Abstract
Prepulse inhibition (PPI) is an example of sensorimotor gating and deficits in PPI have been demonstrated in schizophrenia patients. Phencyclidine (PCP) suppression of PPI in animals has been studied to elucidate the pathological elements of schizophrenia. However, the molecular mechanisms underlying PCP treatment or PPI in the brain are still poorly understood. In this study, quantitative phosphoproteomic analysis was performed on the prefrontal cortex from rats that were subjected to PPI after being systemically injected with PCP or saline. PCP down-regulated phosphorylation events were significantly enriched in proteins associated with long-term potentiation (LTP). Importantly, this dataset identifies functionally novel phosphorylation sites on known LTP-associated signaling molecules. In addition, mutagenesis of a significantly altered phosphorylation site on xCT (SLC7A11), the light chain of system xc-, the cystine/glutamate antiporter, suggests that PCP also regulates the activity of this protein. Finally, new insights were also derived on PPI signaling independent of PCP treatment. This is the first quantitative phosphorylation proteomic analysis providing new molecular insights into sensorimotor gating.
Introduction
Schizophrenia is a complex neuropsychiatric disorder with symptoms including hallucinations, delusions, paranoia, disorganized thoughts, social isolation, and cognitive deficits. About 1% of the US population have schizophrenia and currently there is no cure, with drugs only controlling a few specific symptoms (1). In addition to the core symptoms, schizophrenia is also associated with impairments in attentional and pre-attentional processes, such as sensorimotor gating. Sensorimotor gating is defined as the ability of a sensory event to suppress a motor response and is fundamental to cognition (2). A laboratory paradigm of sensorimotor gating is prepulse inhibition (PPI) of acoustic startle, where a non-startling “pre-stimulus” inhibits the response to a startling stimulus. Decreased PPI has been reported in humans with neuropsychiatric disorders compared to the normal population, but it is best characterized and most widely replicated in schizophrenia patients (3).
It has been hypothesized that schizophrenia results from hypo-function of N-methyl-D-aspartate glutamate receptors (NMDAR), which are ion channels permeable to Ca+2. NMDAR are abundant throughout the brain and are essential for neuronal differentiation, neuronal migration, synaptogenesis, synaptic remodeling, forms of synaptic plasticity (i.e. long-term potentiation (LTP)), and cognitive functions including learning and memory (4-7). The driving force behind the NMDA hypo-function theory of schizophrenia is that NMDAR antagonists, such as phencyclidine (PCP), can induce a psychotic state in normal humans and exacerbate symptoms in schizophrenia patients (8-10). PCP also produces PPI deficits in animals(11). Antipsychotic drugs, effective in controlling schizophrenia symptoms in humans, can reverse the PCP-induced PPI deficits in animals (12). Numerous other reports support this hypo-function theory of schizophrenia. Transgenic mice with reduced NMDAR expression display schizophrenia-like behaviors (13). The majority of genes that are associated with an increased risk for schizophrenia can influence NMDAR function or signaling (14-16). Dysregulation of NMDAR has been documented in postmortem brain tissue from schizophrenia patients(17-21). Finally, an imaging study has demonstrated a reduction of NMDAR in schizophrenia patients in vivo (22).
The PPI circuit is comprised of the limbic cortex (including prefrontal cortex [PFC] and temporal cortex), ventral striatum, ventral pallidum, and pontine tegmentum which converge on the primary startle circuit at the level of the nucleus reticularis pontis caudalis (11). It has been proposed that sub-anestheticdoses of PCP preferentially regulate specific neurons expressing gamma aminobutyric acid (GABA) receptors in the PFC, which disrupts the cortical inhibition and results in excessive glutamate activity (23). Indeed, it has been demonstrated that systemic administration of PCP does activate discrete brain regions including the PFC (24, 25), and the blockade of NMDAR can result in excessive glutamate activity in the PFC(9, 26). Nevertheless, the biochemical signaling events underlying PCP-induced behavioral alterations or PPI itself are poorly understood. A better understanding of the biochemical signaling events associated with the performance of PPI and with the effects of PCP may lead to more effective drug targets in the treatment of schizophrenia.
Besides glutamate, other neurotransmitters including GABA (26), serotonin (27), acetylcholine (28, 29) and dopamine (30) have all been reported to be involved in the regulation of PPI, but the intracellular signaling molecules are unknown. In contrast, NMDAR signaling has been extensively studied. NMDAR signaling involves numerous signal transduction pathways which are heavily regulated by phosphorylation (31, 32). Accordingly, PCP alters the activity of a wide variety of kinases, including CAMKII, ERK, PKB, and GSK (33-36). With a potentially multifaceted effect on brain signaling, an unbiased quantitation of protein phosphorylation could reveal a greater understanding of the signaling events underlying PCP-induced behavioral effects.
In this study, SILAM (Stable Isotope Labeling in Mammals) analysis was employed to identify quantitative differences in protein phosphorylation induced by PCP and PPI in the rat PFC. SILAM was chosen because it has successfully identifying protein expression changes in the habenula responsible for the depressive behavior in rats (37). In SILAM, rats are fed a specialized diet consisting of spirulina enriched with heavy nitrogen (15N) as the only protein source. This results in rats completely enriched in 15N and the 15N tissues can be employed as an internal standard for quantitative MS analysis. The PFC was chosen due to the known abnormalities in this region in schizophrenia and its role in the behavioral effects of PCP and the circuitry of PPI (11, 38-40). This phosphoproteome dataset demonstrated the complexity of phosphorylation signaling in the PFC. Phosphorylation events that were altered upon PCP administration were significantly enriched in proteins that regulate neuronal morphology. One of these phosphorylation events on the stathmin protein was previously described in postmortem schizophrenia brain tissue strengthening the validity of the PCP animal model of schizophrenia. Phosphorylation events on proteins known to be associated with LTP were down-regulated upon PCP treatment, suggesting that the dataset is a source of novel phosphorylation events that regulate LTP. One significant change induced by PCP on the light chain of the cystine/glutamate transporter, xCT, was further investigated. Mutation of the phosphorylation site demonstrated for the first time that the transporter's activity can be modulated by a post-translational modification. Finally, the entire dataset is available on an interactive website: http://sealion.scripps.edu:8080/pint/?project=04cf043e645ab98e5214396575d271ff#query. Overall, this study identifies the phosphorylation events underlying an animal model of schizophrenia and sensorimotor gating.
Experimental Procedures
PPI analysis (Drugs, Animals, behavioral testing)
A total of 16 male Sprague-Dawley rats (Harlan Laboratories, San Diego, CA), weighing 300 to 400 g, were used in these studies. Animals were housed in pairs in clear plastic cages located inside a temperature- and humidity-controlled animal colony and were maintained on a reversed day/night cycle (lights on from 7:00 P.M. to 7:00 A.M.). Food (Harlan Teklad, Madison, WI) and water were available continuously except during behavioral testing, which occurred between 9:00 A.M. and 5:00 P.M. Upon arrival in the colony, all animals were handled gently by the experimenter in order to minimize stress during behavioral testing. Animal facilities were AAALAC-approved, and protocols were in accordance with the “Guiding Principles in the Care and Use of Animals” (provided by the American Physiological Society) and the guidelines of the National Institutes of Health.
Acoustic startle testing was conducted using SR-LAB startle chambers (San Diego Instruments, San Diego, USA). Each chamber consisted of a clear Plexiglass cylinder (8.8 × 19.5 cm) mounted on a solid Perspex base situated inside the ventilated sound attenuating and well-lit (15 W) chamber (39 × 38 × 58 cm). A high frequency loudspeaker inside the chamber produced the background noise of 65 dB and acoustic stimuli 120 dB and 68, 71 and 77 dB pre-pulse stimuli. One week after arrival, all rats underwent a brief baseline startle/PPI session consisting of 120 dB PULSE-ALONE trials and PREPULSE+PULSE trials in which a 12 dB above background noise was presented 100 ms before the onset of the 120 dB pulse. These data were used to assign rats to one of 4 groups (Saline+No PPI test; Saline+PPI test; PCP+No PPI test; PCP+PPI test) matching for baseline startle and PPI. During the acoustic startle/PPI experimental session, rats were presented with five different trial types: pulse alone trials comprised of 40-ms 120dB pulse; prepulse + pulse trials in which 120 dB pulse was preceded (100 ms) by a 20-ms sub-threshold 68, 71 or 77 dB noises; and a no stimulus trial which included a background 65 dB noise. All trial types were presented in a pseudorandom order for a total of 120 trials (12 Pulse-alone trials, 12 of each prepulse+pulse trials, and 60 hidden nostim trials). Six pulse-alone trials were presented at the beginning of the session and another 6 pulse-alone trials were presented at the end of the session and used to assess startle habituation. The amount of PPI was calculated as a percentage score for each prepulse + pulse trial type: %PPI=100-{[(startle response for prepulse+pulse trial)/(startle response for Pulse-alone trial)]×100}.
On the test day, rats were injected with either saline or PCP (1.25 mg/kg; Sigma Aldrich) at a volume of 1ml/kg and placed either 5 minutes later in the startle chambers for measurement of acoustic startle response and prepulse inhibition, which lasted 21 min (PPI tested, see above; for details of methods see (41, 42)) or placed back in a holding cage for 26 min until time of dissection (PPI naïve). Following behavioral testing or the 26 min waiting period, rats (n=3/group) were lightly anesthetized with isoflurane and sacrificed by decapitation. Brains were removed and the frontal cortex was dissected, placed in dry ice-cooled isopentane for 5 sec, and stored at -80° C.
Metabolic 15N Labeling of Rat brains
Sprague-Dawley rats were labeled with 15N as previously described(43). The labeled rats at p45 were sacrificed using halothane and the brains were quickly removed and then frozen with liquid nitrogen. Eight 15N labeled whole brains were homogenized together in buffer H (4mM HepespH 7.5, 0.32M sucrose) and protease and phosphatases inhibitors (Roche, Indianapolis, IN). The 15N labeling efficiency was determined to be 95% using a previous described method(44).
Protein Digestion
After a BCA protein assay (Pierce, Rockford, IL), 500μg of prefrontal cortex from rats from the PPI paradigm were mixed with 500μg of 15N brain. The 14N/15N mixture was precipitated with methanol and chloroform. The precipitated pellet was dissolved in 100μl of 0.2% ProteaseMAX (Promega, Madison, WI) and 100μl of 8M urea. Next the sample was alkylated and reduced as previously described (45). Then, 300μl of 50mM ammonium bicarbonate, 5ul of 1% ProteaseMAX, and 20μg of sequence-grade trypsin (Promega, Madison, WI) were added. The sample was digested in a 37 °C shaking incubator for 3 hours. After the digestion, the samples were frozen at -80 °C until phosphopeptide enrichment.
Phosphopeptide Enrichment
Phosphopeptides were enriched by modifying a previously published protocol using hydroxyapatite (HAP)(46). 30mg of HAP (Bio-Rad, Hercules, CA) was dissolved in 1ml of 40%ACN, 20mM Tris, pH7.4 to make the HAP stock solution. The tryptic digest was thawed and centrifuged at 18,000 × g for 30minutes at room temperature. Acetonitrile was added to the supernatant for a final concentration of 40% and then, 100μl of the HAP stock solution was added. The sample was incubated on a rotator for 1 hour at room temperature. The sample was centrifuged at low speed to pellet the HAP. The pellet was washed three times with 1ml of 60%ACN, 20mM Tris, pH7.4. The phosphopeptides were eluted three times from the HAP with three different buffers in the following order: 20mM K2PO4, pH 7.8, 100mM K2PO4, pH 7.8, and 1M K2PO4, pH 7.8. The elutions were stored at -80°C until MudPIT analysis.
Multidimensional Protein Identification Technology (MudPIT)
A minimum of three technical replicates were analyzed for each biological sample. Each phosphopeptide elution was thawed, 5% formic acid was added, was pressure-loaded onto a fused silica capillary desalting column containing 2cm of 10 μm Jupiter material (Phenomenex, Ventura, CA) followed by 2cm of 3 cm 5-μm Partisphere strong cation exchanger (SCX) (Whatman, Clifton, NJ) into a 250-μm i.d capillary fritted with immobilized Kasil 1624 (PQ Corperation, Valley forge, PA) following the SCX. After the sample was loaded, the desalting column was washed with buffer containing 95% water, 5% acetonitrile, and 0.1% formic acid. Then, a 100-μm i.d capillary with a 5-μm pulled tip packed with 15 cm 4-μm Jupiter material (Phenomenex, Ventura, CA) was attached to a ZDV union and the entire split-column (desalting column–union–analytical column) was placed inline with an Agligent 1200 pump (Agilent Technologies, Santa Clara, CA) and analyzed using a modified 6-step separation described previously(47). As peptides eluted from the microcapillary column, they were electrosprayed directly into an Velos mass spectrometer (ThermoFinnigan, Palo Alto, CA) programmed as previously described (48).
Interpretation of Tandem mass spectra
Data from technical replicates were combined prior to database searching. Both MS1 and MS2 (tandem mass spectra) were extracted from the XCalibur data system format (.RAW) into MS1 and MS2 formats(49) using in house software (RAW_Xtractor). At this point, the MS1 and MS2 files for duplicate analyses were combined. Tandem mass spectra were interpreted by SEQUEST(50), which was parallelized on a Beowulf cluster of 100 computers(51) and results were filtered, sorted, and displayed using the DTA Select 2 program using a decoy database strategy(52). For each MudPIT analysis, the protein false positive rate was < 1%. Specifically, the distribution of SEQUEST values (i.e. Xcorr, DeltaCN, and DeltaMass) for (a) direct and (b) decoy database hits were obtained, and the two subsets were separated by quadratic discriminant analysis. Outlier points in the two distributions (for example, matches with very low Xcorr but very high DeltCN) were discarded. Full separation of the direct and decoy subsets is not generally possible; therefore, the discriminant score was set such that a false positive rate of 5% was determined based on the number of accepted decoy database peptides. In addition, a minimum sequence length of 7 amino acid residues was required, and each protein on the list was supported by at least two peptide identifications. These additional requirements - especially the latter - resulted in the elimination of most decoy database and false positive hits, as these tended to be overwhelmingly present as proteins identified by single peptide matches. After this last filtering step, the false identification rate was reduced to below 1%. Searches were performed against the rat International Protein Index (IPI) database (IPI, v3.05).
Quantitative Analysis using Census
After filtering the results from SEQUEST using DTASelect2, ion chromatograms were generated using an updated version of a program previously written in our lab (53). This software, called Census(54), is available from the authors for individual use and evaluation through an Institutional Software Transfer Agreement (see http://fields.scripps.edu/census for details).
First, the elemental compositions and corresponding isotopic distributions for both the unlabeled and labeled peptides were calculated and this information was then used to determine the appropriate m/z range from which to extract ion intensities, which included all isotopes with greater than 5% of the calculated isotope cluster base peak abundance. MS1 files were used to generate chromatograms from the m/z range surrounding both the unlabeled and labeled precursor peptides.
Census calculates peptide ion intensity ratios for each pair of extracted ion chromatograms. The heart of the program is a linear least squares correlation that is used to calculate the ratio (i.e., slope of the line) and closeness of fit (i.e., correlation coefficient (r)) between the data points of the unlabeled and labeled ion chromatograms. Census allows users to filter peptide ratio measurements based on a correlation threshold because the correlation coefficient (values between zero and one) represents the quality of the correlation between the unlabeled and labeled chromatograms, and can be used to filter out poor quality measurements. In this study, only peptide ratios with correlation values greater than 0.5 were used for further analysis. For normalization, the median of the natural log of the ratios from each Census analysis was calculated, and then, all the medians were shifted to zero(55). The calculated medians were very similar, and it was assumed any shift in the median was due to human error in the protein mixing.
Bioinformatic and Statistical Analysis
A-score was employed to assign a confidence level to the accuracy of site localization of the phosphorylated moiety (56). Ingenuity software was employed to determine the enrichment of protein function and pathways with a particular dataset (57). The p-value measures how likely the observed association between a specific function or disease and our dataset would be if it was only due to random chance. The two major factors in this calculation are the number of proteins annotated to specific function or pathway from our dataset and the total number of proteins annotated to the function or disease in the Ingenuity Knowledge Base. For determining the functional differences and networks between brain regions, the input was the statistically significant changes between brain regions.
Glutamate Uptake Assay
HT22 cells were grown at 37 °C in a 10% CO2 atmosphere in DMEM containing 10%FCS with 100 IU/ml penicillin and 100 μg/ml streptomycin. Mouse xCT cDNA was tagged with EGFP at the C-terminus in the GW1 vector. The serine 26 (S26) was mutated to aspartic acid (D) or alanine (A) by Eton Biosciences (San Diego, CA). Each construct was transfected into HT22 cells using Lipofectamine 2000 (Life Technologies) on 500 000 cells in a 60mm dish plated the day before. The next day the cells were harvested and plated in 24 well plates with 50 000 cells per well with each construct plated in 6 wells. The glutamate uptake assay was performed the next day. For each construct, 10 μM 3H-glu (Perkin Elmer NEN) was added to 3 wells and 3H-glu + 1mM HCA, an inhibitor of system xc-, was added to the other 3 wells for 20min at 37 °C. After washing the cells with ice cold HBSS, 400 μl of 0.2N NaOH was added to each well and the cells were incubated overnight at 37°C. 3H-glu radioactivity from each well was detected with a scintillation counter and the Bradford protein assay was also performed on the cells from each well. The radioactivity (i.e. cpm) was normalized to protein and the HCA-insensitive uptake was subtracted from total uptake for the specific system xc- activity.
Results
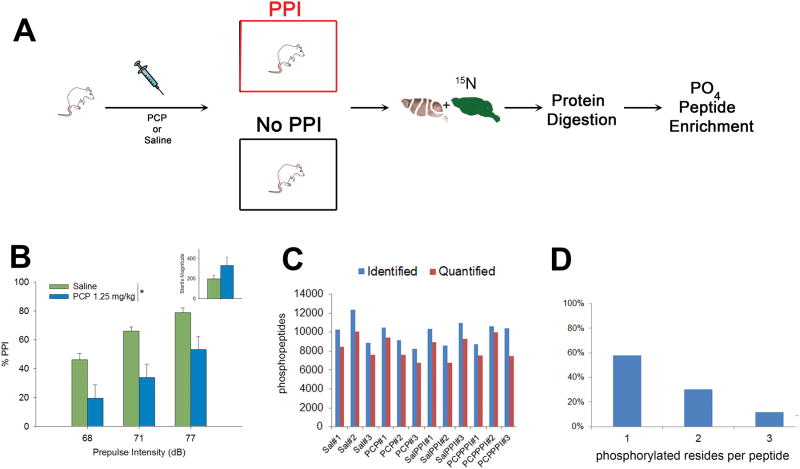
The experimental design for the study is outlined in Figure 1A. Prior to PPI testing, rats were either administered saline or PCP and half of the rats under went behavioral testing while the other half were left undisturbed for the length of the PPI session (26 minutes total). Brains were removed 26 minutes after the injection. A low PCP dose (1.25mg/kg) produced a significant decreased in PPI as previously reported (Figure 1B;(41)). In total, three rats were analyzed in each condition: Saline (Sal), PCP, Saline + PPI (SalPPI), and PCP + PPI (PCPPPI). The PFC was mixed 1:1 (wt:wt) with 15N brain homogenate, which serves as an internal standard to quantify between the biological conditions (43). The 14N/15N mixtures were digested, then enriched for phosphopeptides using hydroxyapatite (HAP) (46) prior to MS analysis. The advantage of SILAM compared to other MS quantitative techniques is that the internal standard is added prior to any sample processing and thus, controls for systematic errors in sample processing (i.e digestion and phosphopeptide enrichment). The identical 14N and 15N phosphopeptides behave the same during the peptide separation and co-elute into the mass spectrometer. All the abundances of 14N and 15N paired peptides are extracted from the MS data and a 14N/15N ratio is calculated by the algorithm Census (58, 59). Since the same 15N material was added to all conditions, a ratio-over-ratio analysis is used and the differences between the biological conditions can be quantitated. Overall, 118995 phosphopeptides were identified with a peptide FDR < 1% and 99810 phosphopeptides were confidently quantified (Figure 1C). This represents 10173 unique phosphopeptides from 1672 unique genes. Fifty-eight percent of the quantified peptides were singly phosphorylated (Figure 1D).
Figure 1.
A) Schematic of the experimental design. Unlabeled 14N rats were injected with saline or PCP then either underwent PPI analysis or mock PPI analysis. The brains of the 14N rats were dissected then mixed with 15N rat brain. The 14N/15N mixtures were digested and then phosphopeptides were enriched for analysis. B) PCP (1.25 mg/kg) significantly disrupted PPI and non-significantly increased startle magnitude (inset). * main effect of drug on PPI, [F(1,12)=14.6, p<0.01].C) The total number of phosphopeptides (y-axis) identified and quantified. X-axis represents each rat in the study. D) The percentage of phosphorylation sites per quantified peptide.
PFC phosphoproteome
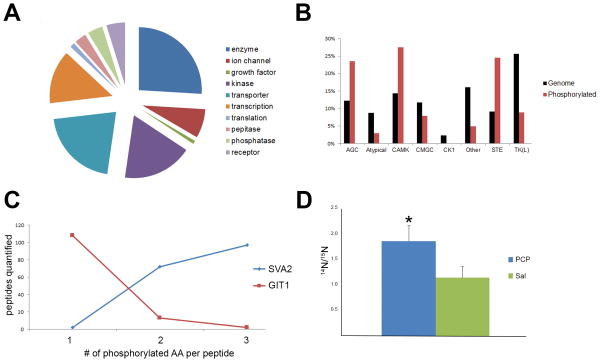
The molecular functions of the quantified phosphoproteins were distributed widely among known protein functions (Figure 2A). Kinases represented 18% of the phosphoproteins quantified demonstrating the vital function phosphorylation plays in kinase regulation. Kinases from all the known families were represented except the Ck1 family, which only represents 2% of known kinases (Figure 2B). The role of phosphorylation in kinase regulation has been most extensively studied in the AGC family (60). This family is named after its founding members: PKA, PKC, and PKG. Phosphorylation of three conserved motifs are required for full kinase activation: the activation loop in the catalytic kinase domain, the hydrophobic motif outside the kinase domain, and the turn motif in the c-terminal tail (60). Out of the 18 phosphorylated AGC kinases identified in this study, 12 kinases were quantified with these conserved phosphorylation events, suggesting a high level of AGC activity (Supplementary Table 1). Interestingly, 29 other phosphorylation events were quantified and only one of these has a reported function. The abundance of phosphorylation events on kinases indicates a tremendous amount of cross-talk between kinase signaling pathways in the brain.
Figure 2.
A) The distribution of the functions of the quantified phosphoproteins. B) Distribution of kinases. The red bars represents the percentage of kinases family members quantified by phosphopeptides and the black bars represent the percentage of each genes in each kinase family of the known 514 kinases. For example, AGC family represents 12% of all the known kinases but represents 24% of all the kinases quantified by phosphopeptides in this study. C) Two examples of multistate phosphorylation in the dataset. The peptide NQSDLDDQHDYDSVASDEDTDQEPLPSAGATR from GIT1 (red) was quantified over 100 times with one phosphorylation event, but much less with two or three phosphorylation events. The peptide GEGAQDEEEGGASSDATEGHDEDDEIYEGEYQGIPR from SVA2 (blue) was rarely quantified with one phosphorylation event, but more often was quantified with two and three phosphorylation events. D) The peptide ASGQAFELILpSPR from stathmin was significantly increased upon PCP treatment. * p value < 0.05
Multiple phosphorylation states were quantified for 825 unique peptide sequences. For example, an N-terminal triply phosphorylated peptide from synaptic vesicle glycoprotein 2A (SV2a) was identified more frequently than the same peptide doubly or singly phosphorylated (Figure 2C and Supplementary Table 2). Phosphorylation of the SV2a N-terminus has been demonstrated to be required for its interaction with synaptotagmin, but the exact sites are unknown (61). A peptide from the G protein-coupled receptor kinase-interactor 1(GIT1) exhibited the opposite trend (Figure 2C). GIT1 is involved in synaptic formation and these observed phosphorylation sites are localized to the region responsible for its synaptic localization(62) The phenomenon of multistate phosphorylation has previously been described as bistability or convergence(63, 64). Our dataset suggests that multiple phosphorylation is also a prominent signaling mechanism in the PFC.
PCP Enrichment Analysis
All the quantified phosphopeptides with a fold-change greater that than 1.5 with a RSD < 50% were analyzed to determine if there was significant enrichment in any protein function or signaling pathway. Comparing the Sal and PCP datasets, there were 232 unique phosphopeptides up-regulated upon PCP treatment and 257 phosphopeptides down-regulated upon PCP treatment (Supplementary Table 3). Proteins annotated to “regulation and organization of the plasma projections” were significantly enriched in both up-regulated (p < 5.83 × 10-9) and down-regulated (p < 2.48 × 10-17) phosphoproteins. Comparing the SalPPI and PCPPPI datasets, there were 2843 unique phosphopeptides in common comprising 943 phosphoproteins. There were 142 phosphopeptides up-regulated in the PCPPPI group and 93 phosphopeptides down-regulated in the PCPPPI group (Supplementary Table 4). The most significantly enriched function (9.3 × 10-13) of the phosphoproteins up-regulated in the PCPPPI group was also the regulation and organization of plasma membrane projections but in the down-regulated dataset, synaptic transmission was the most significantly (2.1 × 10-8) enriched protein function. The regulation and organization of plasma projections includes dendrites, dendritic spines, and axons and relies heavily on cytoskeleton proteins. Four phosphoproteins (CAMK2A, MAG, SYNGAP1, and PPP19RA) in this functional class were up-regulated in both PCP and PCPPPI. Seven phosphoproteins (BSN, MAP1B, MAP2, MAPT, NEFM, NEFH, PRKCE) were up-regulated in PCP and PCPPPI and down-regulated in Sal, but with different phosphopeptides indicating the complexity of cytoskeleton phosphorylation. Changes in neuronal architecture are an important aspect of synaptic plasticity and an altered cytoskeleton has been implicated in numerous cognitive disorders (65). One phosphorylation site (S25) of the cytoskeletal regulating protein, stathmin-1, has previously been analyzed in schizophrenic postmortem brains. The phosphorylation of S25 was found significantly increased in the schizophrenia postmortem PFC (66). The data set presented here showed a significant increase ofpS25 upon PCP treatment (Figure 2D). Overall, this functional analysis suggests a complex phosphorylation signaling network of cytoskeleton proteins underlies the effect of PCP and PPI may alter this signaling network independently of PCP.
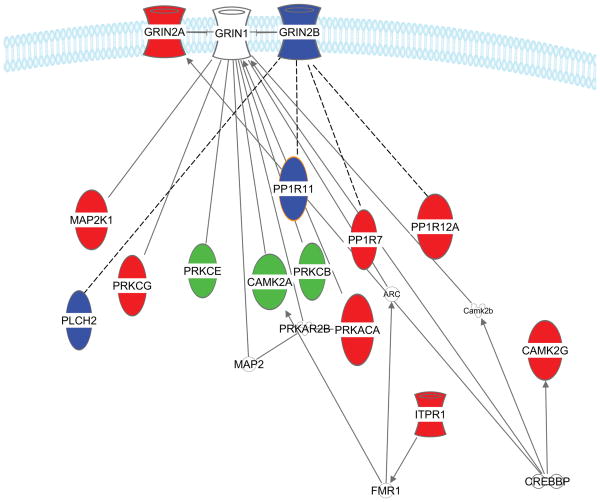
A test for enrichment of specific signal transduction pathways showed there was a difference between the PCP up-regulated and down-regulated datasets. Phosphoproteins annotated to LTP signaling (p value 2.9 ×10-8) was the most significantly enriched pathway in the down-regulated PCP dataset and also significantly down-regulated in the PCPPPI dataset (p value 8.06 × 10-6) when compared to Sal and SalPPI datasets, respectively (Figure 3). Although there was an overlap between phosphoproteins, there was no overlap in exact phosphopeptides. There was also an overlap in protein families. For example, PPP1R11 (protein phosphatase 1, regulatory (inhibitor) subunit 11) was down-regulated in PCPPPI and PPP1R12A (Protein phosphatase 1 regulatory subunit 12A) was down-regulated in PCP. Both of these proteins directly interact with protein phosphatase 1 (PP1 family). The PP1 family is also involved in LTP and inhibition of PP1 enhances learning in rats(67, 68).
Figure 3.
Phosphoproteins down-regulated in PCP and PCPPPI compared to Sal and SalPPI, respectively, which were significantly enriched in LTP signaling pathway. The blue phosphoproteins were down-regulated in PCPPPI, the red phosphoproteins were down-regulated in PCP, and the green phosphoproteins were down-regulated in both PCP and PCPPPI rats. Uncolored proteins were not quantified in our study. The solid lines represent direct relationship have been reported and the dashed lines represent relationships inferred from the literature based on the relationships of proteins from the same family.
Under-studied proteins
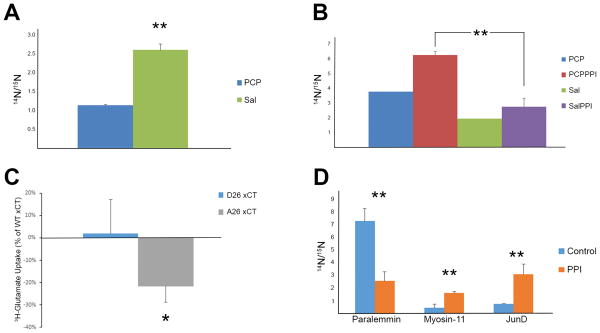
One caveat of these enrichment analyses is that they are biased to proteins or pathways that are heavily reported in the literature. In other words, our dataset may contain novel proteins that are involved in LTP signaling but overlooked by these analyses. Alternatively, it is possible only one protein of a particular pathway is in the dataset. This pathway would not be enriched, but still could be important to PCP signaling. For this reason, statistical analysis (i.e. t-test) was performed to determine statistically significant phosphopeptides regardless of their function. One such phosphoprotein is a brain-enriched guanylate kinase-associated protein (Begain). This brain specific protein was discovered in the yeast-two hybrid screen using PSD-95 as bait (69). PSD-95 is the core scaffolding protein of the NMDAR complex and facilitates glutamate signal transduction at the post-synapse. It has been reported that NMDAR antagonists can prevent the localization of Begain to the synapse (70). The function of this protein and the biochemical mechanism underlying this altered localization are unknown. The phosphorylation of S494 was significantly decreased after PCP treatment (Figure 4A). With a similar phosphorylation pattern as other known LTP signaling proteins, it is possible that S494 phosphorylation plays a role in LTP.
Figure 4.
A) The peptide ADSpFSEGDDLSQGHLAEPCFLR of Begain was significantly down-regulated after PCP treatment. B) The peptide LPpSVGDQEPPGHEK of xCT was significantly up-regulated after PCPPPI. The same trend was trend observed for the PCP and Sal comparison but not enough measurements were collected for statistical analysis. C) Abolishing the phosphorylation site in B) decreases 3H-glutamate uptake through xCT in HT22 cells. Cells were transfected with WT, D26, or A26 xCT cDNA and glutamate uptake assays was performed. The data represents three independent experiments. The y-axis is the 3H-glutamate measurements of D26 and A26 as a percentage of the WT xCT glutamate uptake measurements. D) Phosphopeptides significantly altered in the PFC between rats that underwent PPI or mock PPI (control). Paralemmin (SETMVNAQQpTPLGpTPK), myosin-11(VIENTDGpSEEEMDAR), JunD (LAALKDEPQTVPDVPSFGDpSPPLpSPIDMDTQER). * p value < 0.05, ** p value< 0.01.
Another phosphoprotein that eluded the enrichment analysis was solute carrier family 7 member 11 (SLC7A11 or xCT), which is the rate limiting subunit of the cystine/glutamate antiporter, system xc- (71). Phosphorylation of S26 was significantly up-regulated in PCPPPI compared with SalPPI (Figure 4B). To determine if phosphorylation at this site is able to alter the activity of xCT, cDNAs with mutations in residue 26 were generated. The xCT cDNAs (wild-type (S26), mutation of serine to aspartate (D26), or mutation of serine to alanine (A26)) were transfected into the mouse hippocampal cell line HT22. The A26 mutant, which does not contain the phosphorylation site, showed significantly decreased glutamate uptake as compared to mimicking phosphorylation with the D26mutant (Figure 4C). The alanine mutant, however, still increased glutamate transport above mock transfected cells (data not shown). Changes in system xc- activity have been previously linked to transcriptional control (71, 72), but this is the first direct evidence that phosphorylation can alter the activity of xCT.
It is interesting that phosphoproteins involved in synaptic transmission were significantly enriched when SalPPI was compared to PCPPI but this was not observed comparing Sal and PCP datasets. This suggests that the PPI paradigm could be altering the phosphoproteome independent of PCP, so the PPI dataset was analyzed for altered phosphopeptides from the non-PPI dataset. Comparing the PPI and No-PPI datasets, there were 4072 unique phosphopeptides in common compromising 1188 phosphoproteins. There were 236 phosphopeptides up-regulated in PPI and 206 phosphopeptides up-regulated in no PPI (Supplementary Table 5). There was no significant enrichment in any well-characterized protein function or signaling pathway. Nevertheless, there were large, significant changes between PPI and No-PPI datasets (Figure 4D).
Discussion
The dataset presented here demonstrates the extraordinary complexity of the rat PFC phosphoproteome. There are 514 human kinase genes that have been classified into 8 distinct families based on sequence homology (73), and phosphorylation events were identified in seven families in this study. Phosphorylated CAMK, AGC, and STE kinases were enriched in the brain compared to the overall percentage of these kinases in the whole genome. The CAMK family has previously been reported to be enriched in the brain (74). Alternatively, this enrichment could indicate that these kinases are more active in the brain since many phosphorylation events are linked to kinase activity. One of the biggest obstacles in phosphoproteomics is interpreting the data, since the function of the majority of phosphorylation events are unknown. Even within the well-studied AGC family, the majority of the phosphorylation events have no known function, suggesting many additional layers of kinase regulation exist in the brain beyond what is already described in the literature. While the known AGC phosphorylation events are necessary for a fully active protein conformation, other events may dictate localization and protein interactions to impart substrate specificity in the complex brain milieu. One signaling mechanism that also may aid in signaling specificity is bistability or convergence, as demonstrated by unique peptide sequences identified with one, two and three phosphorylation events. It has been postulated that these multiple phosphorylations do not exhibit a linear relationship with regard to protein activity, but convert graded cell signals into ultrasensitive switches in response to small changes within a cell to produce a binary response. The necessity for this process is born from multiple signaling pathways converging on the same signaling molecule, producing negative and positive feedback loops. The requirement by AGC kinases for multiple phosphorylation events for full activity would fall into this category of a graded response. At the cellular level, this phenomenon has been demonstrated to be necessary for cell cycle progression (75). Although this phenomenon has been demonstrated at the protein level, this dataset demonstrates it at the peptide level with residues in close proximity. A previous report demonstrated the HAP phosphopeptide enrichment strategy doubled the percentage of multiple phosphorylated peptides identified compared to gallium metal affinity enrichment in cell culture (46). More than 40% of the quantified peptides in this study had multiple phosphorylation events, which is three times as many as were observed in a recent brain phosphoproteome report that employed titanium oxide (76). Thus, HAP phosphopeptide enrichment strategy should allow further investigation of this signaling mechanism.
The idea of using both phosphorylated and identical unmodified peptide in quantitative phosphoproteome analysis has been reported for accurate interpretation of a dataset (77). For example, a large difference in a phosphorylation site identified in a wild-type and transgenic knockout mouse brain may solely be due to a difference in protein expression while the extent of protein phosphorylation between the tissues could be identical. There is an inherent technical difficulty, however, in attempting to use unmodified peptides in the analysis. First, phosphopeptides are undetectable without phosphorylation enrichments, which strive to remove unmodified proteins. In our dataset, the unmodified peptides that were identified overlapped with less than 30% of the modified peptides. Thus, two separate MS studies are required: phosphorylation analysis and whole proteome analysis. Phosphopeptide enrichment strategies, however, enrich for low abundant proteins that are undetectable to whole proteome analysis even with extensive fractionation due to limited dynamic range of the MS instruments. For example, comparison of a comprehensive tissue phosphorylation and whole proteome analyses resulted in an peptide overlap of only 36% (78). This potential problem, however, may not apply or have minimal effect on our dataset because the biological question analyzed in this report is mechanistically unique compared to previously published quantitative phosphorylation proteome reports. In this study, differences were examined between identical brain regions induced by a drug treatment and behavior test which lasted for less than 30 minutes. This time frame is too short for changes in translation and the earliest changes, i.e. immediate early genes, are only detected after one hour (79). Supporting this idea, no changes in unmodified proteins were detected in primary cultured neurons after 15 min of PCP, but changes in phosphorylated proteins were detected in this short time frame (80). The fact that these behavioral changes do manifest so quickly after a PCP injection strongly suggest the role of post-translational modifications and this was the impetus for this study.
This study represents the first quantitative phosphoproteomic analysis of PCP and PPI in the rodent brain. PCP-induced PPI deficits are widely used to produce a schizophrenia-like phenotype in rats to investigate the pathophysiology of schizophrenia and develop novel drug targets(81). There have been, however, other phosphoproteomic analyses on the effects of PCP. One study reported quantitative changes in the phosphoproteome induced by PCP in primary cortical neuronal culture (80). Another reported quantitative phosphoproteome changes in the rat frontal cortex using a high PCP concentration (i.e. ten times higher used in this study) that produces ataxia (82) and neurotoxicity (76). Although both are informative studies on PCP signaling in neurons, neither study uses an experimental preparation with as much relevance to an animal model of schizophrenia as that employed in our study. For example, it was demonstrated that the brain phosphoproteome was distinct from the phosphoproteome of primary cultured neurons(80). We chose a low dose of PCP (1.25 mg/kg) because it has minimal effects on motor activity (83, 84) but does reduce PPI in rats, providing a more relevant model for schizophrenia. PPI was employed to verify that the low dose PCP had a behavioral effect relevant to schizophrenia. Consistent with other evidence supporting PCP as a useful strategy to understand the complexities of schizophrenia, PCP reproduced a phosphorylation change in the stathmin protein previously reported in schizophrenic postmortem brains(66). The stathmin protein controls microtubule dynamics through its direct interaction with tubulin. When bound to tubulin, it prevents microtubule formation and promotes disassembly, and upon phosphorylation, it releases tubulin allowing microtubule formation(85). PCP treatment increased phosphorylation on serine25 (pS25) on the stathmin protein in the PFC in this study. A previous report observed an increase in pS25 in the PFC of schizophrenia patients compared to controls but the change was not observed in the brains of people with Alzheimer's disease, indicating some specificity of the effects to schizophrenia. Although the function of pS25 is not understood, data suggest it is phosphorylated by members of the MAPK family, which is crucial to LTP (86). Interestingly, more recent studies have reported abnormal microtubule organization in cultured olfactory epithelium neurons from schizophrenia patients (87, 88). Additionally, stathmin knockout (KO) mice develop normal brains, including normal neuronal morphology, despite abnormal microtubule organization (89). Stathmin KO mice showed no changes in basic synaptic transmission, but did display deficient LTP. Behaviorally, stathmin KO mice have memory deficits in a conditioned fear paradigm, display disrupted maternal care and abnormal adult social interactions, and memory extinction (90-92). Thus, our study demonstrates the acute effect of PCP on the stathmin protein and provides additional evidence that stathmin plays a role in complex cognitive behaviors.
Using protein function enrichment analysis, phosphopeptides that were altered upon PCP treatment were significantly enriched in the regulation and organization of neuronal morphology. Phosphoproteins were both up and down regulated after PCP treatment in this functional category, which reiterates the intricate regulation of cytoskeletal phosphorylation previously observed. For example, there are 238, 174, and 84 phosphorylation sites reported for microtubule-associated protein 1B (MAP1B), MAP2, and neurofilament-H, respectively (93). To date, this remarkable “phospho-code” of cytoskeleton proteins remains a mystery. In the brain, cytoskeleton proteins define the morphology of the neurons. One aspect of dynamic neuronal morphology that has been implicated in schizophrenia is dendritic spines. Dendritic spines protrude from dendrites and often form excitatory synapses which may have an important role in regulating the excitability of a neuron. The loss of dendritic spines has been correlated with cognitive deficits(94). Multiple independent studies have reported a decrease in spine density in postmortem schizophrenia brains (95). In the PFC, chronic PCP administration for seven days resulted in a decrease of spines(96), but it was also reported that one month of PCP administration produced an increase in spine density(97). Thus, it appears PCP does regulate spine density but different laboratory paradigms affect the outcome. Our data demonstrates that acute administration of PCP alters cytoskeletal phosphoproteome, which may be the underlying mechanism of its regulation of dendritic spines.
Pathway analysis revealed that phosphopeptides that were down-regulated in response to PCP were significantly enriched to the LTP signaling pathway. For example, four phosphopeptides of three PKC family members were down-regulated upon PCP treatment. Protein kinase C (PKC) has been demonstrated to regulate LTP and activation of PKC can even rescue LTP from the blockade of NMDAR (98, 99). The phosphorylation of T500 of PKC-beta is located in its kinase catalytic region and is critical for its activity, suggesting this kinase is less active after PCP treatment (100). A decrease in activity was observed in dual specificity mitogen-activated protein kinase 1(MEK1) after phosphorylation on T386. This regulatory site is phosphorylated by extracellular signal-regulated kinase-2 (ERK2) (101, 102). When the site is mutated to block phosphorylation, the unphosphorylated MEK1 is more active than wild-type. The MEK/ERK pathway is essential to LTP and a decrease in ERK phosphorylation and activity have been reported after NMDAR blockade (36, 103). Therefore, the decrease in phosphorylation of T386 in this dataset suggests ERK2 is less active after PCP treatment. A decrease of phosphorylation at S1756 was observed for inositol 1,4,5-trisphosphate receptor (ITPR1), which is an ER calcium channel. This site has been reported to increase the activity of the receptor exporting calcium to the cytoplasm, modulating LTP (104, 105)(106, 107). Although many other phosphoproteins are well-known proteins involved in LTP, the majority of phosphorylation sites quantified do not have a known biological function. The phosphorylation sites with known biological consequences suggest that PCP decreases the activity of these proteins that would reduce or prevent LTP, consistent with the literature (108).
Enrichment analyses are routinely applied to proteomic datasets and can provide important insights to identify biological processes modulated by novel experimental conditions. The decrease in phosphorylation events in response to PCP on known LTP-associated proteins might be expected, but it is a crucial validation for the dataset and identifies novel phosphorylation sites that might play a role in regulating LTP. On the other hand, these analyses rely on proteins and signaling pathways that have been heavily studied. This can be counterintuitive when performing a global unbiased proteomic analysis to discover the involvement of novel proteins in known signaling pathways. However, statistically significant phosphopeptide changes induced by PCP may contain previously uncharacterized LTP signaling molecules. For example, begain protein, whose function is unknown, was decreased in a similar pattern as known LTP molecules and has previously been implicated in NMDAR signaling. Another protein that escaped enrichment analysis but had a significant increase in phosphorylation after PCP was xCT. xCT is the rate limiting subunit of the sodium independent cysteine/glutamate antiporter, system xc-. In the brain, it has been demonstrated that inhibition of this antiporter reduces extracellular levels of glutamate (109, 110). Mutagenesis of the S26 phosphorylation site to A26 shows that this mutant is less active than the wild-type or the D26 mutant suggesting that PCP should increase extracellular glutamate through system xc-. It has been reported that PCP increases extracellular glutamate in the PFC, and group II mGluR agonists can reduce this elevation in extracellular glutamate along with PCP-induced behavioral deficits (30). The molecular mechanism of the PCP induced glutamate release is unknown. Thus, PCP increasing the phosphorylation of xCT couldresult in an increase in extracellular glutamate. Consistent with this theory, xCT expression has been reported to be increased in schizophrenia brains compared to control brains (111). Alternatively, changes in the the phosphorylation of xCT could be a compensatory mechanism to PCP induced extracellular glutamate. Interestingly, Baker et al. reported that manipulation of xCT could block behavioral deficits induced by PCP (111). Thus, the fact that our study and Baker et al. have both implicated system xc- in PCP signaling strongly suggests that further investigation is needed to determine the exact molecular details of the regulation of system xc- function in the brain.
Although PPI was initially employed to verify the effects of PCP, statistical changes in phosphoproteins were observed between PPI-tested and PPI-non tested rats. PPI is an example of sensorimotor gating. Although many drugs have been reported to modulate PPI and relevant circuitry has been dissected through lesion and pharmacological studies, to our knowledge this is the first study to investigate the signaling which underlies sensorimotor gating. The phosphopeptide, SETMVNAQQpTPLGpTPK, of the paralemmin protein was significantly down-regulated greater than 2.5 fold after exposure to the PPI paradigm. Paralemmin controls dendritic spine dynamics, which regulates plasticity and cognition (112). It was also demonstrated that neuronal activity alters its plasma membrane localization, but the molecular mechanism is unknown. We also observed a four-fold increase in the actin-dependent motor protein myosin-11(VIENTDGpSEEEMDAR) after PPI. Although this particular myosin has not been studied in the brain, other myosin proteins have been demonstrated to regulate higher brain function(113). Finally, we observed a greater than 3.5 fold statistical increase in the phosphopeptide, (LAALKDEPQTVPDVPSFGDpSPPLpSPIDMDTQER) of JunD, which is a component of the AP-1 transcription factor family. AP-1 is a large group of dimer complexes composed of Jun, Fos, or ATF DNA binding proteins. Although the biological significance of these quantitated phosphorylation sites are unknown, phosphorylation has been demonstrated to be crucial to all Jun proteins, regulating their expression, activity, and interactions (114). For example, stimulation of glutamate receptors has been reported to recruit JunD to the AP-1 complex, and enhanced JunD interaction with ERK has been demonstrated to be necessary for the learning process of contextual fear extinction (115, 116). These data suggest that these phosphoproteins are involved in the processing of motor and sensory information in the PFC. Since these studies were not designed to dissect differences in phosphoproteins associated specifically with PPI vs startle response, we cannot rule out the possibility that some of these changes in phosphoprotiens in the PFC were due to exposure to the startling stimuli (117).
Conclusions
The known protein signaling molecules modulated by PCP to generate a schizophrenia phenotype are scarce but are invaluable to the field to provide potential drug targets for this devastating disorder. The dataset presented here is a leap forward for this field to begin the exploration of the involvement of these novel signal transduction molecules (http://sealion.scripps.edu:8080/pint/?project=04cf043e645ab98e5214396575d271ff#query). We highlighted specific proteins that have been previously implicated in this model or schizophrenia to increase confidence in this novel dataset. Although this is a big step, there are more to follow. First, these changes have been localized to the PFC, but cell type localization would provide more information about the functional consequences. Second, the ability of phosphorylation to modulate a protein's activity, localization or interactions complicates the functional analysis of this post-translational modification. There are over 200 000 unique phosphorylation sites in the human proteome, but less than 10% have a known biological function (93). For example, we demonstrated for the first time that a phosphorylation modulated by PCP on the xCT protein alters its activity. One method to assign function is to quantitate phosphorylation levels and compare them in different biological conditions. In this study, the sensitivity of quantitative proteomic technology was challenged to identify changes in the PFC induced a subtle but reproducible behavioral alteration triggered by a low dose of PCP. With a time scale of less the 30 minutes, it was postulated changes in phosphorylation is essential for understanding the biological underpinnings of this behavioral paradigm as protein and gene expression changes requires hours. A subset of phosphorylation events in the vast brain phosphoproteome were altered in the PFC after modulating a key regulator of LTP, the NMDAR. With many of these phosphorylation sites on known LTP-associated signaling proteins, this dataset also provides great potential to promote the further understanding of the complex and crucial LTP signaling pathway. Finally, this is the first study to identify potential phosphorylation events underlying sensory and motor processing in the PFC.
Supplementary Material
Supplementary Table 1: Quantified phosphorylation sites from the AGC kinase family. The phosphorylated amino acids are list for the known site required for activation (i.e. “activation” loop, “hydrophobic” motif, “turn” motif), other phosphorylation sites with known function, and phosphorylation sites with no known function. Sites highlighted in red possess and A-score of 18 or greater.
Supplementary Table 2: Multi-state phosphorylated peptides. Unique peptide sequences that were observed to have at least two different phosphorylation states.
Supplementary Table 3-5. Altered phosphorylation peptides. Phosphopeptides are listed that were observed to be altered. Each phosphopeptide is listed with condition specific 14N/15N average ratio, localization score (Ascore > 13 = high), enrichment annotation, IPI identifier, and protein name. Table 3 = Sal vs. PCP, Table 4 = SalPPI vs PCPPPI, Table 5 = No PPI vs. PPI. This data with more detailed protein information is also available at http://sealion.scripps.edu:8080/pint/?project=04cf043e645ab98e5214396575d271ff#query
Acknowledgments
Funding has been provided by National Institutes of Health grants P41 GM103533, R01 MH067880, and R01 DA002925.
Footnotes
Conflict of Interest: Authors declare no competing financial interests in relation to the work described.
References
- 1.Regier DA, Narrow WE, Rae DS, Manderscheid RW, Locke BZ, Goodwin FK. The de facto US mental and addictive disorders service system. Epidemiologic catchment area prospective 1-year prevalence rates of disorders and services. Arch Gen Psychiatry. 1993;50(2):85–94. doi: 10.1001/archpsyc.1993.01820140007001. [DOI] [PubMed] [Google Scholar]
- 2.Perry W, Braff DL. Information-processing deficits and thought disorder in schizophrenia. Am J Psychiatry. 1994;151(3):363–7. doi: 10.1176/ajp.151.3.363. [DOI] [PubMed] [Google Scholar]
- 3.Powell SB, Zhou X, Geyer MA. Prepulse inhibition and genetic mouse models of schizophrenia. Behav Brain Res. 2009;204(2):282–94. doi: 10.1016/j.bbr.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacological reviews. 1999;51(1):7–61. [PubMed] [Google Scholar]
- 5.Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Science's STKE : signal transduction knowledge environment. 2004;2004(255):re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- 6.Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44(1):5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 7.Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nature reviews Neuroscience. 2007;8(6):413–26. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- 8.Krystal JH, Karper LP, Seibyl JP, Freeman GK, Delaney R, Bremner JD, et al. Subanesthetic effects of the noncompetitive NMDA antagonist, ketamine, in humans. Psychotomimetic, perceptual, cognitive, and neuroendocrine responses. Arch Gen Psychiatry. 1994;51(3):199–214. doi: 10.1001/archpsyc.1994.03950030035004. [DOI] [PubMed] [Google Scholar]
- 9.Lahti AC, Koffel B, LaPorte D, Tamminga CA. Subanesthetic doses of ketamine stimulate psychosis in schizophrenia. Neuropsychopharmacology. 1995;13(1):9–19. doi: 10.1016/0893-133X(94)00131-I. [DOI] [PubMed] [Google Scholar]
- 10.Javitt DC, Zukin SR. Recent advances in the phencyclidine model of schizophrenia. Am J Psychiatry. 1991;148(10):1301–8. doi: 10.1176/ajp.148.10.1301. [DOI] [PubMed] [Google Scholar]
- 11.Swerdlow NR, Geyer MA, Braff DL. Neural circuit regulation of prepulse inhibition of startle in the rat: current knowledge and future challenges. Psychopharmacology (Berl) 2001;156(2-3):194–215. doi: 10.1007/s002130100799. [DOI] [PubMed] [Google Scholar]
- 12.Braff DL. Prepulse inhibition of the startle reflex: a window on the brain in schizophrenia. Curr Top Behav Neurosci. 2010;4:349–71. doi: 10.1007/7854_2010_61. [DOI] [PubMed] [Google Scholar]
- 13.Mohn AR, Gainetdinov RR, Caron MG, Koller BH. Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell. 1999;98(4):427–36. doi: 10.1016/s0092-8674(00)81972-8. [DOI] [PubMed] [Google Scholar]
- 14.Moghaddam B, Jackson ME. Glutamatergic animal models of schizophrenia. Annals of the New York Academy of Sciences. 2003;1003:131–7. doi: 10.1196/annals.1300.065. [DOI] [PubMed] [Google Scholar]
- 15.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10(1):40–68. doi: 10.1038/sj.mp.4001558. image 5. [DOI] [PubMed] [Google Scholar]
- 16.Kantrowitz JT, Javitt DC. N-methyl-d-aspartate (NMDA) receptor dysfunction or dysregulation: the final common pathway on the road to schizophrenia? Brain research bulletin. 2010;83(3-4):108–21. doi: 10.1016/j.brainresbull.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akbarian S, Sucher NJ, Bradley D, Tafazzoli A, Trinh D, Hetrick WP, et al. Selective alterations in gene expression for NMDA receptor subunits in prefrontal cortex of schizophrenics. J Neurosci. 1996;16(1):19–30. doi: 10.1523/JNEUROSCI.16-01-00019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao XM, Sakai K, Roberts RC, Conley RR, Dean B, Tamminga CA. Ionotropic glutamate receptors and expression of N-methyl-D-aspartate receptor subunits in subregions of human hippocampus: effects of schizophrenia. Am J Psychiatry. 2000;157(7):1141–9. doi: 10.1176/appi.ajp.157.7.1141. [DOI] [PubMed] [Google Scholar]
- 19.Kristiansen LV, Huerta I, Beneyto M, Meador-Woodruff JH. NMDA receptors and schizophrenia. Current opinion in pharmacology. 2007;7(1):48–55. doi: 10.1016/j.coph.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Geddes AE, Huang XF, Newell KA. Reciprocal signalling between NR2 subunits of the NMDA receptor and neuregulin1 and their role in schizophrenia. Progress in neuro-psychopharmacology & biological psychiatry. 2011;35(4):896–904. doi: 10.1016/j.pnpbp.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 21.Weickert CS, Fung SJ, Catts VS, Schofield PR, Allen KM, Moore LT, et al. Molecular evidence of N-methyl-D-aspartate receptor hypofunction in schizophrenia. Mol Psychiatry. 2013;18(11):1185–92. doi: 10.1038/mp.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilowsky LS, Bressan RA, Stone JM, Erlandsson K, Mulligan RS, Krystal JH, et al. First in vivo evidence of an NMDA receptor deficit in medication-free schizophrenic patients. Mol Psychiatry. 2006;11(2):118–9. doi: 10.1038/sj.mp.4001751. [DOI] [PubMed] [Google Scholar]
- 23.Olney JW, Farber NB. Glutamate receptor dysfunction and schizophrenia. Arch Gen Psychiatry. 1995;52(12):998–1007. doi: 10.1001/archpsyc.1995.03950240016004. [DOI] [PubMed] [Google Scholar]
- 24.Behrens MM, Ali SS, Dao DN, Lucero J, Shekhtman G, Quick KL, et al. Ketamine-induced loss of phenotype of fast-spiking interneurons is mediated by NADPH-oxidase. Science. 2007;318(5856):1645–7. doi: 10.1126/science.1148045. [DOI] [PubMed] [Google Scholar]
- 25.Gao XM, Hashimoto T, Tamminga CA. Phencyclidine (PCP) and dizocilpine (MK801) exert time-dependent effects on the expression of immediate early genes in rat brain. Synapse. 1998;29(1):14–28. doi: 10.1002/(SICI)1098-2396(199805)29:1<14::AID-SYN2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 26.Enomoto T, Tse MT, Floresco SB. Reducing prefrontal gamma-aminobutyric acid activity induces cognitive, behavioral, and dopaminergic abnormalities that resemble schizophrenia. Biol Psychiatry. 2011;69(5):432–41. doi: 10.1016/j.biopsych.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 27.Martin P, Carlsson ML, Hjorth S. Systemic PCP treatment elevates brain extracellular 5-HT: a microdialysis study in awake rats. Neuroreport. 1998;9(13):2985–8. doi: 10.1097/00001756-199809140-00012. [DOI] [PubMed] [Google Scholar]
- 28.Savage S, Mattsson A, Olson L. Cholinergic denervation attenuates phencyclidine-induced c-fos responses in rat cortical neurons. Neuroscience. 2012 doi: 10.1016/j.neuroscience.2012.04.064. [DOI] [PubMed] [Google Scholar]
- 29.Lodge D, Johnson KM. Noncompetitive excitatory amino acid receptor antagonists. Trends Pharmacol Sci. 1990;11(2):81–6. doi: 10.1016/0165-6147(90)90323-z. [DOI] [PubMed] [Google Scholar]
- 30.Moghaddam B, Adams BW. Reversal of phencyclidine effects by a group II metabotropic glutamate receptor agonist in rats. Science. 1998;281(5381):1349–52. doi: 10.1126/science.281.5381.1349. [DOI] [PubMed] [Google Scholar]
- 31.Sanderson JL, Dell'Acqua ML. AKAP signaling complexes in regulation of excitatory synaptic plasticity. Neuroscientist. 2011;17(3):321–36. doi: 10.1177/1073858410384740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scannevin RH, Huganir RL. Postsynaptic organization and regulation of excitatory synapses. Nature reviews Neuroscience. 2000;1(2):133–41. doi: 10.1038/35039075. [DOI] [PubMed] [Google Scholar]
- 33.Lei G, Xia Y, Johnson KM. The role of Akt-GSK-3beta signaling and synaptic strength in phencyclidine-induced neurodegeneration. Neuropsychopharmacology. 2008;33(6):1343–53. doi: 10.1038/sj.npp.1301511. [DOI] [PubMed] [Google Scholar]
- 34.Xia Y, Wang CZ, Liu J, Anastasio NC, Johnson KM. Lithium protection of phencyclidine-induced neurotoxicity in developing brain: the role of phosphatidylinositol-3 kinase/Akt and mitogen-activated protein kinase kinase/extracellular signal-regulated kinase signaling pathways. J Pharmacol Exp Ther. 2008;326(3):838–48. doi: 10.1124/jpet.107.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouri A, Noda Y, Noda A, Nakamura T, Tokura T, Yura Y, et al. Involvement of a dysfunctional dopamine-D1/N-methyl-d-aspartate-NR1 and Ca2+/calmodulin-dependent protein kinase II pathway in the impairment of latent learning in a model of schizophrenia induced by phencyclidine. Mol Pharmacol. 2007;71(6):1598–609. doi: 10.1124/mol.106.032961. [DOI] [PubMed] [Google Scholar]
- 36.Molteni R, Pasini M, Moraschi S, Gennarelli M, Drago F, Racagni G, et al. Reduced activation of intracellular signaling pathways in rat prefrontal cortex after chronic phencyclidine administration. Pharmacol Res. 2008;57(4):296–302. doi: 10.1016/j.phrs.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Li K, Zhou T, Liao L, Yang Z, Wong C, Henn F, et al. beta CaMKII in lateral habenula mediates core symptoms of depression. Science. 2013;341(6149):1016–20. doi: 10.1126/science.1240729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis DA, Gonzalez-Burgos G. Neuroplasticity of neocortical circuits in schizophrenia. Neuropsychopharmacology. 2008;33(1):141–65. doi: 10.1038/sj.npp.1301563. [DOI] [PubMed] [Google Scholar]
- 39.McIntosh AM, Owens DC, Moorhead WJ, Whalley HC, Stanfield AC, Hall J, et al. Longitudinal volume reductions in people at high genetic risk of schizophrenia as they develop psychosis. Biol Psychiatry. 2011;69(10):953–8. doi: 10.1016/j.biopsych.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Moghaddam B, Krystal JH. Capturing the angel in “angel dust”: twenty years of translational neuroscience studies of NMDA receptor antagonists in animals and humans. Schizophr Bull. 2012;38(5):942–9. doi: 10.1093/schbul/sbs075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barr AM, Powell SB, Markou A, Geyer MA. Iloperidone reduces sensorimotor gating deficits in pharmacological models, but not a developmental model, of disrupted prepulse inhibition in rats. Neuropharmacology. 2006;51(3):457–65. doi: 10.1016/j.neuropharm.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Mansbach RS, Brooks EW, Sanner MA, Zorn SH. Selective dopamine D4 receptor antagonists reverse apomorphine-induced blockade of prepulse inhibition. Psychopharmacology (Berl) 1998;135(2):194–200. doi: 10.1007/s002130050501. [DOI] [PubMed] [Google Scholar]
- 43.McClatchy DB, Liao L, Park SK, Venable JD, Yates JR. Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Res. 2007;17(9):1378–88. doi: 10.1101/gr.6375007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClatchy DB, Dong MQ, Wu CC, Venable JD, Yates JR., 3rd 15N metabolic labeling of mammalian tissue with slow protein turnover. J Proteome Res. 2007;6(5):2005–10. doi: 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen EI, McClatchy D, Park SK, Yates JR., 3rd Comparisons of mass spectrometry compatible surfactants for global analysis of the mammalian brain proteome. Anal Chem. 2008;80(22):8694–701. doi: 10.1021/ac800606w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fonslow BR, Niessen SM, Singh M, Wong CC, Xu T, Carvalho PC, et al. Single-step inline hydroxyapatite enrichment facilitates identification and quantitation of phosphopeptides from mass-limited proteomes with MudPIT. J Proteome Res. 2012;11(5):2697–709. doi: 10.1021/pr300200x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 48.Venable JD, Wohlschlegel J, McClatchy DB, Park SK, Yates JR., 3rd Relative quantification of stable isotope labeled peptides using a linear ion trap-Orbitrap hybrid mass spectrometer. Anal Chem. 2007;79(8):3056–64. doi: 10.1021/ac062054i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, et al. MS1, MS2, and SQT- Three Unified, Compact, and Easily Parsed File Formats for the Storage of Shotgun Proteomic Spectra and Identifications. Rapid Commun Mass Spectrom. 2004;18:2162–8. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 50.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5(11):976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 51.Sadygov RG, Eng J, Durr E, Saraf A, McDonald H, MacCoss MJ, et al. Code developments to improve the efficiency of automated MS/MS spectra interpretation. Journal of Proteome Research. 2002;1(3):211–5. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- 52.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 53.MacCoss MJ, Wu CC, III, Y JR. A correlation algorithm for the automated analysis of quantitative ‘shotgun’ proteomics data. Analytical Chemistry. 2003;75(24):6912–21. doi: 10.1021/ac034790h. [DOI] [PubMed] [Google Scholar]
- 54.Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008 doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ting L, Cowley MJ, Hoon SL, Guilhaus M, Raftery MJ, Cavicchioli R. Normalization and statistical analysis of quantitative proteomics data generated by metabolic labeling. Mol Cell Proteomics. 2009;8(10):2227–42. doi: 10.1074/mcp.M800462-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature biotechnology. 2006;24(10):1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 57.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437(7061):1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 58.Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nature methods. 2008;5(4):319–22. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park SK, Yates JR., 3rd Census for proteome quantification. Current protocols in bioinformatics / editoral board, Andreas D Baxevanis [et al] 2010;Chapter 13 doi: 10.1002/0471250953.bi1312s29. Unit 13 2 1-1. [DOI] [PubMed] [Google Scholar]
- 60.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nature reviews Molecular cell biology. 2010;11(1):9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 61.Pyle RA, Schivell AE, Hidaka H, Bajjalieh SM. Phosphorylation of synaptic vesicle protein 2 modulates binding to synaptotagmin. J Biol Chem. 2000;275(22):17195–200. doi: 10.1074/jbc.M000674200. [DOI] [PubMed] [Google Scholar]
- 62.Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25(13):3379–88. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Markevich NI, Hoek JB, Kholodenko BN. Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J Cell Biol. 2004;164(3):353–9. doi: 10.1083/jcb.200308060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kapuy O, Barik D, Sananes MR, Tyson JJ, Novak B. Bistability by multiple phosphorylation of regulatory proteins. Prog Biophys Mol Biol. 2009;100(1-3):47–56. doi: 10.1016/j.pbiomolbio.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benitez-King G, Ramirez-Rodriguez G, Ortiz L, Meza I. The neuronal cytoskeleton as a potential therapeutical target in neurodegenerative diseases and schizophrenia. Curr Drug Targets CNS Neurol Disord. 2004;3(6):515–33. doi: 10.2174/1568007043336761. [DOI] [PubMed] [Google Scholar]
- 66.Hayashi K, Pan Y, Shu H, Ohshima T, Kansy JW, White CL, 3rd, et al. Phosphorylation of the tubulin-binding protein, stathmin, by Cdk5 and MAP kinases in the brain. J Neurochem. 2006;99(1):237–50. doi: 10.1111/j.1471-4159.2006.04113.x. [DOI] [PubMed] [Google Scholar]
- 67.Blitzer RD, Connor JH, Brown GP, Wong T, Shenolikar S, Iyengar R, et al. Gating of CaMKII by cAMP-regulated protein phosphatase activity during LTP. Science. 1998;280(5371):1940–2. doi: 10.1126/science.280.5371.1940. [DOI] [PubMed] [Google Scholar]
- 68.Genoux D, Haditsch U, Knobloch M, Michalon A, Storm D, Mansuy IM. Protein phosphatase 1 is a molecular constraint on learning and memory. Nature. 2002;418(6901):970–5. doi: 10.1038/nature00928. [DOI] [PubMed] [Google Scholar]
- 69.Deguchi M, Hata Y, Takeuchi M, Ide N, Hirao K, Yao I, et al. BEGAIN (brain-enriched guanylate kinase-associated protein), a novel neuronal PSD-95/SAP90-binding protein. J Biol Chem. 1998;273(41):26269–72. doi: 10.1074/jbc.273.41.26269. [DOI] [PubMed] [Google Scholar]
- 70.Yao I, Iida J, Nishimura W, Hata Y. Synaptic and nuclear localization of brain-enriched guanylate kinase-associated protein. J Neurosci. 2002;22(13):5354–64. doi: 10.1523/JNEUROSCI.22-13-05354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, et al. The cystine/glutamate antiporter system x(c)(-) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal. 2013;18(5):522–55. doi: 10.1089/ars.2011.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lewerenz J, Maher P. Basal levels of eIF2alpha phosphorylation determine cellular antioxidant status by regulating ATF4 and xCT expression. J Biol Chem. 2009;284(2):1106–15. doi: 10.1074/jbc.M807325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 74.Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, et al. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5(6):1158–70. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 75.Yang L, MacLellan WR, Han Z, Weiss JN, Qu Z. Multisite phosphorylation and network dynamics of cyclin-dependent kinase signaling in the eukaryotic cell cycle. Biophys J. 2004;86(6):3432–43. doi: 10.1529/biophysj.103.036558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palmowski P, Rogowska-Wrzesinska A, Williamson J, Beck HC, Mikkelsen JD, Hansen HH, et al. Acute phencyclidine treatment induces extensive and distinct protein phosphorylation in rat frontal cortex. J Proteome Res. 2014;13(3):1578–92. doi: 10.1021/pr4010794. [DOI] [PubMed] [Google Scholar]
- 77.Wu R, Dephoure N, Haas W, Huttlin EL, Zhai B, Sowa ME, et al. Correct interpretation of comprehensive phosphorylation dynamics requires normalization by protein expression changes. Mol Cell Proteomics. 2011;10(8):M111 009654. doi: 10.1074/mcp.M111.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–89. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kargieman L, Santana N, Mengod G, Celada P, Artigas F. Antipsychotic drugs reverse the disruption in prefrontal cortex function produced by NMDA receptor blockade with phencyclidine. Proc Natl Acad Sci U S A. 2007;104(37):14843–8. doi: 10.1073/pnas.0704848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liao L, Sando RC, Farnum JB, Vanderklish PW, Maximov A, Yates JR. 15N-labeled brain enables quantification of proteome and phosphoproteome in cultured primary neurons. J Proteome Res. 2012;11(2):1341–53. doi: 10.1021/pr200987h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Powell SB, Geyer MA. Overview of animal models of schizophrenia. Curr Protoc Neurosci. 2007;Chapter 9 doi: 10.1002/0471142301.ns0924s39. Unit 9 24. [DOI] [PubMed] [Google Scholar]
- 82.Melnick SM, Rodriguez JS, Bernardi RE, Ettenberg A. A simple procedure for assessing ataxia in rats: effects of phencyclidine. Pharmacology, biochemistry, and behavior. 2002;72(1-2):125–30. doi: 10.1016/s0091-3057(01)00727-4. [DOI] [PubMed] [Google Scholar]
- 83.Krebs-Thomson K, Lehmann-Masten V, Naiem S, Paulus MP, Geyer MA. Modulation of phencyclidine-induced changes in locomotor activity and patterns in rats by serotonin. Eur J Pharmacol. 1998;343(2-3):135–43. doi: 10.1016/s0014-2999(97)01557-4. [DOI] [PubMed] [Google Scholar]
- 84.Ogren SO, Goldstein M. Phencyclidine- and dizocilpine-induced hyperlocomotion are differentially mediated. Neuropsychopharmacology. 1994;11(3):167–77. doi: 10.1038/sj.npp.1380103. [DOI] [PubMed] [Google Scholar]
- 85.Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84(4):623–31. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- 86.Manna T, Thrower DA, Honnappa S, Steinmetz MO, Wilson L. Regulation of microtubule dynamic instability in vitro by differentially phosphorylated stathmin. J Biol Chem. 2009;284(23):15640–9. doi: 10.1074/jbc.M900343200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Solis-Chagoyan H, Calixto E, Figueroa A, Montano LM, Berlanga C, Rodriguez-Verdugo MS, et al. Microtubule organization and L-type voltage-activated calcium current in olfactory neuronal cells obtained from patients with schizophrenia and bipolar disorder. Schizophr Res. 2013;143(2-3):384–9. doi: 10.1016/j.schres.2012.11.035. [DOI] [PubMed] [Google Scholar]
- 88.Brown AS, Borgmann-Winter K, Hahn CG, Role L, Talmage D, Gur R, et al. Increased Stability of Microtubules in Cultured Olfactory Neuroepithelial Cells from Individuals with Schizophrenia. Progress in neuro-psychopharmacology & biological psychiatry. 2013 doi: 10.1016/j.pnpbp.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schubart UK, Yu J, Amat JA, Wang Z, Hoffmann MK, Edelmann W. Normal development of mice lacking metablastin (P19), a phosphoprotein implicated in cell cycle regulation. J Biol Chem. 1996;271(24):14062–6. doi: 10.1074/jbc.271.24.14062. [DOI] [PubMed] [Google Scholar]
- 90.Shumyatsky GP, Malleret G, Shin RM, Takizawa S, Tully K, Tsvetkov E, et al. stathmin, a gene enriched in the amygdala, controls both learned and innate fear. Cell. 2005;123(4):697–709. doi: 10.1016/j.cell.2005.08.038. [DOI] [PubMed] [Google Scholar]
- 91.Martel G, Nishi A, Shumyatsky GP. Stathmin reveals dissociable roles of the basolateral amygdala in parental and social behaviors. Proc Natl Acad Sci U S A. 2008;105(38):14620–5. doi: 10.1073/pnas.0807507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martel G, Hevi C, Wong A, Zushida K, Uchida S, Shumyatsky GP. Murine GRPR and stathmin control in opposite directions both cued fear extinction and neural activities of the amygdala and prefrontal cortex. PLoS One. 2012;7(2):e30942. doi: 10.1371/journal.pone.0030942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40(Database issue):D261–70. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peters A, Sethares C, Luebke JI. Synapses are lost during aging in the primate prefrontal cortex. Neuroscience. 2008;152(4):970–81. doi: 10.1016/j.neuroscience.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Elsworth JD, Morrow BA, Hajszan T, Leranth C, Roth RH. Phencyclidine-induced loss of asymmetric spine synapses in rodent prefrontal cortex is reversed by acute and chronic treatment with olanzapine. Neuropsychopharmacology. 2011;36(10):2054–61. doi: 10.1038/npp.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Flores C, Wen X, Labelle-Dumais C, Kolb B. Chronic phencyclidine treatment increases dendritic spine density in prefrontal cortex and nucleus accumbens neurons. Synapse. 2007;61(12):978–84. doi: 10.1002/syn.20452. [DOI] [PubMed] [Google Scholar]
- 98.Malenka RC, Madison DV, Nicoll RA. Potentiation of synaptic transmission in the hippocampus by phorbol esters. Nature. 1986;321(6066):175–7. doi: 10.1038/321175a0. [DOI] [PubMed] [Google Scholar]
- 99.Kleschevnikov AM, Routtenberg A. PKC activation rescues LTP from NMDA receptor blockade. Hippocampus. 2001;11(2):168–75. doi: 10.1002/hipo.1034. [DOI] [PubMed] [Google Scholar]
- 100.Orr JW, Newton AC. Requirement for negative charge on “activation loop” of protein kinase C. J Biol Chem. 1994;269(44):27715–8. [PubMed] [Google Scholar]
- 101.Brunet A, Pages G, Pouyssegur J. Growth factor-stimulated MAP kinase induces rapid retrophosphorylation and inhibition of MAP kinase kinase (MEK1) FEBS Lett. 1994;346(2-3):299–303. doi: 10.1016/0014-5793(94)00475-7. [DOI] [PubMed] [Google Scholar]
- 102.Saito Y, Gomez N, Campbell DG, Ashworth A, Marshall CJ, Cohen P. The threonine residues in MAP kinase kinase 1 phosphorylated by MAP kinase in vitro are also phosphorylated in nerve growth factor-stimulated rat phaeochromocytoma (PC12) cells. FEBS Lett. 1994;341(1):119–24. doi: 10.1016/0014-5793(94)80252-1. [DOI] [PubMed] [Google Scholar]
- 103.Kelleher RJ, 3rd, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004;116(3):467–79. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 104.Fernandez de Sevilla D, Nunez A, Borde M, Malinow R, Buno W. Cholinergic-mediated IP3-receptor activation induces long-lasting synaptic enhancement in CA1 pyramidal neurons. J Neurosci. 2008;28(6):1469–78. doi: 10.1523/JNEUROSCI.2723-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harnett MT, Bernier BE, Ahn KC, Morikawa H. Burst-timing-dependent plasticity of NMDA receptor-mediated transmission in midbrain dopamine neurons. Neuron. 2009;62(6):826–38. doi: 10.1016/j.neuron.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.DeSouza N, Reiken S, Ondrias K, Yang YM, Matkovich S, Marks AR. Protein kinase A and two phosphatases are components of the inositol 1,4,5-trisphosphate receptor macromolecular signaling complex. J Biol Chem. 2002;277(42):39397–400. doi: 10.1074/jbc.M207059200. [DOI] [PubMed] [Google Scholar]
- 107.Wagner LE, 2nd, Li WH, Joseph SK, Yule DI. Functional consequences of phosphomimetic mutations at key cAMP-dependent protein kinase phosphorylation sites in the type 1 inositol 1,4,5-trisphosphate receptor. J Biol Chem. 2004;279(44):46242–52. doi: 10.1074/jbc.M405849200. [DOI] [PubMed] [Google Scholar]
- 108.Stringer JL, Greenfield LJ, Hackett JT, Guyenet PG. Blockade of long-term potentiation by phencyclidine and sigma opiates in the hippocampus in vivo and in vitro. Brain Res. 1983;280(1):127–38. doi: 10.1016/0006-8993(83)91180-0. [DOI] [PubMed] [Google Scholar]
- 109.Baker DA, Xi ZX, Shen H, Swanson CJ, Kalivas PW. The origin and neuronal function of in vivo nonsynaptic glutamate. J Neurosci. 2002;22(20):9134–41. doi: 10.1523/JNEUROSCI.22-20-09134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.De Bundel D, Schallier A, Loyens E, Fernando R, Miyashita H, Van Liefferinge J, et al. Loss of system x(c)- does not induce oxidative stress but decreases extracellular glutamate in hippocampus and influences spatial working memory and limbic seizure susceptibility. J Neurosci. 2011;31(15):5792–803. doi: 10.1523/JNEUROSCI.5465-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baker DA, Madayag A, Kristiansen LV, Meador-Woodruff JH, Haroutunian V, Raju I. Contribution of cystine-glutamate antiporters to the psychotomimetic effects of phencyclidine. Neuropsychopharmacology. 2008;33(7):1760–72. doi: 10.1038/sj.npp.1301532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arstikaitis P, Gauthier-Campbell C, Carolina Gutierrez Herrera R, Huang K, Levinson JN, Murphy TH, et al. Paralemmin-1, a modulator of filopodia induction is required for spine maturation. Mol Biol Cell. 2008;19(5):2026–38. doi: 10.1091/mbc.E07-08-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rudolf R, Bittins CM, Gerdes HH. The role of myosin V in exocytosis and synaptic plasticity. J Neurochem. 2011;116(2):177–91. doi: 10.1111/j.1471-4159.2010.07110.x. [DOI] [PubMed] [Google Scholar]
- 114.Hernandez JM, Floyd DH, Weilbaecher KN, Green PL, Boris-Lawrie K. Multiple facets of junD gene expression are atypical among AP-1 family members. Oncogene. 2008;27(35):4757–67. doi: 10.1038/onc.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guedea AL, Schrick C, Guzman YF, Leaderbrand K, Jovasevic V, Corcoran KA, et al. ERK-associated changes of AP-1 proteins during fear extinction. Mol Cell Neurosci. 2011;47(2):137–44. doi: 10.1016/j.mcn.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schwarzschild MA, Cole RL, Hyman SE. Glutamate, but not dopamine, stimulates stress-activated protein kinase and AP-1-mediated transcription in striatal neurons. J Neurosci. 1997;17(10):3455–66. doi: 10.1523/JNEUROSCI.17-10-03455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Takahashi K, Nagai T, Kamei H, Maeda K, Matsuya T, Arai S, et al. Neural circuits containing pallidotegmental GABAergic neurons are involved in the prepulse inhibition of the startle reflex in mice. Biol Psychiatry. 2007;62(2):148–57. doi: 10.1016/j.biopsych.2006.06.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Quantified phosphorylation sites from the AGC kinase family. The phosphorylated amino acids are list for the known site required for activation (i.e. “activation” loop, “hydrophobic” motif, “turn” motif), other phosphorylation sites with known function, and phosphorylation sites with no known function. Sites highlighted in red possess and A-score of 18 or greater.
Supplementary Table 2: Multi-state phosphorylated peptides. Unique peptide sequences that were observed to have at least two different phosphorylation states.
Supplementary Table 3-5. Altered phosphorylation peptides. Phosphopeptides are listed that were observed to be altered. Each phosphopeptide is listed with condition specific 14N/15N average ratio, localization score (Ascore > 13 = high), enrichment annotation, IPI identifier, and protein name. Table 3 = Sal vs. PCP, Table 4 = SalPPI vs PCPPPI, Table 5 = No PPI vs. PPI. This data with more detailed protein information is also available at http://sealion.scripps.edu:8080/pint/?project=04cf043e645ab98e5214396575d271ff#query