR in our study was 23.2% (42/181). This is consistent with specific national studies reporting the frequency of E. dispar ranging from 15%–17% [30, 41]. The finding of only one case of E. histolytica could be due to sample size as the reported prevalence in our country does not exceed 1.5% when antigen detection methods are used [30, 41]. Intriguingly, in our study the frequency of E. moshkovskii was a bit higher than E. dispar frequency suggesting that the circulation of this species is extant and not detected in previous examinations.
To our knowledge this is the second study in South America that sought to identify E. moshkovskii by PCR and is the first report of human infection by E. moshkovskii in Colombia [21]. Despite the mentioned above, the fact of detecting in our country by molecular methods E. moshkovskii in wastewater used for agriculture (unpublished data, pending response pairs) and the recent evidence that supports pathogenicity of this amoeba [42] suggests the likely possibility that this amoeba species is circulating in humans in our country. Also, we need further studies with larger samples to determine the epidemiology of this infection. This finding presents epidemiological significance as it is the first report of E. moshkovskii in Colombia and the second in South America and suggests the existence of this species as a possible emerging pathogen in developing countries. We corroborated these findings conducting DNA sequencing of the 18S rRNA markers observing 97–100% similarity with the E.dispar, E. histolytica, E. moshkovskii sequences from GenBank (Fig 1). Additionally, we detected a low amount of drift in our samples (n103, n100, n009, n129 and n181) that need to be further analyzed by high-resolution molecular markers.
Another factor to consider is that the species of Entamoeba that are not part of the complex E. histolytica/E.dispar/E.moshkvoskii as E. hartmanii, E. bangladeshi and E. poleckii sometimes can be morphologically similar to the afore-mentioned complex [3] and even for people trained in the diagnosis of amoebiasis can generate false positives. This problem is further accentuated in laboratories that are not reference centers where structures such as macrophages or lymphocytes may result in false positives [19]. The main advantage of using PCR is the ability to distinguish the different species of the complex, especially where there is a high prevalence of E. dispar or E. moshkovskii, making it the method of choice to understand the epidemiology of this infection [12]. However, in endemic areas this test cannot be applied routinely by the difficulties in extracting DNA from stool samples and the cost in time and money that led into the performance of this technique.
Microscopy still has an important place in the diagnosis context because identifies a variety of intestinal parasites that cause disease and often appear in co-infections with E. histolytica, E.dispar, E. moshkovskii, as was the case in our study where we detected E. dispar and E. moshkovskii infections with other protozoan pathogens (i.e. Giardia, Blastocystis) and helminths (i.e. Ascaris lumbricoides, Trichiuris trichiura, hookworms) but with no significant association (Table 1; Table 2). Regarding the risk factors for infection with E. dispar and E. moshkovskii neither age nor gender had relationship with this infection, which shows that the main determinants for acquiring this infection are sanitation of the population [43] (Table 1; Table 2). Within the limitations of this study is the selection by volunteer participants but when compared on the basis of the records of the schools of the population, showed no significant differences among students when compared by age, gender and place of origin. One would think that in a random selection, the number of participants with gastrointestinal symptoms, and therefore the number of cases of E. histolytica be increased so that only one patient with this parasite was found in this study. While it is important to note that when different authors have selected only patients with gastrointestinal symptoms, E. dispar is more prevalent than E. histolytica [44, 45, 46] and in many cases E. histolytica infection is presented in co-infection with other intestinal parasites or bacteria such as Escherichia coli or Shigella spp that may explain gastrointestinal symptomatology [45, 46].
Conclusions
In conclusion, this study demonstrated that PCR is a useful diagnostic tool to determine the molecular epidemiology of Entamoeba species. Also, herein we report the first evidence of E. moshkovskii infection in humans from Colombia suggesting the need to pursue more studies to understand the transmission dynamics of this species and more importantly determine its pathogenic role.
Acknowledgments
We thank the children who participated in the work, the community of La Virgen and Rubén Dario Heredia, Nicolás Lemus, Nicolas Castillo and Professor Carlos Clavijo. JDR is principal professor at Universidad del Rosario.
Data Availability
All relevant data are available via GenBank (accession numbers KT825974-KT825993).
Funding Statement
This work was funded by the Research Division Headquarters Bogota (DIB) from Universidad Nacional de Colombia (code 16395).
References
- 1. WHO/PAHO/UNESCO report of a consultation with experts on amoebiasis. Epidemiol Bull. 1997; 18:13–4 [PubMed] [Google Scholar]
- 2. Stanley SL Jr. Amoebiasis. Lancet. 2003;361: 1025–1034. [DOI] [PubMed] [Google Scholar]
- 3. Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin Microbiol Rev. 2007;20: 511–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pritt BS, Clark CG. Amebiasis. Mayo Clin Proc. 2008;83: 1154–1159 10.4065/83.10.1154 [DOI] [PubMed] [Google Scholar]
- 5. Heredia RD, Fonseca JA, López MC. Entamoeba moshkovskii perspectives of a new agent to be considered in the diagnosis of amebiasis. Acta Trop. 2012;123: 139–145. 10.1016/j.actatropica.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 6. Clark CG, Diamond LS. The Laredo strain and other Entamoeba histolytica-like amoebae are Entamoeba moshkovskii . Mol Biochem Parasitol. 1991;46: 11–18. [DOI] [PubMed] [Google Scholar]
- 7. Haque R, Ali IK, Clark CG, Petri WA Jr. A case report of Entamoeba moshkovskii infection in a Bangladeshi child. Parasitol Int. 1998;47: 201–202. [Google Scholar]
- 8. Ali IK, Hossain MB, Roy S, Ayeh-Kumi PF, Petri WA Jr, Haque R, et al. Entamoeba moshkovskii infections in chil- dren, Bangladesh. Emerg Infect Dis. 2003;9: 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tanyuksel M, Ulukanligil M, Guclu Z, Araz E, Koru O, Petri W A Jr. Two cases of rarely recognized infection with Entamoeba moshkovskii . Am J Trop Med Hyg. 2007;76: 723–724. [PubMed] [Google Scholar]
- 10. Santos HL, Bandea R, Martins LA, de Macedo HW, Peralta RH, Peralta JM, et al. Differential identification of Entamoeba spp. based on the analysis of 18S rRNA. Parasitol Res. 2010. March;106(4):883–888. 10.1007/s00436-010-1728-y [DOI] [PubMed] [Google Scholar]
- 11. Parija SC, Khairnar K. Entamoeba moshkovskii and Entamoeba dispar-associated infections in Pondicherry, India. J Health Pop Nutr. 2005;23: 292–295. [PubMed] [Google Scholar]
- 12. Ximénez C, Morán P, Rojas L, Valadez A, Gómez A. Reassessment of the epidemiology of amebiasis: state of the art. Infect Genet Evol. 2009;9: 1023–1032. 10.1016/j.meegid.2009.06.008 [DOI] [PubMed] [Google Scholar]
- 13. Fontecha GA, García K, Rueda MM, Sosa-Ochoa W, Sánchez AL, Leiva B. A PCR-RFLP method for the simultaneous differentiation of three Entamoeba species. Exp Parasitol. 2015: 151–152. 10.1016/j.exppara.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 14. Sharbatkhori M, Nazemalhosseini-Mojarad E, Cheraghali F, Maghsoodloorad FS, Taherkhani H, Vakili M. Discrimination of Entamoeba Spp. in children with dysentery. Gastroenterol Hepatol Bed Bench. 2014;7(3): 164–167. [PMC free article] [PubMed] [Google Scholar]
- 15. Lau l Y, Anthony C, Fakhrurrazi SA, Ibrahim J, Ithoi I, Mahmud R. Real-time PCR assay in differentiating Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii infections in Orang Asli settlements in Malaysia. Parasites & Vectors. 2013;6:250 10.1186/1756-3305-6-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Santos HL, Bandyopadhyay K, Bandea R, Peralta RH, Peralta JM, Da Silva AJ.LUMINEX®: a new technology for the simultaneous identification of five Entamoeba spp. commonly found in human stools. Parasites & Vectors 2013, 6:69 10.1186/1756-3305-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nair G, Rebolledo M, White AC Jr, Crannell Z, Kortum-Richards, et al. Detection of Entamoeba histolytica by recombinase polymerase amplification. Am J Trop Med Hyg. 2015; in press: 15–0276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anuar TS, Al-Mekhlafi HM, Abdul Ghani MK, Abu Bakar E, Azreen SN, Salleh FM, et al. Evaluation of formalin-ether sedimentation and trichrome staining techniques: its effectiveness in detecting Entamoeba histolytica/dispar/moshkovskii in stool samples. J Microbiol Methods. 2013;92: 344–348. 10.1016/j.mimet.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 19. Leiva B, Lebbad M, Winiecka-Krusnell J, Altamirano I, Tellez A, Linder E. Overdiagnosis of Entamoeba histolytica and Entamoeba dispar in Nicaragua: a microscopic, triage parasite panel and PCR study. Arch Med Res. 2006;37: 529–34. [DOI] [PubMed] [Google Scholar]
- 20. Santos HL, Peralta RH, de Macedo HW, Barreto MG, Peralta JM. Comparison of multiplex-PCR and antigen detection for differential diagnosis of Entamoeba histolytica . Braz J Infect Dis. 2007;11: 365–370. [DOI] [PubMed] [Google Scholar]
- 21. Levecke B, Dreesen L, Barrionuevo-Samaniego M, Ortiz WB, Praet N, Brandt J, et al. Molecular differentiation of Entamoeba spp. in a rural community of Loja province, South Ecuador. Trans R Soc Trop Med Hyg. 2011;105: 737–739. 10.1016/j.trstmh.2011.08.010 [DOI] [PubMed] [Google Scholar]
- 22. Ramos F, Morán P, González E, García G, Ramiro M, Gómez A. Entamoeba histolytica and Entamoeba dispar: prevalence infection in a rural Mexican community. Exp Parasitol. 2005; 110: 327–30. [DOI] [PubMed] [Google Scholar]
- 23. Rivero Z, Bracho A, Calchi M, Díaz I, Acurero E, Maldonado A, et al. Detection and differentiation of Entamoeba histolytica and Entamoeba dispar by polymerase chain reaction in a community in Zulia State, Venezuela. Cad Saúde Pública. 2009;25: 151–159. [DOI] [PubMed] [Google Scholar]
- 24. Bracho-Mora A, Rivero de Rodríguez Z, Arraiz N, Villalobos R, Urdaneta H. Detection of Entamoeba histolytica and Entamoeba dispar by PCR in children less than five years of age with diarrhea, in Maracaibo, Venezuela. A preliminary study. Invest Clin. 2013;54(4): 373–381. [PubMed] [Google Scholar]
- 25.Porras A, Buitrago E M. Perfil Epidemiológico 2010, Municipio de Quipile, Cundinamarca. Alcaldía municipal Quipile Cundinamarca [internet] 2011. [Consultado en febrero 25 de 2014]. Disponible en: http://quipile-cundinamarca.gov.co/apc-aafiles/32663438366565323765313464653330/Perfil_epidemiologico_2010.pdf.
- 26.Sierra C. Plan de desarrollo municipio de Quipile—Cundinamarca “Unidos por el desarrollo social 2012–2015”, Quipile, Cundinamarca [internet] 2012. [consultado en febrero 25 de 2014]. Disponible en: http://quipile-cundinamarca.gov.co/apc-aa-files/37663935366463613630323063366162/plan-desarrollo-municipio-quipile-2012-2015.pdf
- 27. Katz N, Chaves A, Pellegrino J: A simple device for quantitative stool thick-smear technique in Schistosomiasis mansoni . Rev Inst Med Trop Sao Paulo 1972;14: 397–400. [PubMed] [Google Scholar]
- 28. Beck JW, García Laverde A, Hartog EM, Shaner AL. Use of the Ritchie-Frick egg counting technic in the study of the effectiveness of the anthelmintic Monopar (stilbazium iodide). Rev Fac Med Univ Nac Colomb. 1965;33: 33–36. [PubMed] [Google Scholar]
- 29. Hamzah Z, Petmitr S, Mungthin M, Leelayoova S, Chavalitshewinkoon-Petmitr P. Differential detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii by a single-round PCR assay. J Clin Microbiol. 2006;44: 3196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. López OY, López MC, Corredor V, Echeverri MC, Pinilla AE. Diferenciación del complejo Entamoeba histolytica/Entamoeba dispar mediante Gal/GalNAc-lectina y PCR en aislamientos colombianos. Rev Med Chil. 2012;140: 476–83. 10.4067/S0034-98872012000400008 [DOI] [PubMed] [Google Scholar]
- 31. Acuña-Soto R, Samuelson J, De-Girolami P, Zárate L, Millan-Velasco F, Schoolmick G. Application of the polymerase chain reaction to the epidemiology of pathogenic and non-pathogenic Entamoeba histolytica . Am J Trop Med and Hyg. 1993;48: 58–70. [DOI] [PubMed] [Google Scholar]
- 32. Tamura KD, Peterson N, Peterson G, Stecher Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 2011;28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larkin MA, Blackshields G, Brown NP, Chenna R, Mc Gettigan PA, Mc William H, et al. ClustalW and ClustalX version 2.Bioinformatics. 2007;23(21): 2947–2948. [DOI] [PubMed] [Google Scholar]
- 34. Altman DG, Machin D, Bryant TN, Gardner MJ. Eds. Statistics with Confidence. 2nd ed London: BMJ Books; 2000. [Google Scholar]
- 35. Hanna RM, Dahniya MH, Badr SS, El-Betagy A. Percutaneous catheter drainage in drug-resistant amoebic liver abscess. Trop Med Int Health. 2000;5: 578–581. [DOI] [PubMed] [Google Scholar]
- 36. Dooley CP, O'Morain CA. Recurrence of hepatic amebiasis after successful treatment with metronidazole. J Clin Gastroenterol. 1988;10: 339–342. [DOI] [PubMed] [Google Scholar]
- 37. Bansal D, Malla N, Mahajan RC. Drug resistance in amoebiasis. Indian J Med Res. 2006;123: 115–118. [PubMed] [Google Scholar]
- 38. Solaymani-Mohammadi S, Rezaian M, Babaei Z, Rajabpour A, Meamar AR, Pourbabai AA, et al. Comparison of a stool antigen detection kit and PCR for diagnosis of Entamoeba histolytica and Entamoeba dispar infections in asymptomatic cyst passers in Iran. J Clin Microbiol. 2006;44: 2258–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Samie A, Obi LC, Bessong PO, Stroup S, Houpt E, Guerrant RL. Prevalence and species distribution of E. Histolytica and E. Dispar in the Venda region, Limpopo, South Africa. Am J Trop Med Hyg. 2006;75: 565–571. [PubMed] [Google Scholar]
- 40. Stark D, van Hal S, Fotedar R, Butcher A, Marriott D, Ellis J, et al. Comparison of Stool Antigen Detection Kits to PCR for Diagnosis of Amebiasis. J Clin Microbiol. 2008;46(5): 1678–1681. 10.1128/JCM.02261-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guzmán C, López MC, Reyes P, Gómez J, Corredor A, Agudelo CA. Diferenciación de Entamoeba histolytica y Entamoeba dispar en muestras de materia fecal por detección de adhesina de E histolytica mediante ELISA. Biomédica. 2001;21: 167–171. [Google Scholar]
- 42. Shimokawa C, Kabir M, Taniuchi M, Mondal D, Kobayashi S, Ali IK, et al. Entamoeba moshkovskii is associated with diarrhea in infants and causes diarrhea and colitis in mice. J Infect Dis. 2012;206: 744–751. 10.1093/infdis/jis414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Benetton ML, Gonçalves AV, Meneghini ME, Silva EF, Carneiro M. Risk factors for infection by the Entamoeba histolytica/E. dispar complex: an epidemiological study conducted in outpatient clinics in the city of Manaus, Amazon Region, Brazil. Trans R Soc Trop Med Hyg. 2005;99: 532–40. [DOI] [PubMed] [Google Scholar]
- 44. Khairnar K, Parija SC, Palaniappan R. Diagnosis of intestinal amoebiasis by using nested polymerase chain reaction-restriction fragment length polymorphism assay. J Gastroenterol. 2007;42: 631–640. [DOI] [PubMed] [Google Scholar]
- 45. Fotedar R, Stark D, Marriott D, Ellis J, Harkness J. Entamoeba moshkovskii infections in Sydney, Australia. Eur J Clin Microbiol Infect Dis. 2008;27: 133–1337. [DOI] [PubMed] [Google Scholar]
- 46. Beltramino JC, Sosa H, Gamba N, Busquets N, Navarro L, Virgolini S, et al. Sobrediagnostico de amebiasis en niños con disentería. Arch Argent Pediatr. 2009;107: 510–514. 10.1590/S0325-00752009000600007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are available via GenBank (accession numbers KT825974-KT825993).