Abstract
Background
Clostridium difficile is a natural resident of the intestinal microbiota; however, it becomes harmful when the normal intestinal microbiota is disrupted, and overgrowth and toxin production occurs. The toxins can cause bloating and diarrhoea, which may cause severe disease and have the potential to cause outbreaks in hospitals and other healthcare settings. Normally, antibiotic agents are used for treatment, although for some of the patients, these treatments provide only a temporary relief with a recurrence of C. difficile–associated diarrhoea.
Objective
The effects of polydextrose (PDX), Lactobacillus acidophilus NCFM, and L. paracasei Lpc-37 on the growth of C. difficile were investigated in an in vitro model of infected human large intestine.
Design
The semi-continuous colonic model is composed of four connected vessels inoculated with human faecal microbes and spiked with pathogenic C. difficile (DSM 1296). PDX in two concentrations (2 and 4%), NCFM, and Lpc-37 were fed to the system during the 2-day simulation, and the growth of C. difficile and several other microbial groups were monitored using quantitative polymerase chain reaction (qPCR) and 16S rDNA sequencing.
Results
The microbial community structure of the simulation samples was closely grouped according to treatment, and the largest shifts in the microbial composition were seen with PDX. The microbial diversity decreased significantly with 4% PDX, and the OTU containing C. difficile was significantly (p<0.01) decreased when compared to control and lactobacilli treatments. The mean numbers of C. difficile also decreased as detected by qPCR, although the reduction did not reach statistical significance.
Conclusions
The treatments influenced the colonic microbiota, and a trend for reduced numbers of C. difficile as well as alterations of several microbial groups could be detected. This suggests that PDX may be able to modulate the composition and/or function of the colonic microbiota in such manner that it affects the pathogenic C. difficile.
Keywords: Clostridium difficile, pathogenic growth suppression, in vitro model, 16S, qPCR
Clostridium difficile is a natural resident of the intestinal microbiota in infants (up to 70%) (1) and adults (up to 17%) (2), but it is also the most common infectious agent in antibiotic-associated diarrhoea (AAD) (3). AAD occurs in about 10% of patients receiving antibiotics, of which C. difficile infection (CDI) accounts for 20% (4), when the antibiotic treatment offers an opportunity for the pathogen to increase in numbers. In addition, C. difficile produce toxins that can cause bloating and diarrhoea. A dramatic change in the epidemiology of CDI has occurred during the last two decades. From being considered as an easily treated side effect of antibiotic usage, it is now concomitant with severe nosocomial disease outbreaks with increased morbidity and mortality. Most individuals with C. difficile symptoms are elderly (>60 years), and increasing age has been identified as a risk factor for both C. difficile acquisition and C. difficile–associated diarrhoea (CDAD) (5). Moreover, elderly subjects are more likely to require antibiotic treatment, and community-acquired CDAD appears to occur more commonly among elderly (6).
Polydextrose (PDX) is a randomly linked oligomer synthesised by condensation of glucose. PDX is used as a food ingredient classified as soluble fibre by the US Food and Drug Administration (FDA), Health Canada as well as the European Union. It is commonly used to increase the dietary fibre content of food, to provide bulk in the replacement of sugar, and to reduce calories and fat content. PDX is only partially fermented by the gut microbiota and has been shown to develop a favourable intestinal microbiota (bifidobacteria and lactobacilli) as well as induce an increased production of short-chain fatty acids (SCFAs), including butyrate, through microbial fermentation in the colon, and thus, decrease colonic pH (7). Probiotics on the other hand are able to inhibit, displace, and compete with pathogens, although these abilities are strain-dependent. Lactobacillus acidophilus NCFM has been shown to have antimicrobial activity (8), as well as the ability to inhibit the adhesion of and displace pathogens (9). Lactobacillus paracasei Lpc-37 has been shown to pose immunomodulatory properties in adults (10, 11), suggested as being able to alter the activity of intestinal microbiota (6) and reduce the risk for community-acquired diarrhoea in children (12).
Different animal and in vitro models have been providing insight into disease pathophysiology for several decades (13–15). Experimental studies using animal models have major advantages over clinical studies, such as availability of study subjects, standardisation of disease severity, the ability to perform invasive tests, extensive tissue sampling, and the possibility to test novel treatment strategies. However, in vitro models of C. difficile have evolved into a practical alternative to animal models in many respects. These models offer the advantages of a high degree of control, larger numbers of replicates, as well as elimination of ethical issues arising with animal models. However, the interaction of host-related factors, for example, immunological events in the disease process, cannot be represented by the in vitro models.
The European Food Safety Authority (EFSA) has published an opinion that states that reducing the numbers of specific pathogenic microbes in the colon, for example, C. difficile, is a beneficial physiological effect as such and contributes to reduced risk for gastrointestinal infections (16). The objective of the present study was to investigate the effects of two probiotics (L. acidophilus NCFM and L. paracasei Lpc-37) and a prebiotic (PDX) on the growth of pathogenic C. difficile, by using an in vitro model of the human large intestine infected by C. difficile.
Materials and methods
Human colon simulator
The Enteromix model of the human large intestine has been previously used in several different studies that have investigated the fermentation and function of prebiotic and probiotic ingredients in the colon (17–19). The simulator consists of four different units, each of which contains four semi-continuously connected glass vessels. Therefore, it is possible to run quadruple simulations simultaneously (i.e. to analyse four different treatments in parallel). The vessels in one unit (V1–V4) model the different compartments of the human colon from proximal to the distal part, each having a different pH and flow rates. At the start of a simulation, each unit was inoculated with pre-incubated faecal microbes, which form the microbiota of the colonic model. In this study, fresh faecal inoculum from healthy volunteers was spiked with viable C. difficile (DSM 1296; 1.8×106±2.25×105 CFU/ml) before inoculation. Faecal donors had not used antibiotics or laxatives at least 3 months before donation, nor had they used probiotic bacteria 3 weeks before donation. Two concentrations of PDX (DuPont Reigate, UK) (2 and 4%), Lactobacillus acidophilus NCFM (ATCC 700396, 3.6×108±1.5×108 CFU/ml), and L. paracasei Lpc-37 (ATCC SD5275, 2×107±1.9×107 CFU/ml) were tested. The test substrates were added to a synthetic medium (20) and fed in 3-h cycles to simulated colon model for 48 h, during which transition of fermented fluids and microbes and feeding of fresh medium occurred. Anaerobic conditions were maintained by flushing the simulator with gaseous nitrogen. At the end of simulations, the microbial slurry was collected from all vessels and stored frozen until subjected to microbial and metabolic analyses to investigate the composition and function of the microbiota. Simulations were performed at least in triplicate for each test component and control.
Flow cytometry and quantitative polymerase chain reaction
Flow cytometry was utilised as previously described (21) for enumeration of C. difficile cultures used for spiking of the simulations as well as the L. acidophilus and L. paracasei cultures, while quantitative polymerase chain reaction (qPCR) was used for quantification of the following bacterial groups or species as previously described: Bifidobacterium spp, C. difficile, Clostridium leptum subgroup IV, Clostridium coccoides/Eubacterium rectale (clostridium cluster XIVa), Bacteroidetes, Lactobacillus spp., L. acidophilus, L. paracasei, Faecalibacterium prausnitzii, and Roseburia intestinalis(19, 20, 22–25).
Barcoded 16S rRNA amplicon sequencing
Genomic DNA was isolated using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI), and the microbial community composition was analysed using barcoded pyrosequencing. For each simulation, the donor inoculum and a control (no test substrates) were sequenced in addition to samples containing the test substrates. The V4 region of the 16S rRNA gene of Bacteria and Archaea was amplified using primers 515F (5′ GTGCCAGCMGCCGCGGTAA 3′) and R806 (5′ GGACTACVSGGGTATCTAAT 3′) including sequencing adapters (Roche Applied Sciences, Indianapolis, IN) and a unique 10-bp barcode that was added to the reverse primer (26). The 25 µl polymerase chain reaction (PCR) reaction contained 2 µl of genomic DNA, a 2× concentration of HotMaster Taq polymerase (5Prime, Gaithersburg, MD), 10 µM of each primer, and PCR-grade water to 25 µl. The PCR reactions were amplified using the following conditions: initial denaturation at 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 55°C for 60 s, 70°C for 60 s, and final extension at 70°C for 4 min. Triplicate PCR reactions were pooled and checked for amplification by gel electrophoresis, followed by PCR purification using the QIAquick PCR purification kit (QIAGEN, Valencia, CA) and quantification using the Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA). The barcoded PCR products were pooled in equimolar amounts from each sample, gel purified using the QIAquick Gel Extraction kit (QIAGEN) and sequenced at the W. M. Keck Center for Biotechnology at the University of Illinois using the Roche GS-FLX 454 pyrosequencer with Titanium chemistry (Roche Applied Sciences, Indianapolis, IN).
The sequencing data were analysed using the QIIME (v. 1.6) pipeline (27). The sequences were de-multiplexed and filtered for quality using the following criteria: no more than two mismatches in the primer sequence, minimum quality score of 25, maximum of six homopolymers, no ambiguous bases allowed, and a minimum sequence length of 200 bp. The sequences were clustered de novo into operational taxonomic units (OTUs) at a 97% similarity level using UCLUST (28), aligned to the Greengenes core set (gg_12_10_otus; available from www.greengenes.lbl.gov/) using PyNAST (29), filtered for alignment gaps, and assigned a taxonomic identification by retraining the Ribosomal Database Project (RDP) Classifier (30) with the Greengenes core set.
Metabolite analysis
Metabolites produced by the microbiota during the 2-day simulation were analysed with high-performance liquid chromatography (HPLC) and gas chromatography (GC), as previously described (31). The concentrations of SCFA, branched fatty acids (BCFA), lactic acid, and biogenic amines (BA) were determined.
Statistical analyses
The statistical differences between the treatments were determined using paired t-test and obtained p-values of 0.05 or less were considered statistically significant. Comparisons were made against the control simulations. In colon fermentation studies, the samples from all vessels, V1–V4, were treated as one group when comparing the treatments to each other. Statistical analyses for the 16S rDNA pyrosequencing were done in the QIIME pipeline. The relative abundance of the microbial taxa was calculated as a percentage of the total sequence reads for each sample, and a false discovery rate (FDR)–corrected ANOVA (32) was used to identify differences among the groups. Beta diversity was calculated using unweighted UniFrac (33) on a rarefied (5,348 sequences per sample) OTU table and visualised using principal coordinate analysis (PCoA). The significance of sample groupings based on the distance matrix was tested with permutational multivariate analysis of variance (PERMANOVA) (34) using 999 permutations for derivation of p-values. For all tests, p<0.05 was considered statistically significant.
Results
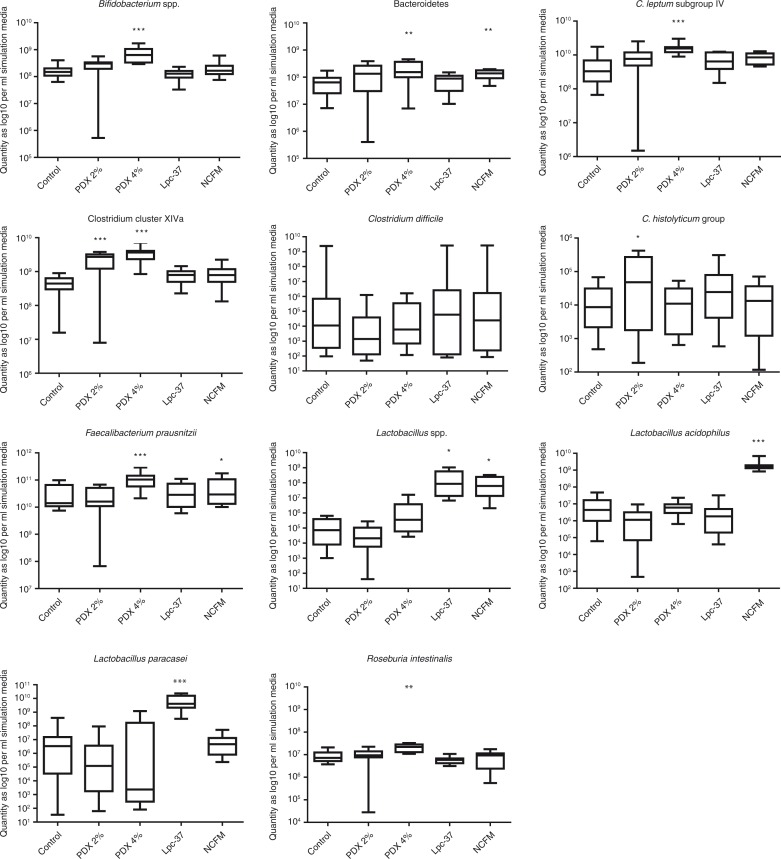
Based on results from the qPCR assays, the two Lactobacillus strains did not have any significant effects on the growth of C. difficile in the colonic model, as the numbers of this pathogenic microbe were similar to the levels measured in the control simulations (Fig. 1). Differences were detected within a simulation unit, especially for Lpc-37 where the numbers of C. difficile were fluctuating between the vessels. PDX had some effects on the growth of this pathogenic microbe: the average numbers of C. difficile decreased with both concentrations (2 and 4%) with more than one log (from 8.5–9.0 log10 to 4.5–6.9 log10/ml simulation media in average) in the distal part of the colonic model (vessels V3 and V4, data not shown). However, this reduction was not statistically significant. The effect of these substrates on the growth of other microbial groups was also investigated. The 4% concentration of PDX had significant effects on the growth of Bifidobacterium (p<0.001), C. leptum subgroup IV (p<0.001), clostridium cluster XIVa (p<0.001), Bacteroidetes (p<0.01), F. prausnitzii (p<0.001), and R. intestinalis (p<0.01). A less pronounced effect was detected for the 2% PDX concentration, for which a small increase in the numbers of Clostridium histolyticum (p<0.05) and clostridium cluster XIVa members (p<0.001) were detected. The C. histolyticum group contains pathogenic C. difficile in addition to the other non-pathogenic clostridia. The tested probiotic lactobacilli had fewer effects than PDX on the colonic model microbiota composition. Both strains increased the numbers of total lactobacilli, as expected, as well as strains resembling L. acidophilus NCFM and L. paracasei Lpc-37, but other microbial groups remained mainly unaffected. Only the abundance of Bacteroidetes (p<0.01) and F. prausnitzii (p<0.05) was increased by NCFM on the whole length of the colon model.
Fig. 1.
The quantity of microbial groups/species as log 10 per ml simulation media measured by qPCR showed as box plots for the V1–V4 vessels combined. The top and the bottom of the box represent upper and lower quartiles, respectively, and the band within the box is the median. The whiskers indicate the minimum and the maximum. Significant data are marked with asterisks, *p<0.05, **p<0.01, and ***p<0.001.
Characterisation of the colonic microbiota
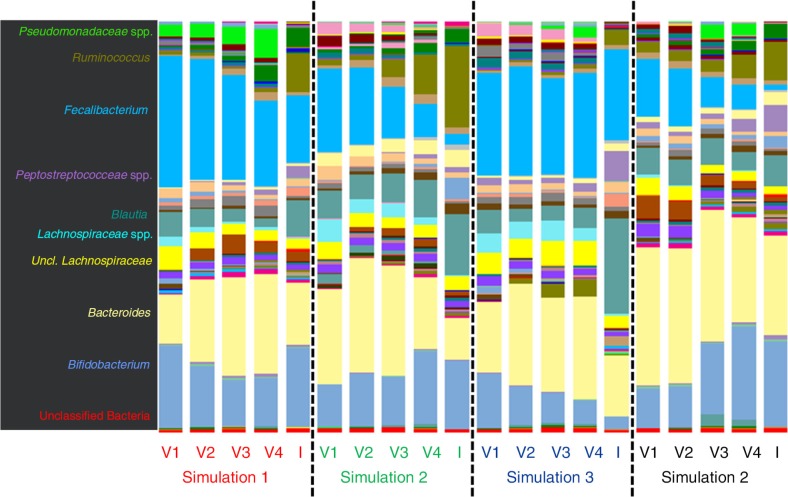
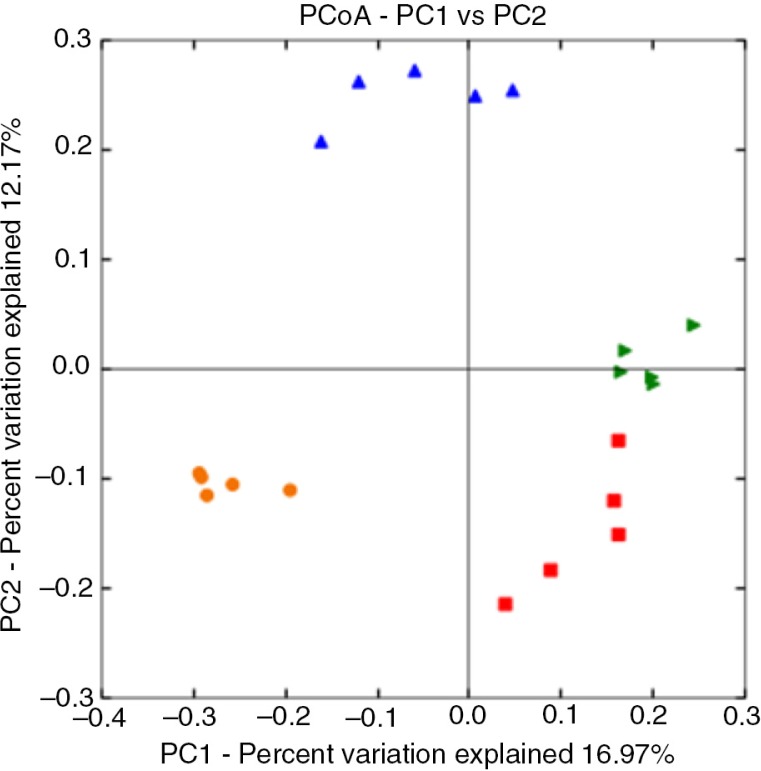
The relative abundance of microbial taxa derived from 16S rDNA pyrosequencing was compared between the donor faecal inoculum samples and the control vessels from each simulation to validate the model system (Fig. 2). Overall, the community composition was relatively consistent between the donor inoculum and control samples from each simulation (Fig. 3), as shown by significant (PERMANOVA, p=0.001) sample clustering by simulation based on the unweighted UniFrac distances. However, it was noted that the Pseudomonadaceae were in higher abundance in the control samples compared to the inoculum, and Ruminococcus was less prevalent in the controls compared to the inoculum. This may indicate that the simulation conditions in the vessels were not ideal for the growth of Ruminococcus species.
Fig. 2.
Donor faecal inoculum (I) samples compared to the control for each vessel (V1–V4) at genus-level identification by 16S rDNA pyrosequencing. For simulations 1 and 4, the same donor sampled 6 months ago was used.
Fig. 3.
Principal coordinate plot of un-weighted UniFrac distances for donor faecal inoculum and control samples. Samples are coloured according to simulation 1 (red), 2 (blue), 3 (orange), and 4 (green). Sample clustering by simulation for inoculum and control samples was significant (PERMANOVA with 999 permutations, p=0.001).
At the phylum level of classification, the faecal microbiota in all samples was dominated by Firmicutes (57.2% abundance), followed by Bacteroidetes (23.7%), Actinobacteria (13.2%), Proteobacteria (4.9%) and less than 1% Verrucomicrobia, Tenericutes, and Lentisphaerae. Interestingly, the Bacteroidetes were less abundant in the ascending colon (4–5% in vessel V1) of the PDX treatments, but were able to proliferate and increase along the vessels of the simulated colon (32–35% in vessel V4). The relative proportions of the other bacterial phyla were relatively stable across all of the samples (data not shown). There were no Archaea sequences detected in any of the samples.
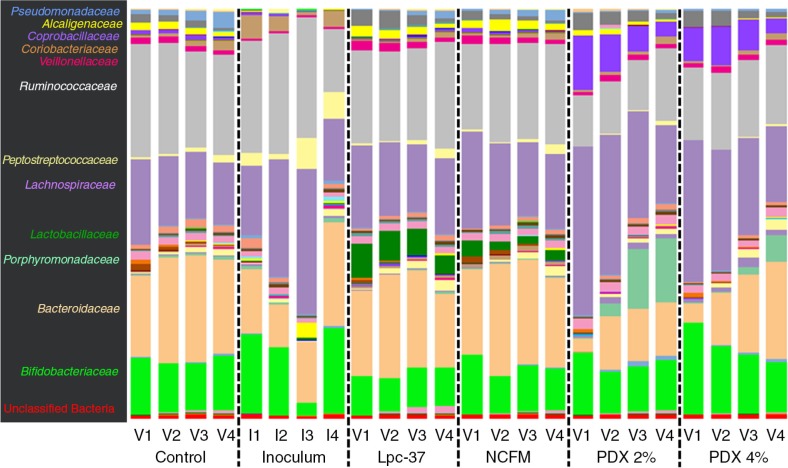
At a finer taxonomic resolution of family level, greater differences amongst the treatments were evident, particularly in the PDX groups (Fig. 4). The proportion of Lactobacillaceae was higher (p<0.001) in the probiotic treatments, as explained by the addition of NCFM and Lpc-37 in those groups. The 16S rDNA sequences from the NCFM and Lpc-37 complete genomes were extracted and compared to those derived from this amplicon study to confirm the presence of both stains in the simulator samples. On average, Lpc-37 comprised 5.66% of the community and NCFM 2.06% in the probiotic groups. Other noted family-level taxonomic shifts were the increased abundance of Coprobacillaceae, Lachnospiraceae, and Porphyromonadaceae and reduced amounts of Ruminococcaceae and Bacteroidaceae in the PDX treatments (p<0.001). The OTUs (minimum 1% abundance) that differed significantly (p<0.05) by group are listed in Table 1. Within the family Lachnospiraceae, several OTUs were increased in PDX including Coprococcus and Blautia spp. along with reduced amounts of Ruminococcus sp. and Bacteroides spp.
Fig. 4.
Family-level taxonomy 16S rDNA pyrosequencing for donor Inoculum samples from simulations 1–4 and control, probiotic (Lpc-37 and NCFM), and prebiotic (PDX2%, PDX 4%) samples from vessels V1–V4.
Table 1.
Relative abundance (%) of OTUs present at least 1% abundance that differed (p<0.05) amongst groups
| OTU | OTU classification | Control | Lpc-37 | NCFM | PDX 2% | PDX 4% |
|---|---|---|---|---|---|---|
| 16155 | Unclassified Coprobacillaceae | 0.52 | 0.19 | 0.42 | 6.80 | 6.37 |
| 6394 | Coprococcus spp. | 0.43 | 0.37 | 0.60 | 3.10 | 4.84 |
| 8547 | Blautia spp. | 0.48 | 0.29 | 0.34 | 1.63 | 3.10 |
| 17045 | Blautia spp. | 2.54 | 2.52 | 2.72 | 5.55 | 2.23 |
| 11070 | Blautia sp. | 0.18 | 0.03 | 0.03 | 0.95 | 1.69 |
| 24251 | Bacteroides spp. | 4.15 | 4.96 | 7.22 | 1.24 | 1.38 |
| 8452 | Parabacteroides sp. | 0.23 | 0.19 | 0.19 | 5.67 | 1.07 |
| 6303 | Ruminococcus sp. | 1.09 | 1.02 | 1.15 | 0.41 | 0.37 |
| 16538 | Lactobacillus sp. (incl. Lpc-37) | 0.01 | 5.66 | 0.01 | 0.01 | 0.01 |
| 14054 | Lactobacillus sp. (incl. NCFM) | 0.01 | 0.00 | 2.06 | 0.00 | 0.00 |
SCFA, BCFA, and BA production
In this study, a significant increase in acetic (p<0.0006), propionic (p<0.0295), and butyric (p<0.0004) acid concentrations could be detected for the lower PDX concentration as compared to the control (Table 2). For the higher PDX concentration, a significant increase could only be detected for butyric acid (p<0.0150). The lower PDX concentration also had a significantly higher (p<0.0076) acetic acid production as compared to the L. acidophilus NCFM. No significant differences could be detected in the concentrations of BA (Table 3). No lactic acid could be detected in any of the samples.
Table 2.
Average (±SD) production of the main SCFAs as well as the branched fatty acids for the different test substances L. acidophilus NCFM; NCFM, L. paracasei Lpc-37; Lpc-37 and polydextrose; PDX during the simulation
| Acetic acid | Propionic acid | Butyric acid | Isobutyric acid | 2-Methylbutyric acid | Isovaleric acid | Valeric acid | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
||||||||
| Sum of vessels | SD | Sum of vessels | SD | Sum of vessels | SD | Sum of vessels | SD | Sum of vessels | SD | Sum of vessels | SD | Sum of vessels | SD | |
| Control | 59.32 | 3.21 | 20.61 | 0.33 | 29.46 | 1.26 | 4.16 | 0.24 | 2.52 | 0.17 | 4.42 | 0.16 | 4.97 | 0.64 |
| NCFM | 90.61a | 4.43 | 30.54 | 0.56 | 61.43 | 0.77 | 4.47 | 0.29 | 2.94 | 0.2 | 5.09 | 0.19 | 4.5 | 0.32 |
| Lpc-37 | 89.38 | 4.62 | 32.78 | 0.72 | 52.93 | 2.46 | 4.53 | 0.09 | 2.87 | 0.22 | 4.80 | 0.22 | 5.32 | 0.25 |
| PDX 2% | 151.40b | 6.32 | 51.65c | 5.45 | 66.79a | 2.38 | 2.26 | 0.31 | 0.47 | 1.66 | 0.19 | 2.82 | 0.52 | |
| PDX 4% | 56.22 | 16.11 | 16.47 | 3.75 | 25.69d | 5.24 | bdl | bdl | bdl | bdl | ||||
Some of the PDX 2% and PDX 4% SCFA were below detection limit (bdl); therefore, no SD values are available. Significant data are marked with letters
PDX 2% vs. NCFM, p<0.001;
PDX 2% vs. control, p<0.001;
PDX 2% vs. control, p<0.05
PDX 4% vs. control, p<0.05.
Table 3.
Average (±SD) production of BA during the colon simulations with the test substances L. acidophilus NCFM; NCFM, L. paracasei Lpc-37; Lpc-37 and polydextrose; PDX
| Methylamine | Putrescine | Cadaverine | Histamine | Tyramine | Spermidine | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|||||||
| Sum of vessels | SD | Sum of vessels | SD | Sum of vessels | SD | Sum of vessels | SD | Sum of vessels | SD | Sum of vessels | SD | |
| Control | 277.9 | 73.0 | 529.3 | 157.4 | 521.6 | 461.1 | 48.5 | 2.1 | 113.8 | 30.5 | 55.5 | 1.9 |
| NCFM | 283.8 | 21.8 | 586.8 | 184.5 | 406.9 | 301.1 | 47.9 | 2.7 | 87.5 | 3.9 | 55.3 | 6.8 |
| Lpc-37 | 282.6 | 160.9 | 535.1 | 311.6 | 625.9 | 532.4 | 48.1 | 30.0 | 116.6 | 64.0 | 58.6 | 36.5 |
| PDX 2% | 225.0 | 28.6 | 194.8 | 73.3 | 483.9 | 183.1 | 43.6 | 6.9 | 129.3 | 35.4 | 79.1 | 8.7 |
| PDX 4% | 216.2 | 569.5 | 91.7 | bdl | 99.9 | bdl | ||||||
For PDX 4%, part of the BA was below detection limit; therefore, no SD values are shown. Some of the PDX 4% BA were below detection limit (bdl).
Discussion
The Enteromix colon simulator mimicking the human large intestine has been developed and used for a broad area of different applications within human health. In the current in vitro study, the effect of two concentrations of PDX (2 and 4%) as well as two Lactobacillus strains (L. acidophilus NCFM and L. paracasei Lpc-37) was examined. The fresh faecal material was spiked with C. difficile and the growth of this pathogen was monitored amongst other microbial groups of the colonic microbiota.
Prebiotic carbohydrates, such as fructo- and galacto-oligosaccharides (FOS and GOS), may have an impact on the resistance to pathogenic microbes and gastrointestinal infections through modifying the intestinal microbiota (35). Many intestinal pathogens utilise mono- or short oligosaccharide sequences on the mucus layer as receptors, and these receptor sites can be mimicked by prebiotic carbohydrates (35). Some studies have demonstrated the effectiveness of prebiotics to reduce the incidence or duration of gastrointestinal infections, such as traveller's diarrhoea and AAD (36–38). In the current study, PDX was shown to reduce the average numbers of pathogenic C. difficile in the distal part of the colonic model; however, the reduction did not reach the level of statistical significance.
Probiotic microbes, such as Lactobacillus and Bifidobacterium strains, may have an impact on the growth of pathogens in a number of different potential mechanisms. First, they are able to produce metabolic end products that lower the gut pH below the levels at which the pathogens are able to effectively compete. Such metabolites are, for example, lactic acid and acetate, but many lactobacilli and bifidobacteria are also able to produce antimicrobial compounds that have direct effects on the pathogenic microbes (35, 39, 40). Bifidobacteria excrete proteinaceous compounds that, alongside with lowered colonic pH, control the growth of a wide range of both Gram-negative and Gram-positive bacteria (41). More recent evidence also demonstrates that especially the acetate produced by bifidobacteria has a significant role in protection against invading pathogenic microbial strains (42). Second, probiotics may also improve the immune stimulation, compete for nutrients, and block binding sites to prevent pathogenic growth (35). In the present study, the probiotic Lactobacillus strains were, however, not able to suppress the growth of C. difficile in the human colon model. This may be due to strain-specific features or possibly too low inoculation levels. However, as the numbers of pathogenic C. difficile were not increased, it may be concluded that the other clostridia in this group were responsible for the increased group numbers measured from qPCR. In previous clinical studies, L. acidophilus NCFM (together with Lactobacillus rhamnosus HN001 in probiotic cheese) was shown to be effective in reducing faecal C. difficile numbers in elderly that harboured this microbe in the beginning of the intervention (22). In a study with L. paracasei Lpc-37 in healthy elderly, although the detected levels of C. difficile were very low, a non-significant decrease could still be detected (25).
PDX in a higher concentration was able to modify the composition of the microbiota significantly, as the growth of several other microbial groups was modulated in the whole length of the colon model. This effect could derive from the slow degradation of PDX; it is known from previous studies that PDX is slowly fermented in the colon and some 10% of the consumed amount is still found in the faeces (43, 44), which differentiates PDX from other rapidly fermented oligosaccharidic prebiotics such as FOS (17). From the 16S pyrosequencing, some species occurred at a higher abundance in the PDX treatment samples including members of Coprobacillaceae, Porphyromonadaceae, and Lachnospiraceae. The OTU that most distinguished the PDX samples belonged to the group Coprobacillaceae. Coprobacillaceae is a proposed new family within the order Erysipelotrichales and little has been published about this group of organisms. The closest taxonomic match to this OTU was Clostridium saccharogumia, but the sequence similarity was less than 90% to the type strain indicating that it may be novel species within this group. C. saccharogumia was originally isolated from human faecal samples and may be involved in the conversion of the secoisolariciresinol diglucoside, a dietary phytoestrogen from plant lignin linked to the prevention of diseases, such as breast and colon cancer, atherosclerosis, and diabetes (45). Porphyromonadaceae was present in the greatest abundance in PDX vessels representing the distal colon (V3–V4). Within this family, an OTU classified as Parabacteroides differed significantly amongst the groups. The closest reference strain match to this OTU was Parabacteroides distasonis, which has shown immunomodulatory effects and decreased intestinal inflammation in DSS-induced murine colitis (46). A significantly higher amount of butyric acid (p<0.01) was detected in the PDX samples, and one of the dominant OTUs differentiating the PDX group was assigned to the genus Coprococcus within Lachnospiraceae. This OTU was found at 3–4% relative abundance in the PDX groups, but at less than 1% abundance in the probiotic and control groups. The closest type strain match to this OTU was Eubacterium hadrum (Anaerostipes hadrus), which is a known butyrate producer. Other members of Lachnospiraceae, such as Blautia were also higher in the PDX treatment. Several recently published papers (47–50) have implicated Lachnospiraceae and butyrogenic bacteria likely play a role in CDI and recovery. In one study, faecal samples from patients with recurring CDI were shown to have reduced amounts of Lachnospiraceae and Ruminococcaceae compared to healthy controls. Following successful faecal microbiota transplantation from a healthy donor, Lachnospiraceae and Ruminococcaceae were restored and comparable to the levels of their healthy counterparts (50). Antharam et al. (47) implicated butyrogenic bacteria belonging to Clostridium clusters IV and XIVa and members of the Lachnospiraceae and Ruminococceae families, as being absent or depleted in patients with nosocomial diarrhoea and CDI. They identified several genera as being the most differentially depleted in the CDI group: Blautia, Pseudobutyrivibrio, Roseburia, Faecalibacterium, Anaerostipes, Subdoligranulum, Ruminococcus, Streptococcus, Dorea, and Coprococcus. Several of these genera also appear to be key taxa in differentiating the PDX treatment in this study. It was also reported that wild-type mice challenged with C. difficile that only developed mild disease were found to have a high proportion of Lachnospiraceae compared to their moribund counterparts. Subsequently, a murine isolate of Lachnospiraceae was able to provide colonisation resistance against C. difficile in germ-free mice (49). Both in vitro and animal studies aimed at directly linking butyrate to the inhibition of C. difficile have yielded mixed results. There are several other plausible modes of action that could explain this correlation, including colonisation resistance (nutrient-niche hypothesis) or production of antimicrobials and metabolites by these closely related organisms. Nonetheless, further research into the role they may play in CDI is warranted, as well as additional studies to determine whether the growth of these bacterial groups would be induced in our model with PDX substrate, but in the absence of C. difficile.
Although the molecular methods (16S rDNA sequencing and qPCR) used here for the microbiota analyses are biased from the PCR and might fail to amplify some targets for unknown reasons, and thus are unable to identify unknown species, the combination of the methods give a good overall picture of the microbiota. Sequencing based on 16S rDNA is a good approach for phylogenetic identification, while certain groups or species can be quantified by qPCR. In this study, although primer sets and target area were different with the methods, a similar prevalence profile of the groups that were detected with both methods was seen, for example, the increase of the lactobacilli for the probiotic treatments. In addition, the increase in L. acidophilus NCFM and L. paracasei Lpc-37 suggests that the strains survived the gastrointestinal model.
Conclusions
The statement from EFSA has highlighted the importance of reducing the numbers of potentially disease-causing microbes or their toxins for the health of the gut. Reducing the numbers of established pathogenic microbes subsequently reduces the risk of gastrointestinal infections and may be a beneficial physiological effect. These in vitro studies give indications that PDX may have beneficial effects on the growth of intestinal C. difficile, as the average numbers of C. difficile were decreased in the last two vessels of the model. Nevertheless, this effect requires further investigations and also possible clinical beneficial outcomes resulting from the reduced numbers of the pathogen should be demonstrated. PDX in the higher concentration (4%) did have significant effects on the composition of the microbiota of the colonic simulator model as the growth of several investigated microbial groups; bifidobacteria, butyrate-producing clostridium groups, R. intestinalis, and F. prausnitzii numbers were significantly increased in the whole length of the colonic model. The lower concentration (2%) of PDX was not as effective in modulating the microbiota composition implying that a high enough dose is needed for PDX to be an active, prebiotic substrate.
Acknowledgements
Markku Saarinen and Krista Salli are thanked for excellent technical assistance.
Conflict of interest and funding
AAH, ACO, and SDF are employees of DuPont. DuPont manufactures and markets the investigated probiotics and polydextrose.
Authors’ contributions
HR and SDF performed the in vitro work and performed the analysis of the results. AAH performed the sequencing part, while ACO participated in planning of the work and analysing the results. All authors have read and approved the final manuscript.
References
- 1.Jangi S, Lamont JT. Asymptomatic colonization by Clostridium difficile in infants: implications for disease in later life. J Pediatr Gastroenterol Nutr. 2010;51:2–7. doi: 10.1097/MPG.0b013e3181d29767. [DOI] [PubMed] [Google Scholar]
- 2.Ozaki E, Kato H, Kita H, Karasawa T, Maegawa T, Koino Y, et al. Clostridium difficile colonization in healthy adults: transient colonization and correlation with enterococcal colonization. J Med Microbiol. 2004;53:167–72. doi: 10.1099/jmm.0.05376-0. [DOI] [PubMed] [Google Scholar]
- 3.Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol. 2004;42:1203–6. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiström J, Norrby SR, Myhre EB, Eriksson S, Granström G, Lagergren L, et al. Frequency of antibiotic-associated diarrhoea in 2462 antibiotic-treated hospitalized patients: a prospective study. J Antimicrob Chemother. 2001;47:43–50. doi: 10.1093/jac/47.1.43. [DOI] [PubMed] [Google Scholar]
- 5.Simor AE, Bradley SF, Strausbaugh LJ, Crossley K, Nicolle LE. Clostridium difficile in long-term-care facilities for the elderly. Infect Control Hosp Epidemiol. 2002;23:696–703. doi: 10.1086/501997. [DOI] [PubMed] [Google Scholar]
- 6.Valerio F, Russo F, de Candia S, Riezzo G, Orlando A, Lonigro SL, et al. Effects of probiotic Lactobacillus paracasei-enriched artichokes on constipated patients: a pilot study. J Clin Gastroenterol. 2010;44:S49–53. doi: 10.1097/MCG.0b013e3181d2dca4. [DOI] [PubMed] [Google Scholar]
- 7.Jie Z, Bang-Yao L, Ming Jie X, Hai-Wei L, Zu-Kang Z, Ting-Song W, et al. Studies on the efffects of polydextrose intake on physiologic functions in Chinese people. Am J Clin Nutr. 2000;72:1503–9. doi: 10.1093/ajcn/72.6.1503. [DOI] [PubMed] [Google Scholar]
- 8.Gilliland SE, Speck ML. Antagonistic action of Lactobacillus acidophilus toward intestinal and foodborne pathogens in associative cultures. J Food Protect. 1977;40:820–3. doi: 10.4315/0362-028X-40.12.820. [DOI] [PubMed] [Google Scholar]
- 9.Collado MC, Meriluoto J, Salminen S. Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Lett Appl Microbiol. 2007;45:454–60. doi: 10.1111/j.1472-765X.2007.02212.x. [DOI] [PubMed] [Google Scholar]
- 10.Roessler A, Friedrich U, Vogelsang H, Bauer A, Kaatz M, Hipler UC, et al. The immune system in healthy adults and patients with atopic dermatitis seems to be affected differently by a probiotic intervention. Clin Exp Allergy. 2008;38:93–102. doi: 10.1111/j.1365-2222.2007.02876.x. [DOI] [PubMed] [Google Scholar]
- 11.Paineau D, Carcano D, Leyer G, Darquy S, Alyanakian M-A, Simoneau G, et al. Effects of seven potential probiotic strains on specific immune responses in healthy adults: a double-blind, randomized, controlled trial. FEMS Immunol Med Microbiol. 2008;53:107–13. doi: 10.1111/j.1574-695X.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 12.Hemalatha R, Ouwehand AC, Forssten SD, Babu Geddan JJ, Sriswan Mamidi R, Bhaskar V, et al. A community-based randomized double blind controlled trial of Lactobacillus paracasei and Bifidobacterium lactis on reducing risk for diarrhea and fever in preschool children in an urban slum in India. Eur J Nutr Food Safety. 2014;4:325–41. [Google Scholar]
- 13.Baines SD, Crowther GS, Todhunter SL, Freeman J, Chilton CH, Fawley WN, et al. Mixed infection by Clostridium difficile in an in vitro model of the human gut. J Antimicrob Chemother. 2013;68:1139–43. doi: 10.1093/jac/dks529. [DOI] [PubMed] [Google Scholar]
- 14.McVay CS, Rolfe RD. In vitro and in vivo activities of nitazoxanide against Clostridium difficile . Antimicrob Agents Chemother. 2000;44:2254–8. doi: 10.1128/aac.44.9.2254-2258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang TW, Bartlett JG, Gorbach SL, Onderdonk AB. Clindamycin-induced enterocolitis in hamsters as a model of pseudomembranous colitis in patients. Infect Immun. 1978;20:526–9. doi: 10.1128/iai.20.2.526-529.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA) Guidance on the scientific requirements for health claims related to gut and immune function. EFSA J. 1984;9:12. doi: http://dx.doi.org/10.2903/j.efsa.2011.1984. Available from: www.efsa.europa.eu/efsajournal. [Google Scholar]
- 17.Mäkeläinen H, Forssten S, Saarinen M, Stowell J, Rautonen N, Ouwehand A. Xylo-oligosaccharides enhance the growth of bifidobacteria and Bifidobacterium lactis in a simulated colon model. Benef Microbes. 2010;1:81–91. doi: 10.3920/BM2009.0025. [DOI] [PubMed] [Google Scholar]
- 18.Mäkeläinen HS, Mäkivuokko H, Salminen SJ, Rautonen NE, Ouwehand AC. The effects of polydextrose and xylitol on microbial community and activity in a 4-stage colon simulator. J Food Sci. 2007;72:M153–9. doi: 10.1111/j.1750-3841.2007.00350.x. [DOI] [PubMed] [Google Scholar]
- 19.Mäkivuokko H, Forssten S, Saarinen M, Ouwehand A, Rautonen N. Synbiotic effects of lactitol and Lactobacillus acidophilus NCFM™ in a semi-continuous colon fermentation model. Benef Microbes. 2010;1:131–7. doi: 10.3920/BM2009.0033. [DOI] [PubMed] [Google Scholar]
- 20.Mäkivuokko H, Nurmi J, Nurminen P, Stowell J, Rautonen N. In vitro effects on polydextrose by colonic bacteria and caco-2 cell cyclooxygenase gene expression. Nutr Cancer. 2005;52:94–104. doi: 10.1207/s15327914nc5201_12. [DOI] [PubMed] [Google Scholar]
- 21.Apajalahti JH, Kettunen H, Kettunen A, Holben WE, Nurminen PH, Rautonen N, et al. Culture-independent microbial community analysis reveals that inulin in the diet primarily affects previously unknown bacteria in the mouse cecum. Appl Environ Microbiol. 2002;68:4986–95. doi: 10.1128/AEM.68.10.4986-4995.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahtinen SJ, Forssten S, Aakko J, Granlund L, Rautonen N, Salminen S, et al. Probiotic cheese containing Lactobacillus rhamnosus HN001 and Lactobacillus acidophilus NCFM(R) modifies subpopulations of fecal lactobacilli and Clostridium difficile in the elderly. Age (Dordr) 2012;34:133–43. doi: 10.1007/s11357-011-9208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuki T, Watanabe K, Fujimoto F, Takada T, Tanaka R. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol. 2004;70:7220–8. doi: 10.1128/AEM.70.12.7220-7228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166–77. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- 25.Forssten SD, Salazar N, López P, Nikkilä J, Ouwehand AC, Patterson Á, et al. Influence of a probiotic milk drink, containing Lactobacillus paracasei Lpc-37, on immune function and gut microbiota in elderly subjects. Eur J Food Res Rev. 2011;1(3):159–72. [Google Scholar]
- 26.Bates ST, Cropsey GWG, Caporaso JG, Knight R, Fierer N. Bacterial communities associated with the lichen symbiosis. Appl Environ Microbiol. 2011;77:1309–14. doi: 10.1128/AEM.02257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 29.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ouwehand AC, Tiihonen K, Saarinen M, Putaala H, Rautonen N. Influence of a combination of Lactobacillus acidophilus NCFM and lactitol on healthy elderly: intestinal and immune parameters. Br J Nutr. 2008;101:367–75. doi: 10.1017/S0007114508003097. doi: http://dx.doi.org/10.1017/S00007114508003097. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- 33.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 35.Gibson GR, McCartney AL, Rastall RA. Prebiotics and resistance to gastrointestinal infections. Br J Nutr. 2005;93(Suppl. 1):S31–4. doi: 10.1079/bjn20041343. [DOI] [PubMed] [Google Scholar]
- 36.Cummings JH, Christie S, Cole TJ. A study of fructo oligosaccharides in the prevention of travellers’ diarrhoea. Aliment Pharmacol Ther. 2001;15:1139–45. doi: 10.1046/j.1365-2036.2001.01043.x. [DOI] [PubMed] [Google Scholar]
- 37.Drakoularakou A, Tzortzis G, Rastall RA, Gibson GR. A double-blind, placebo-controlled, randomized human study assessing the capacity of a novel galacto-oligosaccharide mixture in reducing travellers’ diarrhoea. Eur J Clin Nutr. 2009;64:146–52. doi: 10.1038/ejcn.2009.120. [DOI] [PubMed] [Google Scholar]
- 38.Oli MW, Petschow BW, Buddington RK. Evaluation of fructooligosaccharide supplementation of oral electrolyte solutions for treatment of diarrhea: recovery of the intestinal bacteria. Dig Dis Sci. 1998;43:138–47. doi: 10.1023/a:1018892524790. [DOI] [PubMed] [Google Scholar]
- 39.Martinez FA, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv. 2013;31:482–8. doi: 10.1016/j.biotechadv.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Lew LC, Liong MT. Bioactives from probiotics for dermal health: functions and benefits. J Appl Microbiol. 2013;114:1241–53. doi: 10.1111/jam.12137. [DOI] [PubMed] [Google Scholar]
- 41.Gibson GR, Wang X. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J Appl Bacteriol. 1994;77:412–20. doi: 10.1111/j.1365-2672.1994.tb03443.x. [DOI] [PubMed] [Google Scholar]
- 42.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–7. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 43.Lahtinen SJ, Knoblock K, Drakoularakou A, Jacob M, Stowell J, Gibson GR, et al. Effect of molecule branching and glycosidic linkage on the degradation of polydextrose by gut microbiota. Biosci Biotechnol Biochem. 2010;74:2016–21. doi: 10.1271/bbb.100251. [DOI] [PubMed] [Google Scholar]
- 44.Costabile A, Fava F, Röytiö H, Forssten SD, Olli K, Klievink J, et al. Impact of polydextrose on biomarkers of gut health: a double blind, cross-over, placebo-controlled feeding study in healthy human subjects. Br J Nutr. 2011;108:471–81. doi: 10.1017/S0007114511005782. [DOI] [PubMed] [Google Scholar]
- 45.Clavel T, Lippman R, Gavini F, Dore J, Blaut M. Clostridium saccharogumia sp. nov. and Lactonifactor longoviformis gen. nov., sp. nov., two novel human faecal bacteria involved in the conversion of the dietary phytoestrogen secoisolariciresinol diglucoside. Syst Appl Microbiol. 2007;30:16–26. doi: 10.1016/j.syapm.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 46.Kverka M, Zakostelska Z, Klimesova K, Sokol D, Hudcovic T, Hrncir T, et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin Exp Immunol. 2011;163:250–9. doi: 10.1111/j.1365-2249.2010.04286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol. 2013;51:2884–92. doi: 10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott KP, Martin JC, Duncan SH, Flint HJ. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro . FEMS Microbiol Ecol. 2014;87:30–40. doi: 10.1111/1574-6941.12186. [DOI] [PubMed] [Google Scholar]
- 49.Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun. 2012;80:3786–94. doi: 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song Y, Garg S, Girotra M, Maddox C, von Rosenvinge EC, Dutta A, et al. Microbiota dynamics in patients treated with fecal microbiota transplantation for recurrent Clostridium difficile infection. PLoS One. 2013;8:e81330. doi: 10.1371/journal.pone.0081330. [DOI] [PMC free article] [PubMed] [Google Scholar]