Abstract
This study addresses the individual and combined effects of HIV-1 and methamphetamine (N-methyl-1-phenylpropan-2-amine, METH) on cardiac dysfunction in a transgenic mouse model of HIV/AIDS. METH is abused epidemically and is frequently associated with acquisition of HIV-1 infection or AIDS. We employed microarrays to identify mRNA differences in cardiac left ventricle (LV) gene expression following METH administration (10d, 3mg/kg/d, subcutaneously) in C57Bl/6 wild-type littermates (WT) and Tat-expressing transgenic (TG) mice. Arrays identified 880 differentially expressed genes (expression fold change>1.5, p<0.05) following METH exposure, Tat expression, or both. Using pathway enrichment analysis, mRNAs encoding polypeptides for calcium signaling and contractility were altered in the LV samples. Correlative DNA methylation analysis revealed significant LV DNA methylation changes following METH exposure and Tat expression. By combining these data sets, 38 gene promoters (27 related to METH, 11 related to Tat) exhibited differences by both methods of analysis. Among those, only the promoter for CACNA1C that encodes L-type calcium channel Cav1.2 displayed DNA methylation changes concordant with its gene expression change. Quantitative PCR verified that Cav1.2 LV mRNA abundance doubled following METH. Correlative immunoblots specific for Cav1.2 revealed a 3.5-fold increase in protein abundance in METH LVs. Data implicate Cav1.2 in calcium dysregulation and hypercontractility in the murine LV exposed to METH. They suggest a pathogenetic role for METH exposure to promote LV dysfunction that outweighs Tat-induced effects.
Keywords: HIV-1, AIDS, methamphetamine, heart, epigenetics
INTRODUCTION
Methamphetamine (N-methyl-1-phenylpropan-2-amine, METH) is a widely abused cardiotoxic Schedule II stimulant (Yu, 2003). METH users develop increased heart rate and blood pressure, arrhythmias, cardiomyopathy (CM), and sudden death (Derlet and Horowitz, 1995; Kaye, 2007; Yu, 2003). These are thought to be mediated by catecholamines, stimulation of adrenoreceptors, and resultant alterations in intracellular calcium (Yu, 2003). METH has been shown to modulate mitochondrial function, and mitochondrial dysfunction is recognized as an important feature of human heart failure (Eskandari, 2014; He, 1996b; Neubauer, 2007; Nickel, 2013).
HIV/AIDS is a global epidemic. A higher risk of HIV-1 infection relates to illicit drug use (Bell, 2002; Estrada, 2002). HIV-1 causes CM independent of antiretroviral therapy. Our lab has shown that the HIV-1 viral protein Tat can cause CM in vivo through mitochondrial dysfunction (Fang, 2009; Raidel, 2002; Remick, 2014). Mechanisms for cardiac comorbidities of METH and HIV-1 are not well known. This study addresses the pathogenetic mechanisms of METH and Tat in the heart following short-term METH usage.
While relatively minor mitochondrial dysfunction occurred with the HIV-1 Tat transgene in the absence of METH, significant mitochondrial dysfunction followed 10 day METH. Using microarrays for large scale data collection, we investigated gene expression changes and DNA methylation changes to identify epigenetic effects of Tat and METH on cardiac function. Quantitative RT-PCR (qRT-PCR) and immunoblots were used to validate important cellular targets. Results implicate calcium channel dysregulation, specifically CACNA1C encoding Cav1.2, and related mitochondrial dysfunction in Tat transgenic and METH-treated mice.
While Tat causes cardiac dysfunction, the effects of METH after 10d are more pronounced than Tat-mediated effects alone. Both induce changes in DNA methylation, though only CACNA1C is upregulated by METH. This suggests specific targeting of CACNA1C for long-term gene expression changes in the heart leading to hypercontractility and cardiac damage and offers potential therapeutic targets.
MATERIALS AND METHODS
Reagents
All reagents were analytical grade and purchased from Sigma Aldrich (St. Louis, MO) unless otherwise indicated.
Mouse care
Congenic Tat mice were generated by back crossing (6 generations to C57Bl/6 mice) our original lines created on the inbred FVB/n background and that displayed cardiac-specific Tat expression (Raidel et al., 2002). TG founders were mated with wild-type C57Bl/6 mice (Jackson Labs, Bar Harbor, ME) to produce offspring on a C57Bl/6 background (termed Tat here, internally as Tat-HB) and mice for all experiments were at least six generations from the founding lines. Wild-type (WT) littermates were used as control mice in all experiments. For genotyping, genomic DNA was extracted from mouse tail clippings, and genotype was determined by PCR as described previously (Raidel et al., 2002). All mice were housed at the Emory University Vivarium, in accordance with IACUC protocols, in an AAALAC certified vivarium, and according to NIH guidelines.
Experiments employed mice 8–12 weeks old. In a “two-by two” factorial design, WT and Tat mice were administered 3mg/kg methamphetamine or saline control by IP once daily for 10 days. Physiological, histopathological, and molecular changes were determined after 10 days.
Echocardiography and Electrocardiogram
Mice were anesthetized with Avertin (0.25 mg/g of body weight) and weighed to determine body mass. Echocardiography was performed using VisualSonics Vevo 770 (VisualSonics, Toronto, Ontario, CA). Results from two-dimensional M-mode analysis along the short axis (at the level of the largest LV diameter) were used to determine LV mass, LV end diastolic dimension (LVEDD), posterior wall thickness (PWTH), ejection fraction, and LV fractional shortening. LV mass values were normalized to mouse body weight.
ECG was performed using the AnonyMOUSE ECG screening system (Mouse Specifics Inc., Quincy, MA) with proprietary software. Mice were acclimated to the pad for 10–15 minutes prior to readings, with a minimum of 20 heart rate time points taken for each mouse. ECG parameters included heart rate, heart rate variability, QRS, and QTc. HRV is reported as the standard deviation of the heart rate and is reported in units of beats per minute (BPM).
Histology
Following 10 days of METH, mice were terminated by cervical dislocation under avertin anesthesia, and hearts were removed, sectioned rapidly with a razor blade (2 mm sections), fixed in 10% neutral buffered formalin and embedded in paraffin. Six micrometer histological sections were processed with Masson’s trichrome to highlight myocyte necrosis, and images were collected at 40X magnification using a Nikon Eclipse E800M microscope (Nikon, Melville, NY). Histo-morphometric analysis was performed with the pathologist blinded to treatment or transgenic status. Quantitative microscopic analysis of contraction bands was determined on the stained tissue sections. Five to ten randomly selected photomicrographs (40X) of each LV sample were digitized. Fibrosis was measured using a deconvolution method of color selection with a set tolerance and converted to a binary image using Adobe Photoshop (Adobe, San Jose, CA). Fibrosis was defined as the fraction dense red staining within the total field of view using NIH Image J software (scale = 9.3 pixels/µm). Data are presented as ± SEM.
Mitochondrial DNA (mtDNA) and Nuclear DNA Abundance
Methods utilized are similar to those we described previously (Lewis, 2007). DNA sequences for primers and probes used for quantitation of mitochondrial and nuclear DNA analyzed the COX I gene of the mtDNA and the POLG2 gene of the nuclear DNA. Amplification was performed using the Lightcycler 480 system (Roche, Indianapolis, IN). The number of mtDNA copies was determined by normalizing mtDNA abundance to nuclear DNA abundance using the single-copy nuclear gene POLG2.
Mitochondrial Oximetry
Mitochondrial oximetry was performed as previously described (Koczor, 2013b). Briefly, mitochondria were isolated from mouse hearts immediately following sacrifice using differential centrifugation and a commercial mitochondrial isolation kit (Sigma Aldrich). An aliquot was quantitated for protein using the Bradford assay, and 5µg of protein was placed into a V7 plate (Seahorse Bioscience, Billerica, MA) and centrifuged at 3,400XG for 20 minutes at 4°C. Following centrifugation, pyruvate and malate were added to each well, and the mitochondrial respiration was analyzed in an XF24 flux analyzer (Seahorse Bioscience) using the manufacturer’s protocol. Basal respiration (oxygen consumption rate of the mitochondria immediately following equilibration), State III respiration, State IVo respiration, and respiratory control ratio (RCR) were quantitated. Oximetric results are provided as pmol O2/min/µg protein.
Mitochondrial Electron Transport Chain (ETC) Complex Activity
Mitochondria were isolated from mouse hearts as described above for analysis of electron transport chain function. Citrate synthase and ETC complex 1, 2, 2/3, and 4 assays were obtained from Cayman Chemicals (Ann Arbor, MI), and experiments followed manufacturer protocols using the recommended inhibitors to identify complex-specific activity. Each sample was run in duplicate. For each ETC complex, specific activity was normalized to citrate synthase activity, and results obtained are expressed as a percent of WT vehicle-treated controls.
Gene expression analysis
Gene expression analysis was performed as previously described (Koczor, 2013a). Briefly, total RNA was extracted from at least 3 mouse hearts from each 2X2 groups using the Fibrous Tissue RNeasy kit (Qiagen, Germantown, MD). Double-stranded cDNA was synthesized using the SuperScript Double-Stranded cDNA Synthesis Kit (Life Technologies Corp, Grand Island, NY). cDNA was then labeled with Cy3 and hybridized overnight to a 12x135kb human expression array (Roche Nimblegen). Expression arrays were washed and scanned using Roche Nimblegen MS200 scanner. Images were analyzed using Nimblescan software as directed by the manufacturer, including RMA normalization and generation of expression data. Expression results were analyzed by 2-way ANOVA using Bioconductor for R. Differentially expressed genes were identified as those with a p<0.05 and two-fold change in gene expression compared to controls. Gene ontology was performed using DAVID bioinformatics database (Huang da, 2009). Microarray array data (raw and processed) were deposited into NIH/NCBI Gene Expression Omnibus (GEO) and can be accessed under GEO series GSE64159.
Quantitative RT-PCR
RNA was isolated as described above, and single-stranded cDNA was synthesized using SuperScript III (Life Technologies). Quantitative PCR was performed using SYBR green dye and a LightCycler 480 system (Roche) using manufacturer protocols. Primers for ADCY6, Bcl-XL, CACNA1C, COX10,MYBPC3, and PGC-1α (Integrated DNA Technologies, Coralville, IA) are shown in Table 1. Individual sample results were normalized to controls ACTB and RN18S (Qiagen). Results are expressed as fold change of WT-Saline controls.
Table 1.
Quantitative RT-PCR Primers
| ADCY6 | |
|---|---|
| Forward: | GCGGTGAGGGAGAATCACTG |
| Reverse: | GCCCTGACACGCAGTAGTAA |
| BCLXL | |
| Forward: | AAGTTCCCCCGGTCTCTTCAG |
| Reverse: | CCAAGATAAGGTTCTGGCTGACTG |
| CACNA1C | |
| Forward: | AGTGAGACTAATCCAGCTGAACA |
| Reverse: | AGATGCGGGAGTTCTCCTCT |
| COX10 | |
| Forward: | CCTCCTCACAGGCTCAGTA |
| Reverse: | CCTGTGTGGCAATCCCTGTAT |
| MYBPC3 | |
| Forward: | AAGAGAGACAGTTTCCGGAGG |
| Reverse: | CATGCCTCGAAGGTCTGTGA |
| PGC1A | |
| Forward: | TTGACTGGCGTCATTCGGG |
| Reverse: | ACCAGAGCAGCACACTCTATG |
DNA methylation analysis
DNA methylation analysis was performed as previously described (Koczor, 2013a). Briefly, DNA was extracted as described above. Total cellular DNA was sonicated to an average fragment size of 200–500bp. A sample of DNA was set aside for later normalization (denoted ‘Input’), and then a portion of the sonicated DNA was enriched using the MethylCollector Ultra kit (Active Motif, Carlsbad, CA) following the manufacturer’s directions. Both the methylated and input DNA were amplified using whole genome amplification (Sigma-Aldrich). Samples of the methylated and input DNA were validated for enrichment of methylated DNA using PCR. For DNA methylation analysis, Roche Nimblegen 2.1M Deluxe Promoter Arrays were utilized (Roche Nimblegen). DNA was labeled with Cy3 and Cy5 dyes to distinguish methylated and input DNA, and DNA was allowed to hybridize to arrays overnight. Arrays were scanned using a Roche Nimblegen MS200 scanner. Images were analyzed using Nimblescan software as directed by the manufacturer (which included normalizing to the input DNA), resulting in a final analysis including a P-score of the detected methylated DNA peak and annotation to the probe location. A two-way ANOVA was performed on only the differentially expressed gene promoters to identify DNA methylation changes that correlate with gene expression changes. DNA methylation results were analyzed using Bioconductor for R. Differentially methylated gene promoter regions were identified as those with a p<0.05 and peak score change greater than 1. Gene ontology was performed using DAVID bioinformatics database. Microarray array data (raw and processed) were deposited into NIH/NCBI Gene Expression Omnibus (GEO) and can be accessed under GEO series GSE64159.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism 5.0 (Graphpad, La Jolla, CA). Each experiment was analyzed using a one-way or two-way ANOVA were appropriate, with a p<0.05 deemed statistically significant. In determining interactions between Tat and METH in qRT-PCR and echocardiography results, a two-way ANOVA was utilized. The experiments are displayed as a mean ± SEM, with all experiments performed at least 3 times each.
RESULTS
Mouse physiology
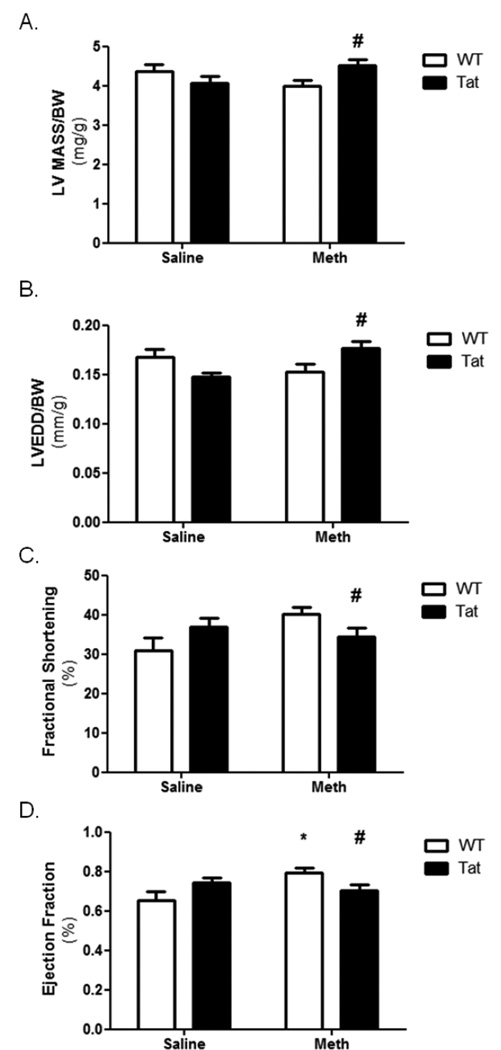
Tat mice express the HIV-1 Tat gene and polypeptide exclusively in the heart using an α-myosin heavy chain promoter (Raidel, 2002). Littermates without the transgene (wild type; WT) were used for controls. Mice were treated with 3mg/kg methamphetamine (METH) at doses lower than those previously shown to elicit heart failure (Matsuo, 2009; Yu, 2002). Echocardiography did not show any significant changes in LV mass, LVEDD, or fractional shortening after 10 days (Figure 1A–C). There was a significant increase in ejection fraction in wild-type METH-treated mice compared to saline-treated (Figure 1D). Interestingly, this effect was not seen in METH-treated Tat mice. Two-way ANOVA results found a significant interaction (p<0.05) between Tat and METH in LV mass, LVEDD, and fractional shortening.
Figure 1. Echocardiography of Tat and METH mice.
Echocardiography was performed following 10 days of METH or saline vehicle in WT or Tat mice. Results identified no change in LV mass (A), LVEDD (B), or fractional shortening (C) that resulted from either Tat or METH. A small but significant increase in ejection fraction (D) was identified in METH-treated WT mice compared to WT-saline mice. A negative interaction between Tat and METH was identified by two-way ANOVA. * denotes p<0.05 compare to WT-saline mice by one-way ANOVA; # denotes a significant interaction (p<0.05) between Tat and METH by two-way ANOVA.
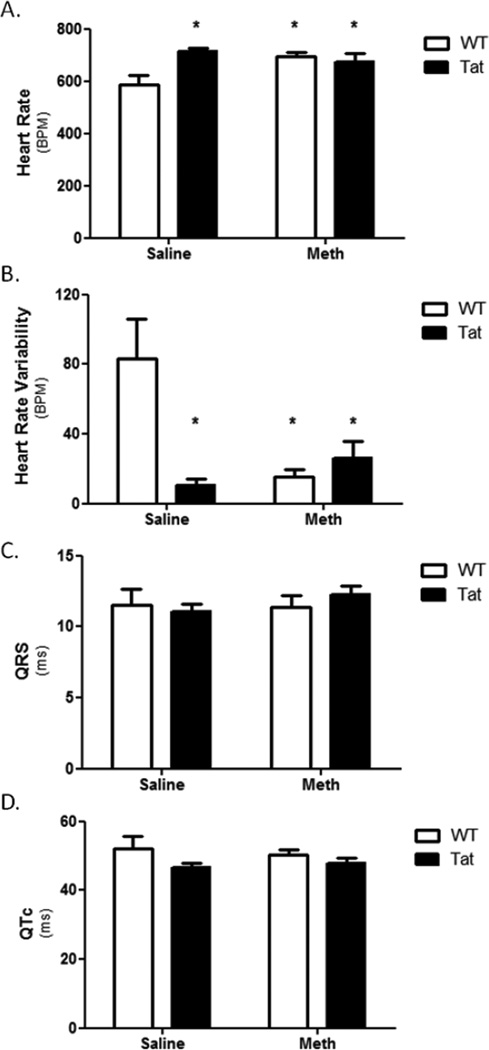
Electrocardiograms (ECG) were performed on mice one hour after final saline or METH injection. ECG results showed significant increase in heart rates of WT-METH, Tat-Saline, and Tat-METH compared to WT-Saline (Figure 2A). Heart rate variability (HRV) is associated with cardiac abnormalities including heart failure (Henry, 2012). This parameter was significantly reduced in WT-METH, Tat-Saline, and Tat-METH compared to WT-Saline (Figure 2B). No change was observed in QRS or QTc intervals (Figure 2C and D, respectively). Together with the echocardiographic data, ECG results show elevated heart rate and possible increased risk of heart failure in METH and Tat but no pathological signs of cardiomyopathy or arrhythmia for the duration of the study.
Figure 2. Electrocardiogram of Tat and METH mice.
ECG was performed one hour after the final METH or vehicle (day 10). Heart rate (A) was elevated in Tat-saline, WT-METH, and Tat-METH mice compared to WT-saline mice, while heart rate variability (B) was reduced in all three experimental groups compared to WT-saline. No changes in QRS or QTc were identified following 10 days of METH or saline. * denotes p<0.05 compared to WT-saline mice.
Mitochondrial function
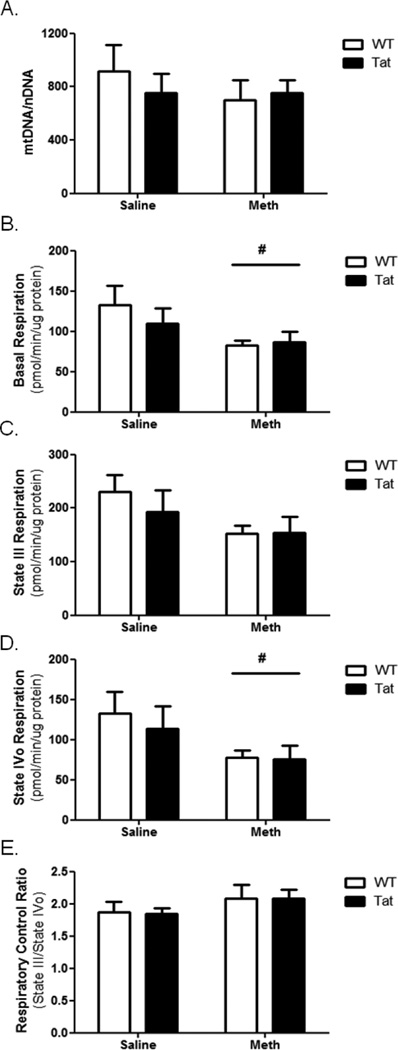
Cardiac mitochondria are organellar targets for both METH and Tat (Flores, 1993; He, 1996a; Liou, 2014; Lord, 2010; Raidel, 2002). Following 10 days of METH or saline vehicle, cardiac LV mtDNA abundance was quantitated. No significant change in mtDNA abundance was caused either by Tat transgene or METH administration (Figure 3A).
Figure 3. Cardiac mitochondrial dysfunction is caused by METH.
(A) Cardiac LV mtDNA abundance was determined for each experimental group and normalized to nuclear DNA. No change in LV mtDNA abundance was observed in any experimental group compared to LV of WT-saline mice. Mitochondria were isolated from LV tissue and analyzed using a Seahorse XF24 oximetric analyzer. Basal respiration (B) was significantly reduced in METH-treated mice. State III respiration (C) was not significantly altered following METH exposure, but State IVo respiration (D) was significantly reduced following METH exposure. The respiratory control ratio (E) was unchanged. Tat transgene did not significantly alter any mitochondrial functional parameter within the experimental timeline. # denotes p<0.05 for METH-treated groups compared to saline-treated groups by two-way ANOVA.
Oximetric analysis of isolated LV mitochondria revealed functional changes in LV mitochondrial electron transport. Basal respiration was significantly diminished in METH-treated WT and Tat mice (Figure 3B). While no significant change was seen in state III respiration, state IVo respiration was significantly reduced in METH-treated mice (Figure 3C and 3D, respectively). There was no significant change in RCR caused by Tat or METH (Figure 3E).
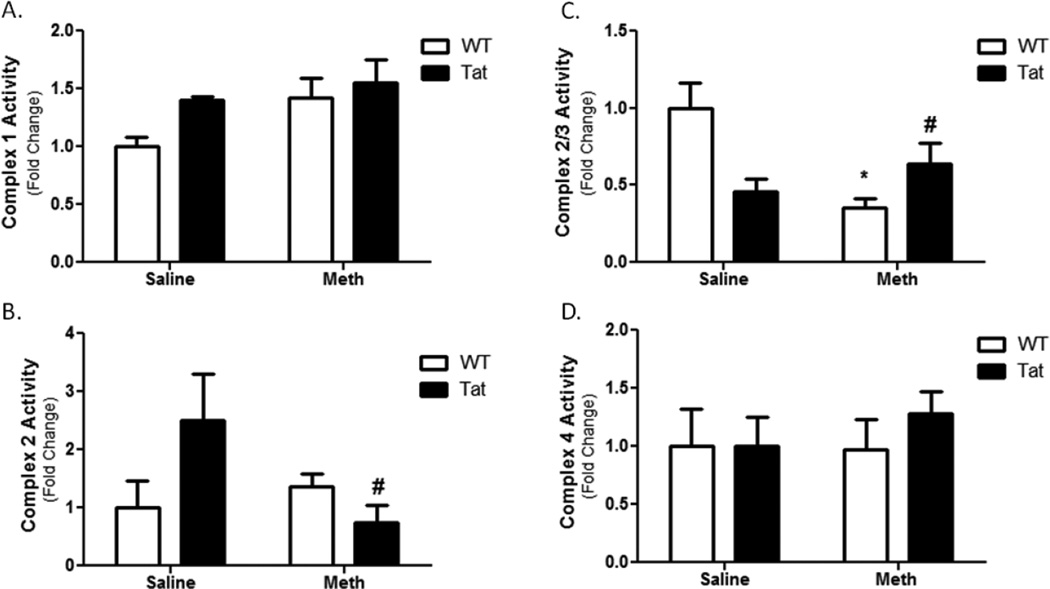
Analysis of ETC complexes localized the mitochondrial dysfunction to specific loci within the ETC. ETC complexes 1 and 4 showed no change in activity (Figure 4A and 4D, respectively). ETC complex 2 activity was not significantly altered by Tat or METH compared to their respective controls (Figure 4B). However, in the presence of the Tat TG and METH, ETC complex 2 activity was reduced. This indicates a significant interaction between METH and Tat on ETC complex 2 activity. In parallel, METH significantly reduced complex 2/3 activity while the combination of METH and in presence of the Tat transgene increased ETC complex 2/3 activity compared to METH alone (Figure 4C). Taken together, METH causes an ETC complex 3 deficit indicated by diminished complex 2/3 activity with no change in the complex 2 activity.
Figure 4. Mitochondrial Electron Transport Chain Complex Activity.
Complexes of the mitochondrial electron transport chain were assessed. No change was observed in Complex 1 (A). No statistical change in complex 2 activity was caused by Tat mice or METH alone, but a significant interaction was observed between METH and Tat during co-administration (B). Though this interaction was significant, there was not a statistically significant change in Tat-METH Complex 2 activity compared to controls. The Complex 2/3 assay showed a significant decrease with the METH-treated WT (C). Interestingly, the combined presence Tat and METH led to an increase in complex 2/3 activity. No change was observed in Complex 4 (D). * denotes p<0.05 compare to WT-saline mice by one-way ANOVA; # denotes a significant interaction (p<0.05) between Tat and METH by two-way ANOVA.
Gene expression
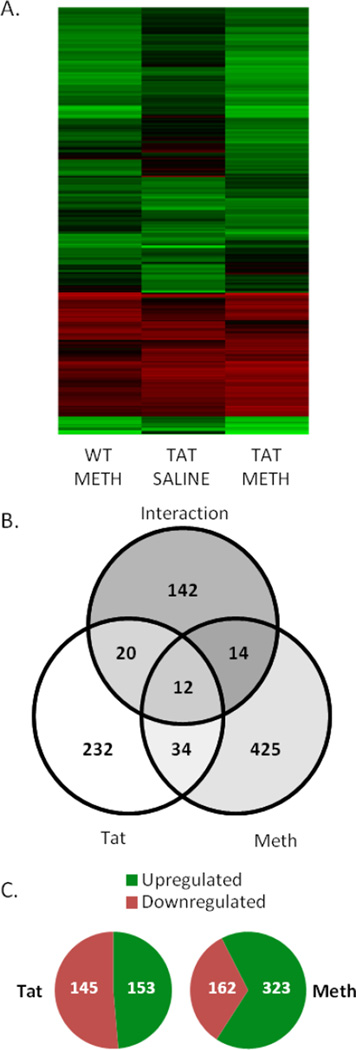
Using microarray technology, we analyzed 17,869 gene expression profiles for changes resulting from the Tat transgene, METH treatment, or both. Statistical analysis revealed 880 genes that were significantly differentially expressed based on Tat, METH, their interaction, or a combination of these (Supplemental Table 1). Heatmaps show that the upregulated and downregulated genes group similarly between WT-METH, Tat-Saline, and Tat-METH groups (Figure 5A). Tat mice exhibited 298 differentially expressed genes, of which 232 were exclusively differentially expressed by Tat (Figure 5B). Tat genes were nearly split evenly between those up-regulated and those down-regulated compared to WT-saline genes (Figure 5C). METH-treated mice exhibited 485 differentially expressed genes, with 67% up-regulated by METH and 33% down-regulated by METH. Analysis also revealed 183 genes that displayed interactions between METH-treatment and the Tat transgene. Of these, 90% (165 of 183) were found to have an antagonistic effect (i.e. the magnitude of gene expression change caused by Tat or Meth was reduced when the Tat transgene and METH-treatment were both present). This is consistent with echocardiographic findings where Tat and METH revealed antagonistic changes (Figure 1).
Figure 5. Gene Expression Microarrays.
Gene expression microarrays were performed on three mice from each experimental group. Results were analyzed by two-way ANOVA, with significantly regulated genes identified as those with a p<0.05 and a fold-change in expression greater than 2. (A) Heat map showing all 880 differentially regulated genes, normalized to WT-saline expression. Red denotes decreased expression and green denotes increased expression. (B) Analysis revealed 880 differentially regulated genes, with 425 differentially regulated only by METH, 232 regulated only by Tat, and 142 found to have a significant interaction between METH and Tat. Some genes were found to overlap between groups. (C) For Tat-regulated genes, the number of downregulated genes nearly equaled the number of upregulated genes. For METH-associated genes, 67% were upregulated and 33% were downregulated.
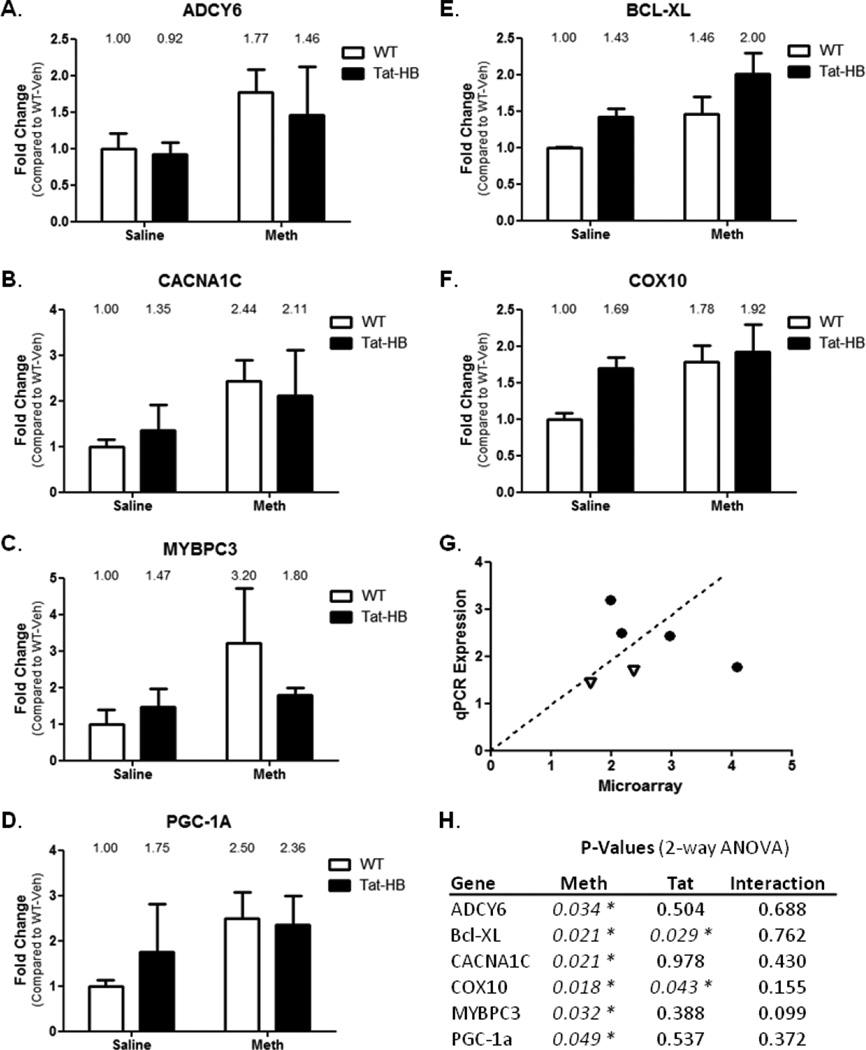
Select genes were analyzed for expression using quantitative RT-PCR. Genes were selected based on ontology and literature searches that implicated the target gene in cardiac dysfunction, calcium handling, and mitochondrial function. On that basis, we analyzed gene expression of ADCY6, CACNA1C, MYBPC3, and PGC-1α in relation to METH-induced changes, and Bcl-XL and COX10 in relation to Tat-induced changes. ADCY6 is an adenylate cyclase that mediates β-adrenergic stimulation (Hodges, 2010); Bcl-XL is an antiapoptotic protein; CACNA1C is the L-type calcium channel important for proper cardiac action potentials (Goonasekera, 2012); COX10 is a subunit of mitochondrial Complex 4; MYBPC3 is a myosin binding protein exclusively expressed in the heart and implicated in familial-inherited CM (Adalsteinsdottir, 2014); and PGC-1α is a key activator of mitochondrial biogenesis (Spiegelman, 2007). METH increased gene expression in ADYC6, CACNA1C, MYBPC3, and PGC-1α genes (Figures 6A–D, respectively). Tat did not statistically contribute to differential gene expression of these genes. Bcl-XL and COX10, which displayed significant expression changes caused by Tat, were verified to be differentially expressed by Tat expression (Figures 6E and F, respectively). Interestingly, METH was found to modulate the expression of these genes by qPCR. QPCR agreed with microarray results when compared “head-to-head” with these 6 genes (Figure 6G). QPCR results and microarray results point to broad cardiac gene expression changes from METH in pathways that regulate cardiac action potentials, calcium uptake, mitochondrial biogenesis, and CM.
Figure 6. Quantitative RT-PCR.
QRT-PCR was performed using primers designed to interrogate important cardiac targets. Results complimented microarray data. ADCY6 (A), CACNA1C (B), MYBP3C (C), and PGC-1α (D) were elevated following METH treatment. Bcl-XL (E) and COX10 (F) were elevated as a result of Tat expression. Interestingly, Bcl-XL and COX10 also were differentially expressed as a result of METH treatment. G) QPCR Expression results were compared graphically to microarray values for validation. Dotted line represents 1:1 ratio. Circles are genes differentially expressed by METH, and open triangles are genes differentially expressed by Tat. H) P-Values from two-way ANOVA analysis of qPCR expression. * denotes p<0.05.
DNA Methylation
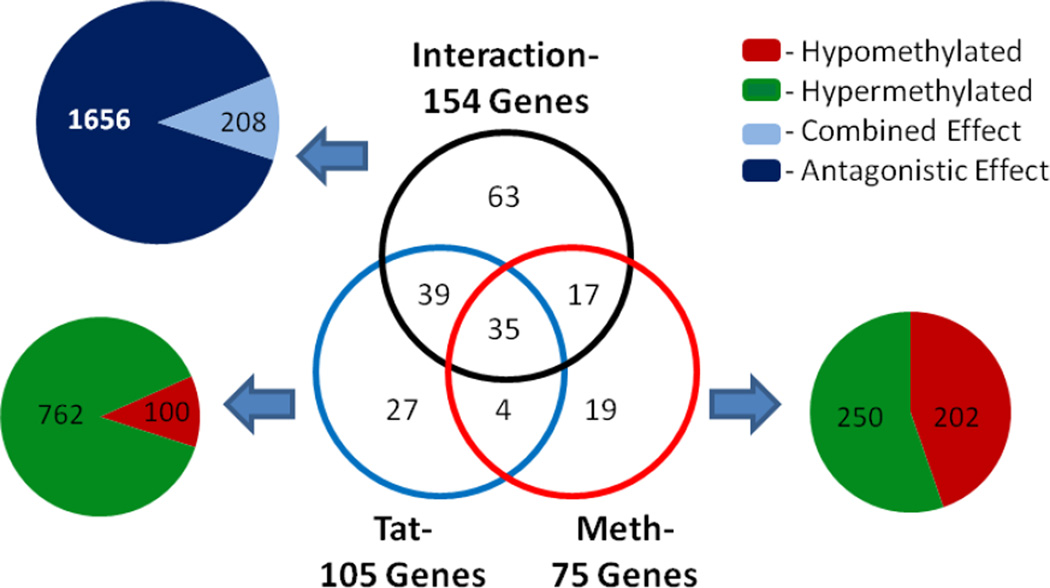
To identify epigenetic changes that may be operable even after 10 days of METH, DNA methylation analyses were performed. Changes in gene expression correlate with epigenetic modifications, including DNA methylation, in the failing heart (Koczor, 2013a; Koczor, 2013c). Using gene promoter microarrays combined with MeDIP, DNA methylation was analyzed in LV tissue to identify changes in methylation associated with gene expression changes. A two-way ANOVA was performed to identify differentially methylated regions associated with the 880 differentially expressed genes. DNA methylation changes were observed for Tat mice and METH-treated groups, in some cases independently for each factor (Figure 7). Differentially methylated gene promoters caused by Tat or METH are in Supplemental Table 2. There were 862 differentially methylated regions in 105 genes in Tat hearts, with 762 peaks hypermethylated and 100 peaks hypomethylated in Tat mice compared to WT mice (Figure 7). There were 452 differentially methylated regions in 75 genes in WT-METH hearts, with 250 peaks hypermethylated and 202 peaks hypomethylated in compared to WT mice. Additionally, promoter regions were identified where Tat and METH significantly interacted to alter DNA methylation (Supplemental Table 3). Analysis revealed 1,864 regions in 154 gene promoters exhibiting Tat and METH interactions, with nearly 90% of these interactions antagonizing single factor effects (Figure 7). The differentially methylated regions were confined, in total, to 204 genes, demonstrating about 23% of the 880 differentially expressed genes have DNA methylation changes as well.
Figure 7. DNA Methylation.
The Tat transgene and METH treatment were both found to alter DNA methylation in the gene promoters of the 880 differentially expressed genes in the heart. In total, 105 gene promoters were differentially methylated in Tat mice, 75 gene promoters were differentially methylated following METH treatment, and 154 gene promoters showed a significant interaction between METH and Tat transgene. For Tat mice, the 105 gene promoters displayed 452 differentially methylated regions, with 250 being hypermethylated and 202 hypomethylated compared to WT-saline. Following METH-treatment, 862 regions were differentially methylated within the 75 gene promoters, with 762 hypermethylated and 100 hypomethylated. Interactions were found between Tat and METH in 1,864 regions of the 154 gene promoters, of which 1,656 interactions were antagonistic and 208 were combined effects greater than either factor.
Individual genes were analyzed to identify those that had differential gene expression resulting from either factor (i.e. Tat or METH) in addition to a change in DNA methylation caused by that same factor. Genes were verified to ensure gene promoter DNA methylation changes altered gene expression according to current paradigms (i.e. hypomethylation of gene promoters increases gene expression, hypermethylation decreases gene expression) (Tao, 2014). We identified 27 genes that had differential DNA methylation correlated to changes in gene expression resulting from METH, while 11 genes were found for Tat (Table 2). Importantly, DNA methylation patterns of the six genes analyzed by qRT-PCR (Figure 6) revealed that only CACNA1C displayed differential methylation in agreement with its gene expression changes following METH.
Table 2.
Differentially Expressed Genes with DNA Methylation Changes
| TatHB | DNA Methylation | Gene Expression (vs WT-Saline) | ||||||
|---|---|---|---|---|---|---|---|---|
| RefSeq ID | Gene Symbol |
Gene Name | Increase | Decrease | TATHB P- Value |
WT- METH |
TATHB- Saline |
TATHB- METH |
| NM_009253 | serpina3m | serine (or cysteine) peptidase inhibitor, clade A, member 3M |
3 | 3 | 0.006 | 0.651 | 0.484 | 0.401 |
| NM_138649 | syt17 | synaptotagmin XVII | 15 | 0 | 0.007 | 1.045 | 0.694 | 0.384 |
| NM_027885 | smug1 | single-strand selective monofunctional uracil DNA glycosylase |
4 | 0 | 0.019 | 0.661 | 0.341 | 0.382 |
| NM_001081023 | CACNA1S | calcium channel, voltage-dependent, L type, alpha 1S subunit |
4 | 0 | 0.020 | 0.649 | 0.466 | 0.588 |
| NM_152803 | HPSE | heparanase | 12 | 0 | 0.021 | 0.633 | 0.569 | 0.497 |
| NM_026261 | UBL4B | ubiquitin-like 4B | 6 | 0 | 0.028 | 0.514 | 0.544 | 0.391 |
| NM_001039586 | glyctk | glycerate kinase | 29 | 0 | 0.033 | 0.728 | 0.764 | 0.495 |
| NM_033612 | CELA1 | chymotrypsin-like elastase family, member 1 |
4 | 0 | 0.034 | 0.447 | 0.433 | 0.344 |
| NM_139151 | SPAG4 | sperm associated antigen 4 | 4 | 2 | 0.038 | 0.576 | 0.426 | 0.339 |
| NM_026981 | DTWD1 | DTW domain containing 1 | 2 | 0 | 0.048 | 0.681 | 0.622 | 0.475 |
| NM_148948 | DICER1 | Dicer1, Dcr-1 homolog (Drosophila) | 12 | 0 | 0.049 | 0.550 | 0.434 | 0.551 |
| Methamphetamine | DNA Methylation | Gene Expression (vs WT-Saline) | ||||||
| RefSeq ID |
Gene Symbol |
Gene Name | Increase | Decrease |
METH P- Value |
WT- METH |
TATHB- Saline |
TATHB- METH |
| NM_021441 | TAF1C | TATA box binding protein (Tbp)- associated factor, RNA polymerase I, C |
12 | 16 | 0.002 | 2.718 | 1.557 | 2.875 |
| NM_001048148 | LIME1 | zinc finger, CCCH-type with G patch domain; Lck interacting transmembrane adaptor 1 |
0 | 7 | 0.007 | 2.249 | 0.839 | 2.597 |
| NM_001159533 | CACNA1C | calcium channel, voltage-dependent, L type, alpha 1C subunit |
0 | 4 | 0.011 | 2.982 | 1.182 | 2.978 |
| NM_001039511 | IVNS1ABP | influenza virus NS1A binding protein | 0 | 4 | 0.012 | 2.829 | 0.958 | 3.059 |
| NM_019701 | CLCNKB | chloride channel Kb | 4 | 4 | 0.013 | 0.518 | 0.490 | 0.407 |
| NM_001162415 | Pfkfb2 | 6-phosphofructo-2-kinase/fructose-2,6- biphosphatase 2 |
0 | 2 | 0.013 | 3.314 | 1.523 | 3.197 |
| NM_001109909 | SRRT | serrate RNA effector molecule homolog (Arabidopsis) |
0 | 3 | 0.014 | 2.687 | 1.331 | 2.641 |
| NM_024229 | PCYT2 | phosphate cytidylyltransferase 2, ethanolamine |
3 | 2 | 0.016 | 2.609 | 1.372 | 2.420 |
| NM_001039586 | glyctk | glycerate kinase | 6 | 0 | 0.017 | 0.728 | 0.764 | 0.495 |
| NM_145439 | TMC6 | transmembrane channel-like gene family 6 |
0 | 3 | 0.018 | 2.610 | 1.050 | 2.004 |
| NM_026585 | D6Wsu116e | DNA segment, Chr 6, Wayne State University 116, expressed |
0 | 16 | 0.018 | 1.926 | 1.089 | 2.208 |
| NM_001038010 | KAT2A | K(lysine) acetyltransferase 2A | 0 | 2 | 0.019 | 2.178 | 1.013 | 2.267 |
| NM_021544 | SCN5A | sodium channel, voltage-gated, type V, alpha |
0 | 1 | 0.020 | 1.352 | 1.158 | 2.467 |
| NM_019444 | Ramp2 | receptor (calcitonin) activity modifying protein 2 |
0 | 7 | 0.020 | 1.731 | 0.653 | 2.254 |
| NM_033398 | JMJD6 | jumonji domain containing 6 | 0 | 5 | 0.028 | 2.781 | 2.634 | 2.579 |
| NM_001033150 | Plekhm2 | pleckstrin homology domain containing, family M (with RUN domain) member 2 |
1 | 2 | 0.028 | 2.105 | 1.403 | 2.024 |
| NM_001081290 | Bat2l2 | BAT2 domain containing 1; predicted gene 4972 |
0 | 9 | 0.028 | 4.793 | 1.132 | 6.642 |
| NM_001136071 | Lsp1 | lymphocyte specific 1 | 0 | 6 | 0.031 | 1.252 | 0.686 | 2.063 |
| NM_001163290 | cabc1 | chaperone, ABC1 activity of bc1 complex like (S. pombe) |
0 | 6 | 0.032 | 3.596 | 1.925 | 3.635 |
| NM_001166506 | sec14l1 | SEC14-like 1 (S. cerevisiae) | 1 | 4 | 0.033 | 2.260 | 1.500 | 2.126 |
| NM_001170744 | CTBP2 | C-terminal binding protein 2 | 0 | 6 | 0.035 | 2.647 | 1.867 | 3.402 |
| NM_148930 | RBM5 | RNA binding motif protein 5 | 0 | 3 | 0.036 | 3.538 | 1.587 | 3.860 |
| NM_024217 | Cmtm3 | CKLF-like MARVEL transmembrane domain containing 3 |
0 | 2 | 0.043 | 1.498 | 1.066 | 2.394 |
| NM_027504 | Prdm16 | PR domain containing 16 | 2 | 4 | 0.044 | 3.507 | 1.726 | 2.903 |
| NM_019676 | Plcd1 | phospholipase C, delta 1 | 0 | 1 | 0.044 | 2.138 | 1.307 | 1.837 |
| NM_145979 | CHD4 | chromodomain helicase DNA binding protein 4 |
0 | 14 | 0.045 | 6.628 | 1.755 | 6.187 |
| NM_146329 | Olfr642 | olfactory receptor 642 | 8 | 0 | 0.046 | 0.489 | 0.525 | 0.339 |
CACNA1C
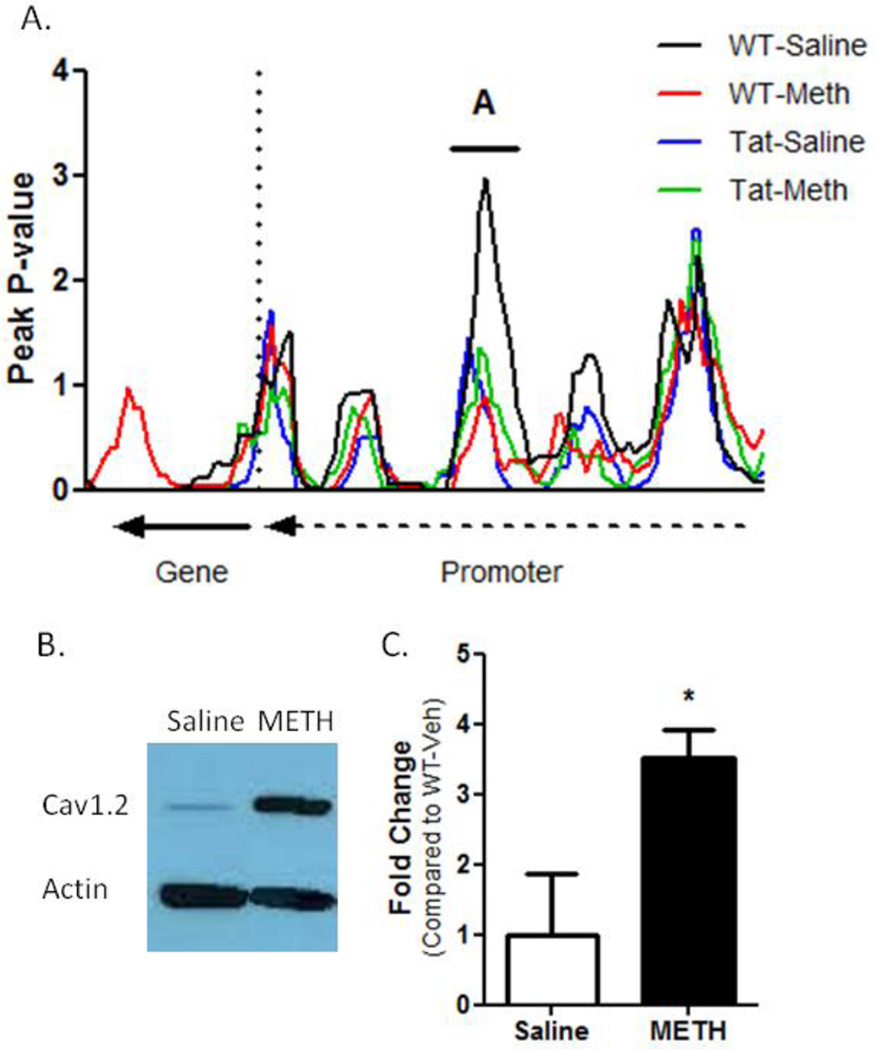
Expression changes were linked to a decrease in DNA methylation resulting from METH treatment (Figure 8A). A promoter region 3.5kb upstream of the CACNA1C transcription start site was hypomethylated following METH treatment in WT and Tat mice, though this peak was only significant for METH groups. Immunoblots were performed using heart protein lysates from WT-saline and WT-METH groups to determine if increased CACNA1C gene expression produced increased Cav1.2 protein abundance (Figure 8B). Cav1.2 protein abundance was elevated in METH-treated WT mice to 350% Cav1.2 abundance compared to WT-saline controls (Figure 8C). METH induces CACNA1C gene expression and protein accumulation in the heart, with reduced DNA methylation observed in the CACNA1C promoter following METH treatment that correlated to gene expression changes.
Figure 8. CACNA1C Regulation.
(A) CACNA1C gene expression increases were associated with a decrease in promoter DNA methylation (denoted as “A” on the figure). This hypomethylation was observed in all experimental groups (though it was only significant in Meth-treated WT and Tat groups) and correlated to qRT-PCR results. (B) Immunoblot results show CACNA1C gene product Cav1.2 was elevated in METH-treated WT mice compared to saline-treated WT mice. (C) Quantitative densitometric analysis of immunoblots show Cav1.2 protein levels are 3.5X greater in METH-treated mice to saline-treated mice.
Histology
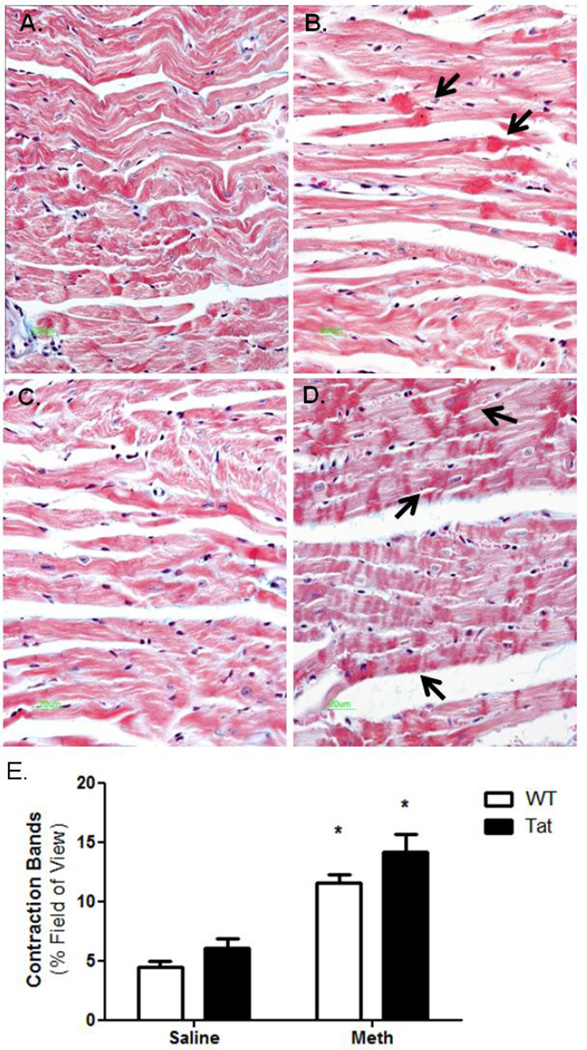
Histopathological analysis was used to identify features of hypercontractility such as contraction bands. Sections of LV tissue were stained with Masson’s trichrome to identify pathohistological changes in Tat and METH-treated mice. Histological analysis revealed extensive foci consistent with contraction bands in METH-treated mice, independent of Tat genotype (Figure 9). Contraction bands have been identified in hypercontracted LV tissue following drugs of abuse such as METH (Islam, 1995; Matoba, 2001). These findings are consistent with previous reports of METH-induced contraction bands and show changes (histological and molecular) associated with METH abuse can be seen prior to significant cardiac dysfunction visualized by echocardiography.
Figure 9. Contraction band necrosis.
Left ventricle sections of experimental mice were sectioned and stained with Masson’s trichrome. A) WT-saline, B) WT-METH, C) Tat-saline, D) Tat-METH. Images revealed a significant increase in the number of identifiable contraction bands in METH-treated mice. Black arrows note areas of contraction bands. Using Image J and computational morphometry, the abundance of contraction bands was determined. METH exposure significantly increased the number of contraction bands in LV sections, while Tat elicited a modest, though insignificant, increase in contraction bands. * denotes p<0.05 compare to WT-saline mice.
DISCUSSION
Our results demonstrate novel molecular changes in a murine model of HIV/AIDS exposed to METH for relatively brief, daily duration (10d). In clinical substance abuse, the problem is significant because METH addicts are at a higher risk for contracting HIV-1 infection and for exhibiting cardiac dysfunction (Bell, 2002; Estrada, 2002). The effects of these two comorbidities are not completely understood clinically, and their comparative and relative impact is both underexplored partially because it is difficult to examine such data meaningfully in that setting. In contrast, our findings from an authentic HIV/AIDS model coupled with controlled administration of METH present a physiological, molecular, and genetic analysis of cardiac dysfunction as may occur in humans with co-morbidities of METH abuse and clinical HIV/AIDS. In a relatively short-term METH administration paradigm (10d), most changes of cardiac dysfunction and gene expression are mediated by METH compared to HIV-Tat. The long-term effects of this interaction remain to be studied in detail.
Short-term METH abuse is associated with arrhythmias and sudden death resulting from overdose; METH also is associated with CM, though this is more likely related to repeated abuse of longer duration (Derlet and Horowitz, 1995; Kaye, 2007; Yu, 2003). In that regard, our mouse model does not develop CM after 10 day METH administration; however, the pathophysiological events observed in METH administration resemble those seen with amphetamine toxicity or abuse, and attest to the chemical and pharmacological similarities between these catecholamine-like drugs (Carvalho, 2012).
As pertinent negative information, echocardiographic results revealed no change in LV mass, LVEDD, or LV fractional shortening (Figures 1A, B, and C, respectively). The small but significant increase in ejection fraction observed in METH-treated WT mice is a reasonable expected outcome in the scenario of METH treatment (Figure 1D). In further support, elevated heart rate (by ECG) was consistently found with METH administration and was correlated with an observed decrease in HRV (Figures 2A and B, respectively). The significance of this latter finding is unclear in absence of overt CM or other diseases. (1996; Radaelli, 2014). Interestingly, the echocardiographic effects of Tat and METH co-administration (10d) appeared to counteract each other suggesting that there are compensatory events ongoing that warrant further exploration (Figures 1A–D). It is worth noting that Tat alone increased heart rate and HRV independent of METH exposure (Figures 2A and B, respectively). This finding would be consistent with additional cardiac stress resulting from Tat-mediated oxidative stress, which is known to cause CM. (Raidel, 2002)
Our results also demonstrate mitochondrial dysfunction in METH-treated mice. Mitochondrial State IVo respiration was diminished in METH-treated mice and was unaltered in face of Tat transgenesis (Figure 3). The effect of METH on mitochondrial respiration does not appear to be mediated by mtDNA genes encoding ETC proteins; no changes were observed in mtDNA abundance (Figure 4A). Analysis of individual ETC complexes revealed significant changes in complexes 2 and 3. Tat stimulates ETC complex 2; superimposition of METH ameliorates this increase (Figure 4B). Results from ETC complex 2/3 assays are contrasting. Tat and METH individually result in reducing complex 2/3 activity; METH treatment of Tat mice increases ETC complex 3 activity. Taking these data in toto, it may be reasonable to consider that that effects observed in ETC complex 2/3 assay indicate a deficiency in ETC complex 3. Mitochondrial dysfunction in ETC complexes 2 and/or 3 here leads to reduced mitochondrial respiration.
Gene expression arrays and DNA methylation arrays together provide novel insights into the molecular pathways affected by METH and HIV/AIDS in the heart. We identified 880 differentially regulated genes (Figure 5). Gene ontology results pointed toward calcium dysfunction in Tat and METH-treated mice, specifically highlighted by an increase in CACNA1C gene expression (Figure 6C). Immunoblots confirmed that Cav1.2 protein abundance was increased in METH-treated WT mice (Figure 8B). DNA methylation arrays point to changes in CACNA1C gene promoter methylation as a contributing factor in CACNA1C gene upregulation.
We localized region in the promoter of CACNA1C that was significantly hypomethylated in METH-treated mice and reduced but not significant in Tat-mice. It is interesting to hypothesize that this region of differential DNA methylation is a regulatory region of CACNA1C gene expression, but we do not have data to adequately support this hypothesis. DNA methylation results show a low correlation between DNA methylation changes and gene expression results observed by microarrays, with only 11 of 105 Tat gene promoters (10%) and 27 of 75 METH gene promoters (36%) showing DNA methylation changes with corresponding gene expression changes. It is possible that DNA methylation changes do not serve as epigenetic regulation of the region but rather are a side-effect of chromatin remodeling and transcriptional changes in these regions or that other epigenetic events (like histone methylation) are modulating DNA methylation here. It is also possible that the DNA methylation observed in the CACNA1C promoter is the highly specific and targeted response following METH. Further investigation into the role of this promoter region is warranted. The reliability of DNA methylation changes found between sample groups suggests the possibility that DNA methylation changes in specific gene promoters may be a useful diagnostic marker of disease or toxicity like those in METH-induced or Tat-induced cardiac changes.
Changes in calcium regulation in the heart appear as a relatively early marker of Tat and METH-induced heart disease in the model system here. It is provocative to note that CACNA1C is a critical moiety for heart development, and conversely, mutations in CACNA1C are associated with dysrhythmic syndromes (Timothy syndrome and Brugada syndrome) characterized by cardiac developmental abnormalities, heart failure, and sudden death. These are features found in METH overdose (Simms, 2014; Splawski, 2005). Findings here highlight the importance of CACNA1C abundance and dysfunction in the adult heart. Mitochondrial dysfunction contribute to alterations in intracellular calcium handling and promotes dysrhythmia and heart failure (Yang, 2014). Increased cytosolic calcium uptake may lead to the dysfunctional cardiac hypercontractility and pathological contraction bands (Figure 9). Our studies implicate CACNA1C as a rational target of short-term METH that exhibits differential expression and coordinated changes in DNA methylation.
In summary, Tat and METH alter gene expression and DNA methylation in the murine heart and serve as an in vivo model of acute, repeated effects of METH on the heart in HIV/AIDS patients. The effects of METH appear more pronounced than those of Tat with respect to gene expression changes and mitochondrial functional changes in the 10d period examined here. Additional experiments are planned to define whether a longer treatment period causes Tat-mediated effects on the heart to overcome METH toxicity or conversely to amplify toxic cardiac dysfunction. Due to METH (and to a lesser extent Tat) effects on calcium uptake in the heart (i.e. CACNA1C), a number of calcium-related intracellular pathways may be therapeutic targets in preventing METH and/or Tat CM and improving health.
Supplementary Material
Highlights.
HIV-1 Tat and methamphetamine (METH) alter cardiac gene expression and epigenetics.
METH impacts gene expression or epigenetics more significantly than Tat expression.
METH alter cardiac mitochondrial function and calcium signaling independent of Tat.
METH alters DNA methylation, expression, and protein abundance of CACNA1C (Cav1.2).
Acknowledgments
None
These studies were supported by DHHS/NIH NIDA DA030996 and NHLBI HL125262 to WL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None
REFERENCES
- 1.Heart rate variability: standards of measurement, physiological interpretation and clinical use. Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation. 1996;93(5):1043–1065. [PubMed] [Google Scholar]
- 2.Adalsteinsdottir B, Teekakirikul P, Maron BJ, Burke MA, Gudbjartsson DF, Holm H, Stefansson K, DePalma SR, Mazaika E, McDonough B, Danielsen R, Seidman JG, Seidman CE, Gunnarsson GT. A Nationwide Study on Hypertrophic Cardiomyopathy in Iceland: Evidence of a MYBPC3 Founder Mutation. Circulation. 2014 doi: 10.1161/CIRCULATIONAHA.114.011207. [DOI] [PubMed] [Google Scholar]
- 3.Bell JE, Arango JC, Robertson R, Brettle RP, Leen C, Simmonds P. HIV and drug misuse in the Edinburgh cohort. Journal of acquired immune deficiency syndromes. 2002;31(Suppl 2):S35–S42. doi: 10.1097/00126334-200210012-00003. [DOI] [PubMed] [Google Scholar]
- 4.Carvalho M, Carmo H, Costa VM, Capela JP, Pontes H, Remiao F, Carvalho F, Bastos Mde L. Toxicity of amphetamines: an update. Archives of toxicology. 2012;86(8):1167–1231. doi: 10.1007/s00204-012-0815-5. [DOI] [PubMed] [Google Scholar]
- 5.Derlet RW, Horowitz BZ. Cardiotoxic drugs. Emergency medicine clinics of North America. 1995;13(4):771–791. [PubMed] [Google Scholar]
- 6.Eskandari MR, Rahmati M, Khajeamiri AR, Kobarfard F, Noubarani M, Heidari H. A new approach on methamphetamine-induced hepatotoxicity: involvement of mitochondrial dysfunction. Xenobiotica; the fate of foreign compounds in biological systems. 2014;44(1):70–76. doi: 10.3109/00498254.2013.807958. [DOI] [PubMed] [Google Scholar]
- 7.Estrada AL. Epidemiology of HIV/AIDS, hepatitis B, hepatitis C, and tuberculosis among minority injection drug users. Public health reports. 2002;117(Suppl 1):S126–S134. [PMC free article] [PubMed] [Google Scholar]
- 8.Fang Q, Kan H, Lewis W, Chen F, Sharma P, Finkel MS. Dilated cardiomyopathy in transgenic mice expressing HIV Tat. Cardiovascular toxicology. 2009;9(1):39–45. doi: 10.1007/s12012-009-9035-5. [DOI] [PubMed] [Google Scholar]
- 9.Flores SC, Marecki JC, Harper KP, Bose SK, Nelson SK, McCord JM. Tat protein of human immunodeficiency virus type 1 represses expression of manganese superoxide dismutase in HeLa cells. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(16):7632–7636. doi: 10.1073/pnas.90.16.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goonasekera SA, Hammer K, Auger-Messier M, Bodi I, Chen X, Zhang H, Reiken S, Elrod JW, Correll RN, York AJ, Sargent MA, Hofmann F, Moosmang S, Marks AR, Houser SR, Bers DM, Molkentin JD. Decreased cardiac L-type Ca(2)(+) channel activity induces hypertrophy and heart failure in mice. The Journal of clinical investigation. 2012;122(1):280–290. doi: 10.1172/JCI58227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He SY, Matoba R, Fujitani N, Sodesaki K, Onishi S. Cardiac muscle lesions associated with chronic administration of methamphetamine in rats. The American journal of forensic medicine and pathology. 1996a;17(2):155–162. doi: 10.1097/00000433-199606000-00014. [DOI] [PubMed] [Google Scholar]
- 12.He SY, Matoba R, Sodesaki K, Fujitani N, Ito Y. Morphological and morphometric investigation of cardiac lesions after chronic administration of methamphetamine in rats. Nihon hoigaku zasshi = The Japanese journal of legal medicine. 1996b;50(2):63–71. [PubMed] [Google Scholar]
- 13.Henry BL, Minassian A, Perry W. Effect of methamphetamine dependence on heart rate variability. Addiction biology. 2012;17(3):648–658. doi: 10.1111/j.1369-1600.2010.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodges GJ, Gros R, Hegele RA, Van Uum S, Shoemaker JK, Feldman RD. Increased blood pressure and hyperdynamic cardiovascular responses in carriers of a common hyperfunctional variant of adenylyl cyclase 6. The Journal of pharmacology and experimental therapeutics. 2010;335(2):451–457. doi: 10.1124/jpet.110.172700. [DOI] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Islam MN, Kuroki H, Hongcheng B, Ogura Y, Kawaguchi N, Onishi S, Wakasugi C. Cardiac lesions and their reversibility after long term administration of methamphetamine. Forensic science international. 1995;75(1):29–43. doi: 10.1016/0379-0738(95)01765-b. [DOI] [PubMed] [Google Scholar]
- 17.Kaye S, McKetin R, Duflou J, Darke S. Methamphetamine and cardiovascular pathology: a review of the evidence. Addiction. 2007;102(8):1204–1211. doi: 10.1111/j.1360-0443.2007.01874.x. [DOI] [PubMed] [Google Scholar]
- 18.Koczor CA, Lee EK, Torres RA, Boyd A, Vega JD, Uppal K, Yuan F, Fields EJ, Samarel AM, Lewis W. Detection of differentially methylated gene promoters in failing and nonfailing human left ventricle myocardium using computation analysis. Physiological genomics. 2013a;45(14):597–605. doi: 10.1152/physiolgenomics.00013.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koczor CA, Torres RA, Fields E, Qin Q, Park J, Ludaway T, Russ R, Lewis W. Transgenic mouse model with deficient mitochondrial polymerase exhibits reduced state IV respiration and enhanced cardiac fibrosis. Laboratory investigation; a journal of technical methods and pathology. 2013b;93(2):151–158. doi: 10.1038/labinvest.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koczor CA, Torres RA, Fields EJ, Boyd A, He S, Patel N, Lee EK, Samarel AM, Lewis W. Thymidine kinase and mtDNA depletion in human cardiomyopathy: epigenetic and translational evidence for energy starvation. Physiological genomics. 2013c;45(14):590–596. doi: 10.1152/physiolgenomics.00014.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis W, Day BJ, Kohler JJ, Hosseini SH, Chan SS, Green EC, Haase CP, Keebaugh ES, Long R, Ludaway T, Russ R, Steltzer J, Tioleco N, Santoianni R, Copeland WC. Decreased mtDNA, oxidative stress, cardiomyopathy, and death from transgenic cardiac targeted human mutant polymerase gamma. Laboratory investigation; a journal of technical methods and pathology. 2007;87(4):326–335. doi: 10.1038/labinvest.3700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liou CM, Tsai SC, Kuo CH, Williams T, Ting H, Lee SD. Chronic methamphetamine exposure induces cardiac fas-dependent and mitochondria-dependent apoptosis. Cardiovascular toxicology. 2014;14(2):134–144. doi: 10.1007/s12012-013-9237-8. [DOI] [PubMed] [Google Scholar]
- 23.Lord KC, Shenouda SK, McIlwain E, Charalampidis D, Lucchesi PA, Varner KJ. Oxidative stress contributes to methamphetamine-induced left ventricular dysfunction. Cardiovascular research. 2010;87(1):111–118. doi: 10.1093/cvr/cvq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matoba R. [Cardiac lesions in methamphetamine abusers] Nihon hoigaku zasshi = The Japanese journal of legal medicine. 2001;55(3):321–330. [PubMed] [Google Scholar]
- 25.Matsuo A, Ikematsu K, Nakasono I. C-fos, fos-B, c-jun and dusp-1 expression in the mouse heart after single and repeated methamphetamine administration. Legal medicine. 2009;11(6):285–290. doi: 10.1016/j.legalmed.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Neubauer S. The failing heart--an engine out of fuel. The New England journal of medicine. 2007;356(11):1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 27.Nickel A, Loffler J, Maack C. Myocardial energetics in heart failure. Basic research in cardiology. 2013;108(4):358. doi: 10.1007/s00395-013-0358-9. [DOI] [PubMed] [Google Scholar]
- 28.Radaelli A, Mancia G, Balestri G, Rovati A, Anzuini A, Di Rienzo M, Paolini G, Castiglioni P. Cardiovascular variability is similarly altered in coronary patients with normal left ventricular function and in heart failure patients. Journal of hypertension. 2014 doi: 10.1097/HJH.0000000000000312. [DOI] [PubMed] [Google Scholar]
- 29.Raidel SM, Haase C, Jansen NR, Russ RB, Sutliff RL, Velsor LW, Day BJ, Hoit BD, Samarel AM, Lewis W. Targeted myocardial transgenic expression of HIV Tat causes cardiomyopathy and mitochondrial damage. American journal of physiology. Heart and circulatory physiology. 2002;282(5):H1672–H1678. doi: 10.1152/ajpheart.00955.2001. [DOI] [PubMed] [Google Scholar]
- 30.Remick J, Georgiopoulou V, Marti C, Ofotokun I, Kalogeropoulos A, Lewis W, Butler J. Heart failure in patients with human immunodeficiency virus infection: epidemiology, pathophysiology, treatment, and future research. Circulation. 2014;129(17):1781–1789. doi: 10.1161/CIRCULATIONAHA.113.004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simms BA, Souza IA, Zamponi GW. Effect of the Brugada syndrome mutation A39V on calmodulin regulation of Cav1.2 channels. Molecular brain. 2014;7:34. doi: 10.1186/1756-6606-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiegelman BM. Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators. Novartis Foundation symposium. 2007;287:60–63. discussion 63-9. [PubMed] [Google Scholar]
- 33.Splawski I, Timothy KW, Decher N, Kumar P, Sachse FB, Beggs AH, Sanguinetti MC, Keating MT. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(23):8089–8096. doi: 10.1073/pnas.0502506102. discussion 8086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao H, Yang JJ, Shi KH, Deng ZY, Li J. DNA methylation in cardiac fibrosis: New advances and perspectives. Toxicology. 2014;323C:125–129. doi: 10.1016/j.tox.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Yang KC, Bonini MG, Dudley SC., Jr Mitochondria and arrhythmias. Free radical biology & medicine. 2014;71:351–361. doi: 10.1016/j.freeradbiomed.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Q, Montes S, Larson DF, Watson RR. Effects of chronic methamphetamine exposure on heart function in uninfected and retrovirus-infected mice. Life sciences. 2002;71(8):953–965. doi: 10.1016/s0024-3205(02)01769-1. [DOI] [PubMed] [Google Scholar]
- 37.Yu Q, Larson DF, Watson RR. Heart disease, methamphetamine and AIDS. Life sciences. 2003;73(2):129–140. doi: 10.1016/s0024-3205(03)00260-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.