Abstract
Direct visualization of bioorthogonal alkyne or azide handles using fluorogenic azide-alkyne cycloaddition conducted on the surface of a blot membrane. The method eliminates the need for separation steps to remove excess small molecule reagents before attachment of antigen molecules or other visualization handles, and is especially useful for the analysis of peptides and small proteins. A variety of potential fluorogenic reagents are assessed, and sensitivity (<0.1 picomole) similar to current commercially available fluorescence imaging methods is possible.
Bioorthogonal chemistry has become an essential tool for interrogation and modulation of biomolecules in diverse and complex environments.1 The azide-alkyne cycloaddition reaction was one of the first and remains among the most important in bioorthogonal chemistry, including the more recent development of fluorogenic click reactions and strained-alkynes for copper-free conjugation.2,3 In practice, a major use of bioorthogonal reactions such as the azide-alkyne cycloaddition is as a secondary modification reaction, appending complex functionality after an initial attachment of a bioorthogonal handle4 by chemical,5 enzymatic,6 genetic,7 or metabolic means.8 In this paper, we present a simple method to visualize proteins with a bioorthogonal handle by fluorogenic chemical reactions on the surface of a polyvinylidene difluoride (PVDF) blot membrane, and we describe efforts to optimize this method.
We initiated efforts on this front after struggling with gel-based analysis of modifications to peptide and small proteins.9,10 Most chemical protein modification reactions require a significant excess of a small-molecule reagent, and separation steps are required to remove by-product and unreacted small-molecules. In our hands, efficient separation of excess small-molecules was challenging, especially for very small proteins/peptides (<10 kDa) and for fairly large multifunctional small-molecule reagents. A conventional analysis of modified small proteins, with secondary solution-phase attachment of an antigen for western blot or of a fluorophore led to undesired fluorescent bands attributed to small-molecule agents that swamped the desired signal and especially to poor quantification of the desired peptide or protein. These problems were magnified for peptides, for which size-exclusion-based separation methods are inefficient or impossible. Among the several analytical methods that might ameliorate this problem, we chose to explore direct visualization of the primary modification by conducting a secondary chemical reaction to attach a fluorophore to biomolecules on the surface of a blot membrane (Figure 1a). Perhaps surprisingly, we are not aware of this technique being used elsewhere, apart from an example of on-membrane antigen attachment for western blotting.11
Fig. 1.
a) Schematic description of chemical blotting. i) Proteins subject to a modification reaction. (ii) The alkyne-labeled protein can be liberated from excess small molecules by SDS-PAGE iii) The alkyne-labeled protein is poised for bioorthogonal detection after it is transferred onto a blot membrane. iv) Fluorogenic, bioorthogonal reaction on the membrane surface reveal the labeled protein. b) Probes examined in this work.
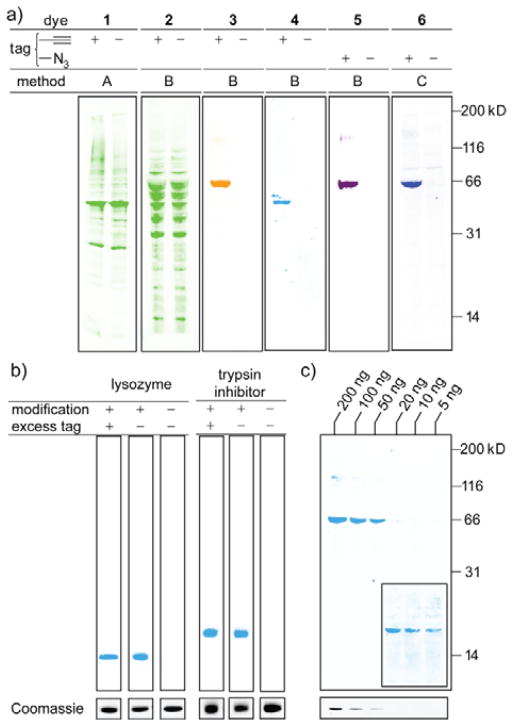
To minimize the background staining, we examined fluorogenic probes, “pro-fluorophores” that are non-emissive or minimally emissive prior to a specific conjugation reaction. The adoption of fluorogenic molecules greatly improved imaging quality, and allowed sensitive and selective detection of even quite small proteins such as terminal-alkyne-labeled Fyn SH3 domain (7 kDa, Fig. 3a). We examined several known fluorogenic reagents.12–15 For example, azido-iridium complex 3 (Ir-N3), recently developed in our labs for its long-lifetime emission, or azido-coumarin 4 did provide a single strong fluorescence band corresponding to modified proteins, in stark contrast to initial efforts with traditional reactive fluorophores (Fig. 2a). The approach is reasonably general and is not limited to azide-based fluorophores. The copper-catalyzed and copper-free detection of azide-labeled proteins were possible with the fluorogenic ethynyl-coumarin 5 (Q-yne) and cyclooctyne 6 (Fl-DIBO), respectively. Copper-free detection of cyclooctyne-labeled proteins (lysozyme and trypsin inhibitor) was also feasible for analyte solutions containing a large excess of cyclooctyne small molecules (Fig. 2b).
Fig. 3.
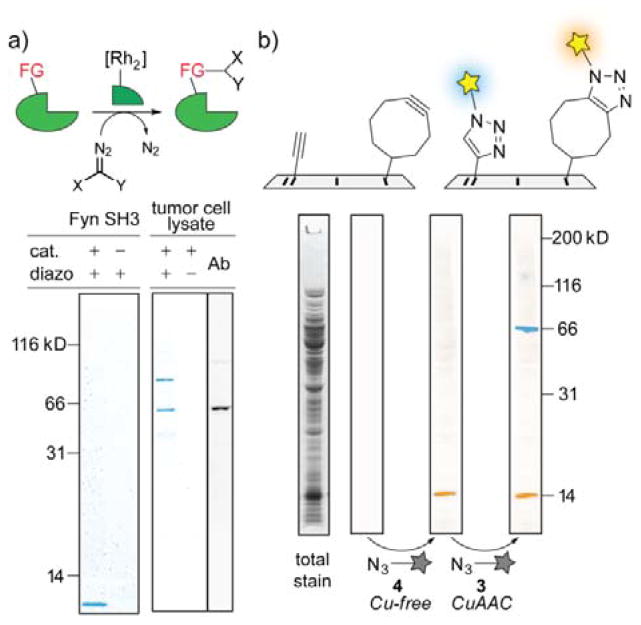
a) Site-specific modification of proteins by rhodium-metallopeptide catalyst (X = alkyne group) and chemical blotting analysis of the modification reaction with azide 4.9 Left: Fyn SH3 in E. coli lysate, right: Yes-kinase in PC3 tumor cell lysate and antibody detection by Yes-antibody (Ab). b) Sequential detection of cyclooctyne-tagged lysozyme and alkyne-tagged BSA in the presence of E. coli lysate through Cu-free and Cu-catalyzed azide-alkyne-cycloaddition, respectively.
Fig. 2.
a) Screening for detection of alkyne- or azide-tagged protein. Lanes 1 and 4: maltose binding protein (MBP), 2 and 3: bovine serum albumin (BSA), 5 and 6: BSA in E. coli lysate. Method A: Cu-free Sonogashira coupling. Method B: Cu-catalyzed cycloaddition. Method C: Cu-free cycloaddition. b) Analysis of cyclooctyne-tagged protein solutions in the presence or absence of large excesses of small-molecule cyclooctyne. c) Sensitivity of 4 for different amount of tagged BSA in E. coli lysate. Inset: image of right-most lanes at longer exposure time. Coomassie blue staining of the same concentration of tagged BSA (in the absence of E. coli lysate) at bottom. Impurity present in BSA (~116 kDa) is observed in some of lanes (See ESI for details).
After confirming that the four fluorogenic probes are amenable for the visualization of the bioorthogonal reporter-tagged proteins on blot membrane, we evaluated the capability in more detail (Table 1). Fluorescence measurement of the probes conjugated to corresponding bovine serum albumin (BSA) on blot membrane reveals that emission maxima of all of the four probes slightly shifts to blue region relative to that measured in solution.12–15 The probes cover a range of emission wavelengths (Figure 2 uses false-color imaging to convey dye emission), including broad emission in red region with iridium complex 3 (λem = 610 nm)—a spectral region minimizes background fluorescence from biomolecules and PVDF membrane.15 Somewhat surprisingly, there was a significant difference in the sensitivity and selectivity among the four probes examined. Successful, robust imaging required micromolar concentrations of most dyes, but azido-coumarin 4 required only 100 nM. At this concentration, imaging of a standard-size blot membrane (w: 7 cm, h: 9 cm) requires only 400 ng of the readily-available azide 4. We also assessed the minimum observable protein concentration. Here too, azide 4 proved substantially better than other dyes. With azide 4, 5 ng (~0.08 pmol) of protein could be imaged (Figure 2c, inset), a level similar to the limits of detection given for commercially available fluorescence-based gel imaging methods.16
Table 1.
Performance of four fluorogenic probes for chemical blotting
| dyes | 3 (Q-N3) | 4 (Ir-N3) | 5 (Q-yne) | 6 (Fl-DIBO) |
|---|---|---|---|---|
| λema [nm] | 471 | 610 | 384 | 435 |
| minimum proteinb [ng] | 5 | 200 | 100 | 100 |
| probe concnc [μM] | 0.1 | 10 | 3.3 | 10 |
| note | azide | azide | alkyne | alkyne (Cu-free) |
Emission maxima of the probes conjugated with alkyne- or azide-tagged BSA on PVDF membrane.
Sensitivity of the probes to lower concentration of alkyne- or azide-tagged BSA in the presence of E. coli lysate.
Lowest concentration of the probes for effective detection of alkyne- or azide-tagged BSA in the presence of E. coli lysate. See ESI for detailed conditions and images.
In our hands, the chemical blotting method does indeed significantly improve imaging peptides and small proteins, the initial impetus for our work. We described a method for site-specific installation of an alkyne moiety by rhodium-metallopeptide catalyzed reactions of diazo compounds (Figure 3a).9 Modification of small protein domain (Fyn SH3, 7 kDa) could be reproducibly assessed with azide 4. In contrast, we failed to image this modification with solution-based fluorophore attachment or by western blotting. Overexpressed proteins are not required; we described9 visualizing modified Yes kinase at natural abundance levels in PC3 tumor cell lysate (Figure 3a, right).
Sequential detection of different modified proteins is also possible (Figure 3b). Cyclooctyne-tagged lysozyme, alkyne-tagged BSA, and E. coli lysate were separated by SDS-PAGE and transferred to blot membrane. First, incubation of the membrane with iridium complex 3 selectively labeled cyclooctyne-labeled lysozyme. Subsequent copper-catalyzed visualization of the alkyne-tagged BSA by coumarin 4 produced two strong fluorescence bands on the membrane. The two bands are readily differentiated by the emission profiles of the two dyes (ESI). Likewise, sequential detection of alkyne-tagged BSA and azide-tagged BSA was accomplished with coumarin reagents 4 and 5, respectively (ESI). The double-blotting example demonstrates a mild and chemoselective biorthogonal reaction that, in practice, minimizes materials usage and minimizes sample preparation.
The methods described here may well be complementary to existing protein-based detection methods. As a secondary functionalization method, the approach should be general for visualization of proteins with bioorthogonal handles installed by varied chemical, enzymatic, genetic, or metabolic methods. The method is inexpensive and operationally simple, requiring none of the blocking and secondary antibody recognition steps common in western blotting, for example. Because of the covalent dye attachment, the treated membrane could be imaged after long storage unlike traditional immunoblotting (ESI). Furthermore, the chemical reagents described here withstand elevated temperature and variation in solvent, or pH. The significant differences in imaging efficiency with the fluorogenic molecules examined here point to the potential for future fluorophore optimization. However, it is noteworthy that azido-coumarin 4 achieves sensitive protein imaging on par with current state-of-art commercial methods. The method obviates the need for solid-phase extraction or size-exclusion chromatography to remove a large excess of byproduct and unreacted small-molecules, which in our hands is problematic for small or heavily modified proteins.
Supplementary Material
Acknowledgments
We acknowledge support from the National Institutes of Health under grant number 5R21CA170625, from the Robert A. Welch Foundation Research Grant C-1680, and from the National Science Foundation (CHE-1055569).
Footnotes
Electronic Supplementary Information (ESI) available: Detailed procedures, optimization data, emission spectra, synthesis and characterization of new compounds. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Sletten EM, Bertozzi CR. Angew Chem Int Ed. 2009;48:6974. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Droumaguet C, Wang C, Wang Q. Chem Soc Rev. 2010;39:1233. doi: 10.1039/b901975h. [DOI] [PubMed] [Google Scholar]
- 3.Debets MF, van Berkel SS, Dommerholt J, Dirks AT, Rutjes FP, van Delft FL. Acc Chem Res. 2011;44:805. doi: 10.1021/ar200059z. [DOI] [PubMed] [Google Scholar]
- 4.Patterson DM, Nazarova LA, Prescher JA. ACS Chem Biol. 2014;9:592. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- 5.Ball ZT. Curr Opin Chem Biol. 2015;25:98. doi: 10.1016/j.cbpa.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Nat Biotechnol. 2007;25:1483. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ngo JT, Tirrell DA. Acc Chem Res. 2011;44:677. doi: 10.1021/ar200144y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Proc Nat Acad Sci USA. 2011;108:3141. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vohidov F, Coughlin JM, Ball ZT. Angew Chem Int Ed. 2015;54:4587. doi: 10.1002/anie.201411745. [DOI] [PubMed] [Google Scholar]
- 10.Kundu R, Ball ZT. Chem Commun. 2013;49:4166. doi: 10.1039/c2cc37323h. [DOI] [PubMed] [Google Scholar]
- 11.Tsai CS, Yen HY, Lin MI, Tsai TI, Wang SY, Huang WI, Hsu TL, Cheng YSE, Fang JM, Wong CH. Proc Nat Acad Sci USA. 2013;110:2466. doi: 10.1073/pnas.1222183110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. Org Lett. 2004;6:4603. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Z, Fahrni CJ. J Am Chem Soc. 2004;126:8862. doi: 10.1021/ja049684r. [DOI] [PubMed] [Google Scholar]
- 14.Friscourt F, Fahrni CJ, Boons GJ. J Am Chem Soc. 2012;134:18809. doi: 10.1021/ja309000s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohata J, Vohidov F, Aliyan A, Huang K, Marti AA, Ball ZT. 2015 doi: 10.1039/c5cc06099k. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bio-Rad. 2015 http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_2032F.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.