Abstract
Phytohormones are central components of complex signalling networks in plants. The interplay between these metabolites, which include abscisic acid (ABA), auxin (IAA), ethylene, jasmonic acid (JA) and salicylic acid (SA), regulate plant growth and development and modulate responses to biotic and abiotic stress. Few methods of phytohormone profiling can adequately quantify a large range of plant hormones simultaneously and without the requirement for laborious or highly specialised extraction protocols. Here we describe the development and validation of a phytohormone profiling protocol, based on methyl-chloroformate derivatisation of the plant metabolites and analysis by gas chromatography/mass spectrometry (GC–MS). We describe the analysis of 11 metabolites, either plant phytohormones or intermediates of phytohormone metabolism; ABA, azelaic acid, IAA, JA and SA, and the phytohormone precursors 1-aminocyclopropane 1-carboxylic acid, benzoic acid, cinnamic acid, 13-epi-12-oxophytodienoic acid (13-epi-OPDA), linoleic acid and linolenic acid, and validate the isolation from foliar tissue of the model legume Medicago truncatula. The preparation is insensitive to the presence of water, facilitating measurement of the volatile metabolites. Quantitation was linear over four orders of magnitude, and the limits of detection between two and 10 ng/mL for all measured metabolites using a single quadrupole GC–MS.
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-015-0837-0) contains supplementary material, which is available to authorized users.
Keywords: Jasmonic acid, Salicylic acid, Ethylene, Abscisic acid, Idole-3-acetic acid, Azelaic acid, Plant defence, Medicago truncatula
Introduction
Phytohormones regulate important plant processes, including growth and development and response to biotic and abiotic stresses. They have been shown to act in complex signalling networks, where crosstalk signals a reaction to stress, such as attack by microbial pathogens and insect pests. Jasmonic acid (JA), salicylic acid (SA) and ethylene (ET) are thought to be central components of signalling pathways ultimately leading to the activation and fine tuning of defence responses; and abscisic acid (ABA) and auxin (indole-3-acetic acid; IAA) have also been implicated in defence against pathogens (Robert-Seilaniantz et al. 2011; Spaepen and Vanderleyden 2011). Both synergistic and antagonistic interactions between hormone signalling pathways have been reported to modulate defence to specific pathogens/pests (reviewed in Kunkel and Brooks 2002). To better understand the mechanisms of phytohormone activity, methods have been developed for the quantitation of these molecules and their regulators (reviewed in Schmelz et al. 2004).
Metabolomics analyses benefit from numerous analytical approaches to achieve maximum metabolome coverage. Volatile metabolites in particular can be lost during preparative stages within broader non-targeted metabolomics studies, if not specifically considered. Even within the complement of the plant metabolome termed phytohormones, the chemical diversity is such that complementary sample preparation and analytical methods be used for global phytohormone measurement (Boiero et al. 2007). The lower molecular weight and particularly higher volatility of ET for example, as compared to other commonly measured phytohormones renders this metabolite in particular, less amenable to global phytohormone quantitation. As a compromise to using an additional and specific method, solely for ET quantitation, Birkemeyer et al. (2003) and Schmelz et al. (2004) instead indirectly measured this phytohormone by way of quantitation of the amino acid precursor 1-aminocyclopropane 1-carboxylate (ACC). ACC has been demonstrated to have a direct correlation to ET production and as such is considered a marker of ET quantitation for global phytohormone measurement (Birkemeyer et al. 2003; Schmelz et al. 2004). ACC is the intermediate in the conversion of l-methionine to ET in the biochemical pathway of ET biosynthesis in plants (Caspi et al. 2014). This metabolite has since been analysed simultaneously with ABA, ABA-β-d-glucopyranosyl ester (ABA-GE), 24-epibrassinolide (BL), gibberellic acid A3 (GA3), IAA, JA and SA, thus allowing for more complete phytohormone coverage using a single gas chromatography/mass spectrometry (GC–MS) method (Birkemeyer et al. 2003). Müller and Munné-Bosch (2011) have used this same approach to measuring ET biosynthesis using liquid chromatography/mass spectrometry (LC–MS), again, to employ methods of phytohormone profiling incompatible with the quantitation of ET directly (Müller and Munné-Bosch 2011).
Chiwocha et al. (2003) also demonstrated the strength of LC/MS based phytohormone measurement by quantifying ABA, along with numerous IAAs, cytokinins and gibberellins, and their biochemical intermediates from lettuce (Chiwocha et al. 2003). More recently, Chen et al. (2012) developed a highly sensitive method to quantify phytohormones from rice leaves. Using a nano-LC approach, the assay benefited from a significant improvement in the lower limits of detection over higher flow LC systems, using as little as five milligrams of leaf tissue (Chen et al. 2012).
Common to both LC/MS and GC/MS metabolomics is the use of chemical modification or derivatisation of target metabolites, prior to instrumental analysis. Methods of phytohormone measurement by GC have particularly benefited from the conversion of these metabolites to their more volatile, thermally stable derivatives, with the most comprehensive GC–MS methods to date reporting the application of silylating agents to phytohormone profiling (Birkemeyer et al. 2003; Boiero et al. 2007; Engelberth et al. 2003; Muller et al. 2002; Schmelz et al. 2003, 2004).
As an alternative to silylation, Villas-Bôas et al. (2003), used the alkylating agent methyl chloroformate (MCF) to form stable derivatives of amino and non-amino organic acids, and validated its application to non-targeted metabolomics. Perrine et al. (2004) first applied this method of derivatisation to the measurement of IAA and tryptophan in rice, though the method was not expanded to include other phytohormones. MCF derivatisation, under alkaline conditions and in the presence of pyridine, was however found to be simple, high-throughput, and unlike silylation, derivatisation can proceed in the presence of water (Husek 1998; Smart et al. 2010; Villas-Bôas et al. 2003).
Although thorough, some of the more complete methods of phytohormone profiling have laborious and time-consuming extraction protocols, reducing throughput and potentially compromising optimal experimental design. So too, the lower limits of quantitation (LOQ) are often compromised in order to profile greater numbers of phytohormones. Thus there is a need for a method which can adequately quantify a large range of plant hormones simultaneously and within biologically relevant detection limits, whilst maintaining a reasonable degree of throughput in terms of instrumental and phytohormone extraction protocols.
A high throughput and sensitive method that would specifically identify a wide range of phytohormones and biochemical intermediates would benefit the plant science community. Toward this goal, we describe a simple GC–MS based approach to phytohormone quantitation, utilising a rapid and high-throughput extraction and derivatisation protocol for the measurement of various plant hormones and their precursors. The method uses MCF to derivatise metabolites and enhance non-polar extraction from plant tissues and was applied to phytohormone quantitation in the model plant Medicago truncatula.
Medicago truncatula has been used extensively to study plant–microbe interactions, not only for beneficial microbes like Rhizobia (Gough and Jacquet 2013) and mycorrhiza (Krajinski and Frenzel 2007), but also pathogens such as Aphanomyces euteiches (Hamon et al. 2010; Hilou et al. 2014), Phoma medicaginis (Kamphuis et al. 2008, 2012) and Rhizoctonia solani (Anderson et al. 2010; Anderson and Singh 2011), as well as insect pests (Kamphuis et al. 2013a; Kamphuis et al. 2013b). In addition, M. truncatula has been used to study certain aspects of plant development (Verdier et al. 2013) and responses to abiotic stress (Li et al. 2011; Wang et al. 2011), where plant hormones are key players and where the method described here will facilitate future studies. We show that the method can simultaneously quantify 11 metabolites, either plant phytohormones or their precursors [ABA, azelaic acid (AZ), IAA, JA and SA, and the phytohormone precursors ACC, benzoic acid (BA), cinnamic acid (CA), 13-epi-12-oxophytodienoic acid (13-epi-OPDA), linoleic acid and linolenic acid] from 100 mg amounts of plant tissue in M. truncatula with limits of detection between 2 and 10 ng mL−1 (7–30 mM).
Materials and methods
Chemicals
Reagents and standards including methyl chloroformate, pyridine, sodium bicarbonate, sodium hydroxide and sodium sulfate (anhydrous), n-alkanes (decane, dodecane, pentadecane, nonadecane, docosane, octacosane, dotriacontane, hexatriacontane), ABA, AZ, BA, CA, deuterated cinnamic acid (CA-d6), idole-3-acetic acid (IAA), JA, ACC, linolenic acid, linoleic acid and SA were purchased from Sigma-Aldrich (St Louis, MO, USA). 13-epi-12-oxophytodienoic acid (13-epi-OPDA) was purchased from Cayman Chemical (Ann Arbor, MI, USA). Chloroform (HPLC grade), and LC–MS grade methanol and water were purchased from Thermo Fischer Scientific (Scoresby, VIC, Australia).
Plant preparation and growth
The M. truncatula accession Jester was used for development and proof of concept in this study. Jester is closely related to A17 with whom it shares 89 % of its genome. A17 is a derivative of the cultivar ‘Jemalong’ and the reference accession for the M. truncatula species. To ensure uniform germination, seeds were scarified using sandpaper and transferred to a Petri dish lined with blotting paper, and irrigated with sterile water. The seeds were kept at room temperature for 48 h; germinated seedlings were planted in Arabidopsis mix (Richgro Garden Products, Jandakot, Western Australia, 6164). Plants were grown in individual 0.9-L pots in growth cabinets with 16 h of light (22 °C) and 8 h of dark (20 °C) under high-pressure sodium and fluorescent lamps at 280 µE m−2 s−1 and tissue was harvested from 4-week-old plants.
Phytohormone isolation and derivatisation
Leaves of M. truncatula were excised and immediately submerged in liquid nitrogen before storage at −80 °C. For the extraction of phytohormones, the leaf tissues were first ground to a fine powder in the presence of liquid nitrogen by mortar and pestle and 100 mg transferred to a two mL microcentrifuge tube. The extraction and combined derivatisation proceeded as described by Villas-Bôas et al. (2003), with some modification. To the ground tissue, 20 µL of 20 µg mL−1 deuterated cinnamic acid (CA-d6; in methanol) was directly added and the sample suspended in 200 μL of a sodium hydroxide (1 % w/v) solution. Added to the suspension were 147 µL of methanol and 34 µL of pyridine, before vigorous mixing by vortex for 25–30 s. Methyl chloroformate (20 µL) was then added and the suspension vigorously mixed for 25–30 s. A second volume of methyl chloroformate (20 μL) was added and the samples again mixed for 25–30 s. Subsequently, chloroform (400 µL) was added, the sample mixed for 10 s and a 50 mM sodium bicarbonate solution (400 µL) added. Following further mixing for 10–15 s, the extract was separated into two phases by centrifugation for 30 s at 16,100×g. The lower organic layer containing the phytohormones was transferred by pipette to a fresh microcentrifuge tube, being careful not to disturb the layer of plant debris separating the aqueous and organic layers. Anhydrous sodium sulphate was then added until the crystalline sodium sulphate appeared dry upon further addition. One hundred microlitre of the water-free solution was transferred to a glass analytical vial for GC–MS analysis.
Calibration standards
Calibration standards were prepared, and subsequently derivatised by MCF as described, with minor modification. Individual standards were dissolved in methanol. A solution containing ABA, ACC, AZ, BA, CA, 13-epi-OPDA, IAA, JA, linolenic acid, linoleic acid and SA, each at 200 µg mL−1 was prepared and subsequently diluted in methanol for calibration stocks at the following concentrations: 0.02, 0.20, 2.00 and 20.00 µg mL−1. Three calibration levels were prepared from each stock solution (Supplementary Table 1).
The calibration solutions were prepared for analysis by addition to the 1 % NaOH solution already described. The metabolite standards and internal standard were added together with methanol to a total volume of 167 µL, and finally pyridine to maintain the 200:167:34 (1 % NaOH solution:methanol:pyridine) ratio described. MCF derivatisation proceeded as already described. Final calibration concentrations were 0.002, 0.005, 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 2, 5, 10, 20 µg mL−1 after derivatisation and extraction (Supplementary Table 1).
Gas chromatography/mass spectrometry
All method development was carried out on an Agilent 6890 gas chromatograph with a split/splitless injector and an Agilent 5973N quadrupole mass spectrometer with an EI source (Agilent, Palo Alto, CA, USA). The GC inlet was equipped with an Agilent focus liner with glass wool insert (SGE, Analytical Science Pty. Ltd.). A Varian Factor Four 5 ms column (30 m × 0.25 mm × 0.25 μm with a 10 m guard column) was used for all analyses (Varian, Palo Alto, CA, USA). Injection temperature and transfer-line were both 250 °C. A three µL injection was used with a 35 psi pressure pulse, held for 1 min. Chromatographic conditions had the column held at 40 °C for 1 min, and then temperature programmed at 20 °C min−1 to 320 °C, held for 2 min (17 min total). Helium was used as the carrier gas (1 mL min−1). The (electron ionisation; EI) ion source was maintained at 200 °C and solvent delay was 4.5 min. The MS was run in a scan mode (m/z 50–400) of acquisition for selection of appropriate EI mass fragments for each analyte. For quantitation methods, the MS was run in selected ion monitoring (SIM) mode with the respective ions presented in Table 1. The dwell time for each ion was 30 ms and each analyte had its own SIM window where possible. Methyl linoleate, methyl linolenate and methyl abscisate had the lowest cycle time of 1.83 cycles s−1. At basal biological levels, as determined during method development (using unchallenged M. truncatula), a minimum of 7–9 points across a peak was achieved and maintained for all analytes.
Table 1.
The identifying features of the analytes resolved by these methods; accounting for the target phytohormones and related metabolites, isomers and multiple derivatisation products
| Abbreviation | Name | CAS (underivatised analyte) | Retention indice | Retention time (min) | Quantifier ion (m/z) | Qualifier ion 1 (m/z) | Qualifier ion 2 (m/z) | SIM cycle (cycle/sec) |
|---|---|---|---|---|---|---|---|---|
| C10 | Decane | 124-18-5 | 1000 | 5.63 | 85 | 142 | 99 | |
| MeBA | Benzoic acid, Me ester | 65-85-0 | 1137 | 6.44 | 105 | 136 | 77 | 3.8 |
| C12 | Dodecane | 112-40-3 | 1200 | 7.08 | 85 | 170 | 99 | |
| MeSA | Salicylic acid, Me ester | 69-72-7 | 1215 | 7.19 | 120 | 152 | 92 | 3.1 |
| MeACC | 1-aminocyclopropane-1-carboxylic acid, carbamate | 22059-21-8 | 1267 | 7.55 | 141 | 109 | 82 | 3.1 |
| MeCA-d6 | d6-Cinnamic acid, Me ester | 91453-04-2 | 1411 | 8.51 | 137 | 168 | 109 | 3.0 |
| MeCA | Cinnamic acid, Me ester | 140-10-3 | 1411 | 8.52 | 131 | 162 | 103 | 3.0 |
| C15 | Pentadecane | 629-62-9 | 1500 | 9.13 | 99 | 212 | 113 | |
| MeMeSA | Salicylic acid, Dimethyl ester | 69-72-7 | 1523 | 9.3 | 135 | 165 | 92 | 2.7 |
| MeAz | Azelaic acid, Me ester | 123-99-9 | 1553 | 9.48 | 152 | 185 | 143 | 2.7 |
| MeJA isomer 1 | Jasmonic acid, Me ester isomer 1 | 77026-92-7 | 1655 | 10.02 | 151 | 224 | 83 | 3.8 |
| MeJA isomer 2 | Jasmonic acid, Me ester isomer 2 | 77026-92-7 | 1673 | 10.12 | 151 | 224 | 83 | 3.8 |
| MeIAA | 3-Indole acetic acid, Me ester | 87-51-4 | 1863 | 11.32 | 189 | 103 | 77 | 3.8 |
| C19 | Nonadecane | 629-92-5 | 1900 | 11.41 | 99 | 268 | 113 | |
| Linoleic Acid, Me Ester | cis,cis-9,12-Octadecadienoic Acid, Me Ester | 60-33-3 | 2106 | 12.40 | 67 | 81 | 94 | 1.8 |
| Linolenic Acid, Me Ester | cis,cis,cis-9,12,15-Octadecatrienoic Acid, Me Ester | 463-40-1 | 2114 | 12.43 | 79 | 95 | 108 | 1.8 |
| MeABA isomer 1 | Abscisic acid, Me Ester | 14375-45-2 | 2129 | 12.51 | 190 | 162 | 134 | 1.8 |
| C22 | Docosane | 629-97-0 | 2200 | 12.86 | 99 | 310 | 113 | |
| MeABA isomer 2 | Abscisic acid, Me Ester | 14375-45-2 | 2205 | 12.89 | 190 | 162 | 134 | 3.8 |
| 13-epi-OPDA, Me Ester isomer 1 | 13-epi-12-oxophytodienoic acid, Me ester isomer 1 | 71606-07-0 | 2293 | 13.37 | 163 | 238 | 206 | 3.8 |
| 13-epi-OPDA, Me Ester isomer 2 | 13-epi-12-oxophytodienoic acid, Me ester isomer 2 | 71606-07-0 | 2306 | 13.45 | 163 | 238 | 206 | 3.8 |
| C28 | Octacosane | 630-02-4 | 2800 | 16.21 | 85 | 99 | 71 |
Method validation
The described method was validated in three ways; extraction efficiency, limit of quantitation and intra and inter-day reproducibility.
The extraction efficiency was assessed by dividing a single ground sample of unchallenged M. truncatula foliar tissue among 12 replicate tubes; half of which were spiked with a phytohormone standard solution, for comparison against the remaining non-spiked controls. One hundred microlitre of a 20 ng µL−1 standard solution were spiked prior to the MCF derivatisation. The basal levels were determined for the non-spiked controls and the mean normalised intensity used as a baseline subtraction against the spiked. Baseline subtracted concentrations were determined against a calibration curve.
The LOQ was calculated from the derivatised standards and defined as the lowest concentration where a percent relative standard deviation (%RSD) of <10 (for n ≥ 6) was obtained, whilst maintaining a signal to noise (S/N) ratio >10. The limit of quantitation of each MCF derivative within M. truncatula sample matrix was determined on a sample by sample basis. This quantitation was defined by the concentration where the quantifier ion of each given MCF derivative measured a signal to noise ratio >10, whilst maintaining the ratio of qualifying ions to within 20 % of the values determined from the pure standard.
To test the reproducibility of all phytohormones, within and between extraction batches, ground M. truncatula foliar tissue was again divided among technical replicates and extractions performed over three consecutive days. Forty-eight individual MCF-preparations and analyses were prepared each day. These included 16 spiked with the phytohormone standards prior to MCF preparation, 16 non-spiked, 10 calibration standards and 8 standard dilutions. The spiked and non-spiked extractions were randomised among the GC–MS sequence with a 0.5 ng µL−1 calibration standard injected every fifth sample. This analytical sequence was repeated for three consecutive days, preparing fresh extractions daily. Before each sequence, a 10 cm length of the GC guard column was trimmed and the GC inlet liner replaced, consistent with internal routine preventative maintenance practices. The lowest level calibration sample was re-injected at the end of each analytical sequence, and again following preventative maintenance. Fifty injections were performed over each 24 h period.
Data analysis
Data analyses were carried out using Agilent Chemstation software (v D.01.00). EI mass fragments for quantitation were selected based on their relative intensity and uniqueness (Table 1). Ion ratios between the main quantifier and qualifier ions were determined using pure standards. One quantifier ion and two qualifier ions were chosen for minimum reporting requirements.
To ensure accuracy at the lower LOQ, two calibration curves were constructed. The lower calibration was of concentrations downward from 5 µg mL−1; and the higher, upwards from 2 µg mL−1. Where an analyte was found to be close to the limit of detection (LOD), the higher calibration points were not used for quantitation.
Results and discussion
Interplay between the biochemical pathways of phytohormone synthesis have long been established (Kunkel and Brooks 2002; Pilet 1965). Methods amenable to measuring all major classes of phytohormones however are essential to further understand their interplay, and robust, easy to use protocols of phytohormone quantitation are integral to ongoing efforts to characterise the role of these metabolites in plant stress responses and various aspects of plant development (Robert-Seilaniantz et al. 2011; Spaepen and Vanderleyden 2011).
Targeted metabolite profiling by GC–MS is efficient and reproducible, though current methods of phytohormone profiling by GC–MS can be complicated by use of specialised equipment (Schmelz et al. 2003, 2004), or are amenable to fewer classes of phytohormones (Table 2). To this end we developed and validated a method of metabolite extraction and derivatisation by MCF (Husek 1998; Smart et al. 2010; Villas-Bôas et al. 2003) for application to phytohormone quantitation. The sample preparation and derivatisation steps are presented schematically in Supplementary Fig. 1.
Table 2.
Comparison of literature-cited methods of phytohormone profiling
| Author, year | Metabolitea | Extraction | Sample matrix | Tissue mass | Instrument | Analytical run length | Extraction time | Notes |
|---|---|---|---|---|---|---|---|---|
| Pan et al. (2008) | Z, GA3, ICA, IAA, BA, ABA, MeIAA, SA, CA, MeBA, IBA, JA, MeSA, MeCA, MeJA, GA4, OPDA | SLEa | Arabidopsis thaliana, shoot | 50-100 mg FW | LC–ESI–MS/MS | 20 min | approximately 2.5 h per batch | Batch size undefined |
| Barkawi et al. (2010) | Auxins | SLE, 2 × SPE and derivatisationb | Arabidopsis thaliana seedling, tomato seedling root, maize inflorescence, Saccharomyces cerivisiae | 25 mg FW | GC–EI–MS | 17.5 min | 3 h per 8 samples (manually) | Alternative automated protocol suggested with use of a robotic liquid handler |
| Schmelz et al. (2004) | SA, CA, JA, IAA, ABA, Linoleic acid, Linolenic acid,OPDA, COR | SLE, derivatisation and VPEc | Arabidopsis thaliana, shoot | 100 mg FW | GC–CI–MS | 25 min | 6 h per 48 samples | Use of specialised equipment required for extraction of the phytohormones. |
| Muller et al. (2002) | SA, JA, IAA, ABA, OPDA | SLE, multiple SPE and derivatisation | Arabidopsis thaliana, shoot | 20–200 mg FW | GC–CI–MS/MS | 21 min | 60 samples per day | |
| Chen et al. (2012) | Gibberellins, ABA, JA, IAA, IBA, SA | SLE, 2 × SPE, LLE and derivatisationd | Oryza sativa, leaf | 5 mg FW | nano-LC–ESI–QToF–MS | 74 min | ND | Highly specialised instrument for quantitation of phytohormones |
| Chiwocha et al. (2003) | Z, ZR, DPA, IAAsp, gibberellins, 2iP, ABA-GE, PA, IPA, 7′-OH-ABA IAA, ABA | SLE and SPE | Lactuva sativa, seed | 50-100 mg DW | LC–ESI–MS/MS | 30 min | approximately 26 h per batch | Batch size undefined, 24 h SLE step. |
| Perrine et al. (2004) | IAA, Trp | Simultaneous derivatisation and LLE | Rhizobium, exudate | 1 mL supernatant | GC–EI–MS | 14 min | approximately 15 min per sample | |
| Durgbanshi et al. (2005) | ABA, IAA, JA | SLE and LLE | Citrus clementina, mature leaf, Hordeum vulgare, seedlings | 250–500 mg FW | LC–ESI–MS/MS | 21 min | ND | |
| This study, 2015 | BA, SA, ACC, CA, AZ, JA, IAA, Linoleic acid, Linolenic acid, ABA, 13-epi-12OPDA. | Simultaneous derivatisation and LLE | Medicago truncatula, shoot | 100 mg FW | GC–EI–MS | 17 min | 20 samples per batch, 30–45 min per batch |
Only methods applied to the quantitation of two or more phytohormone-relevant metabolites were included
Z zeatin, GA3/GA4 gibberellins, ICA indole-3-carboxylic acid, BA benzoic acid, ABA abscisic acid, MeIAA methyl indole-3-acetate, SA salicylic acid, CA cinnamic acid, MeBA methyl benzoate, IBA indole butyric acid, JA jasmonic acid, MeSA methyl salicylate, MeCA methyl cinnamate, OPDA oxophytodienoic acid, COR coranatine, ZR zeatin riboside, ABA-GE abscisic acid glucose ester, 7′-OH-ABA 7′-hydroxy-abscisic acid, PA phaseic acid, DPA dihydorxyphaseic acid, IAAsp indole-3-aspartate, 2iP isopentenyladenine, IPA isopentenyladenosine, Trp tryptophan, ACC 1-aminocyclopropane carboxylic acid, IAA indole-3-acetic acid, AZ azelaic acid, 13-epi-12-OPDA 13-epi-12-oxophytodienoic acid, FW fresh weight, DW dry weight, ESI electrospray ionisation, EI electron ionisation, CI chemical ionisation
aSolid:liquid extraction; b solid-phase extraction; c vapour-phase extraction; d liquid:liquid extraction
Three key areas of method development were assessed; (1) the capacity of MCF to derivatise a number of phytohormones and phytohormone precursors, (2) the suitability of application to plant tissues, and quantitation of MCF-derivatives in a plant sample matrix (in our case foliar tissue from the model legume M. truncatula), and, (3) the cost, through-put and ease at which the method can be applied to routine phytohormone measurement. For comparison, this presented method was compared alongside current literature citations of phytohormone profiling (Table 2).
Extraction and derivatisation
Ensuring the amenability of metabolites to routine analytical methods by way of chemical modification is common practice in targeted and non-targeted metabolite profiling. Silylation is the most common derivatising agent for GC–MS metabolomics, yet more volatile metabolites, for example BA, suffer from the requirement to dry the metabolite extract (data not shown).
We identified a combination of 11 metabolites, phytohormones, or their biochemical pathway intermediates; linolenoate, linoleote, 13-epi-12-oxo phytodienoate, ACC, indole-3-acetate, abscisate, azelate, benzoate, cinnamate, jasmonate and salicylate as amenable to MCF derivatisation and quantitation by GC–MS. The structures of the MCF derivatised metabolites are provided in Supplementary Fig. 2. Two additional phytohormones were of interest to this study, gibberellic acid (GA) and the cytokinin, zeatin (Z). GA, whilst amenable to the methods of isolation and derivatisation, is not well suited to GC analysis under the conditions most favourable to broader profiling (data not shown). Z on the other hand is not efficiently derivatised, likely due to steric hindrance of the functional groups required for MCF modification and as such is not amenable to the described methods. These two metabolites are well suited to measurement by LC–MS. Previous studies observed the efficiency of MCF derivatisation to be related to the alkalinity of the extract, with amino, di- and tricarboxylic acid yields being highest when the reaction mixture were above pH 10 (Villas-Bôas et al. 2003). Smart et al. (2010) recommended a pH higher than 12 for efficient MCF derivatisation. Additionally, Villas-Bôas et al. (2003) observed a general decrease in yield with longer reaction times in a non-targeted analysis of fungal metabolites.
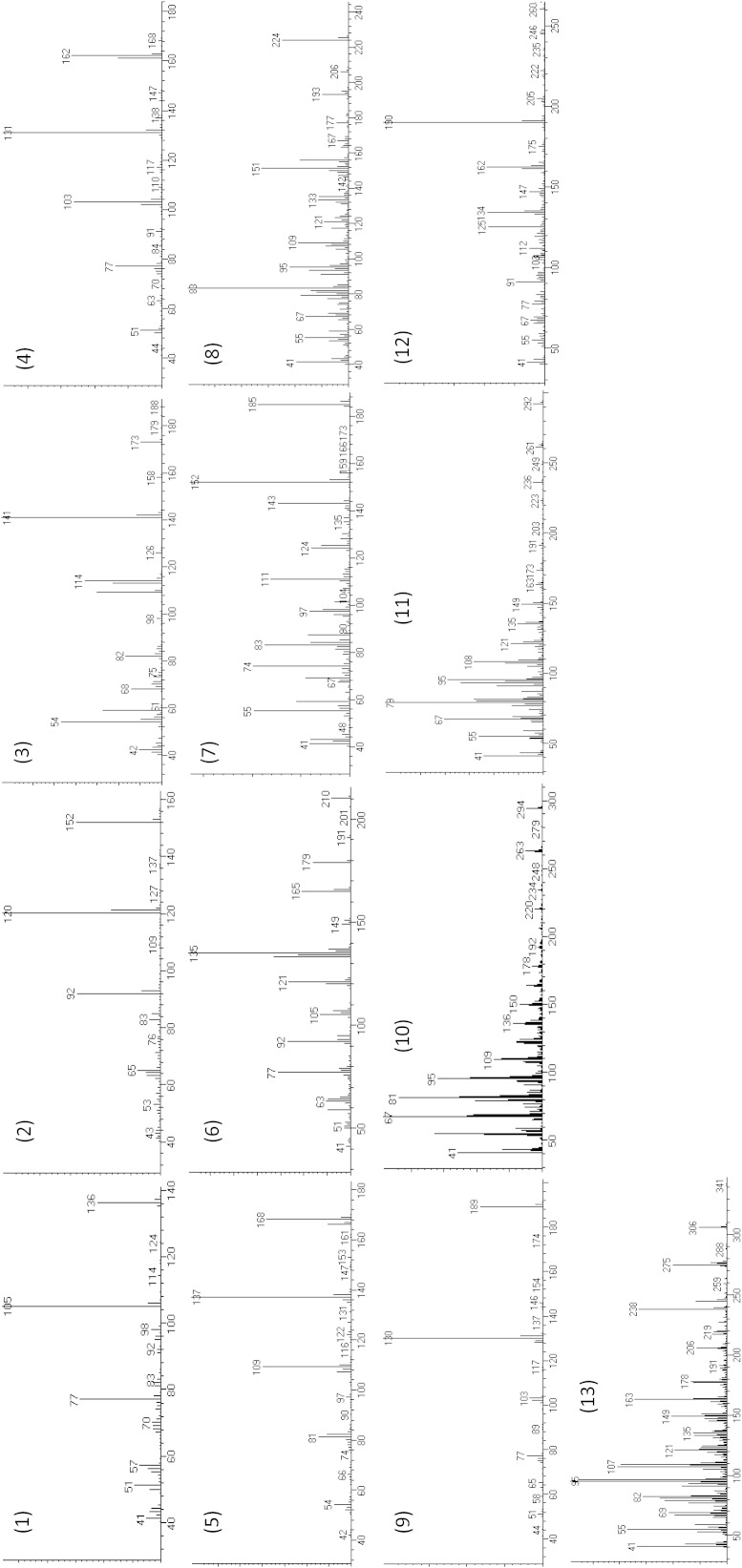
Our data demonstrated that pH and reaction time do not greatly influence the complete derivatisation of these 11 metabolites within the M. truncatula sample matrix, with no measured difference within one and four percent (w/v) NaOH solution, nor reaction times from 30 s to 5 min (data not shown). Of the phytohormones quantified, SA was the only analyte that formed multiple derivatisation products under the tested conditions. The predominant product, in all cases, resulted from the derivatisation of both the carboxylic acid and hydroxyl functional groups with the minor product only derivatising at the carboxylic acid functional group. The mass spectrum for all measured analytes is presented in Fig. 1.
Fig. 1.
Mass spectrum for the methyl chloroformate derivatised products for (1) benzoic acid, (2) salicylic acid (methyl derivative), (3) 1-aminocyclopropane carboxylic acid, (4) cinnamic acid, (5) cinnamic acid-d6, (6) salicylic acid (dimethyl derivative), (7) azelaic acid, (8) jasmonic acid (isomer 1 and 2), (9) indole-3-acetic acid, (10) linoleic acid, (11) linolenic acid, (12) abscisic acid (isomer 1 and 2), (13) 13-epi-12-oxo phytodienoic acid (isomer 1 and 2). Isomers for jasmonic acid, abscisic acid and 13-epi-12-oxo phytodienoic acid resolve chromatographically but present the same mass spectrum in electron ionisation mode, a single mass spectrum is therefore presented
Consistent with these preliminary analyses, and for confidence in achieving quantitative derivatisation of subsequent sample sets within this study, each were prepared with a mid-range calibration standard, and the metabolites extracted and derivatised in groups of no more than 20, allowing no sample to exceed 5 min reaction time.
Quantitation
Technical variability can be introduced into the analysis during extraction, derivatisation or instrumental analysis. Sample matrix may also interfere with the yield of derivatised products. In this method, we use isotopically labelled cinnamic acid (CA-d6) to compensate for these possible inconsistencies. Normalisation of individual analyte responses to this internal standard can partially correct unavoidable variability. CA-d6 was spiked directly into the ground plant foliar tissue, before extraction, to best mimic the recovery and derivatisation of the analysed compounds. The deuterated CA internal standard provided reproducibility with a single MCF-derivatisation product, and mass spectrum easily distinguished from the naturally occurring analogue.
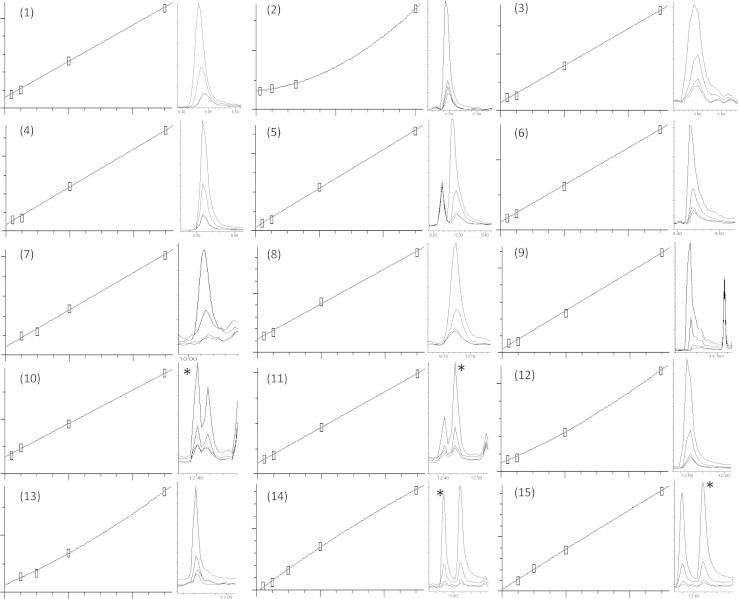
Most analytes were linear over four orders of magnitude or greater (data not shown). The highest calibration standard for MeBA and MeIAA overloaded the MS detector, so quantitation was limited to a maximum concentration of 10 µg mL−1. For accurate quantitation of less abundant phytohormones the calibrations at the lower LODs are of particular importance. Eight of the 11 metabolites had a measured linearity within two and 50 ng mL−1 (r2 ≥ 0.997; Fig. 2).
Fig. 2.
Four lowest calibration standards and the chromatogram overlay for: (1) MeBA—R2 0.999, 0.002–0.05 µg mL−1, (2) MeSA—R2 0.999, 0.005–0.2 µg mL−1, (3) MeACC—R2 0.999, 0.002–0.05 µg mL−1, (4) MeCA—R2 0.998, 0.002–0.05 µg mL−1, (5) MeMeSA—R2 0.999, 0.002–0.05 µg mL−1, (6) MeAz—R2 0.999, 0.002–0.05 µg mL−1, (7) MeJA isomer 1—R2 0.997, 0.005–0.05 µg mL−1, (8) MeJA isomer 2—R2 0.997, 0.002–0.05 µg mL−1, (9) MeIAA—R2 0.999, 0.002–0.05 µg mL−1, (10) linoleic acid, Me Ester—R2 0.999, 0.002–0.05 µg mL−1, (11) linolenic acid, Me Ester—R2 0.999, 0.002–0.05 µg mL−1, (12) MeABA isomer 1—R2 0.999, 0.002–0.05 µg mL−1, (13) MeABA isomer 2—R2 0.999, 0.005–0.05 µg mL−1, (14) Me-13-epi-OPDA isomer 1—R2 0.999, 0.002–0.05 µg mL−1, (15) Me-13-epi-OPDA isomer 2—R2 0.999, 0.005–0.05 µg mL−1. Asterisked peaks represent the analyte of interest for that calibration
All metabolites, except for 13-epi-OPDA were reproducibly detected at 0.005 ng/µL or lower (Table 3). All of the metabolites had a calculated %RSD less than 10 (n ≥ 6). This reproducibility would be improved with the availability of purer metabolite standards. As an example, ABA, JA and 13-epi-OPDA were purchased as a cis- and trans- mix of isomers which chromatographically resolve by these methods (Table 3). However, it is the sum of these isomers which represent the prepared concentration. Isomers of the same metabolite were therefore quantified using the same ions to allow calibration and sample responses to be summed for absolute quantitation.
Table 3.
The determined limits of quantitation (LOQ, ng µL−1 and mM), extraction efficiencies, and the intra- and inter-day reproducibility of the MCF-derivatized phytohormones/phytohormone precursor (in order of elution)
| Compound | LOQ ng μL−1 (mM) | RSD at LOQ (%) | Recovery (%) | Recovery RSD (%) | Intra-day reproducibility RSD (%) | Inter-day reproducibility RSD (%) |
|---|---|---|---|---|---|---|
| MeBA | 0.002 (14.7) | 8.4 | 96.0 | 6.6 | 4.9 | 18.9 |
| MeACC | 0.002 (11.6) | 3.6 | 79.3 | 3.8 | 10.3 | 16.6 |
| MeCA | 0.002 (12.3) | 9.6 | 97.7 | 4.4 | 4.9 | 14.2 |
| MeCA-d6 (IS) | – | – | 99.0 | 1.6 | – | – |
| MeMeSA | 0.005 (30.1) | 9.1 | 67.4 | 5.8 | 12.3 | 22.3 |
| Methyl Azelate | 0.005 (23.1) | 9.1 | 84.5 | 5.2 | 10.9 | 17.4 |
| MeJA Isomer 1 | 0.02 (89.2) | 7.0 | 88.0 | 9.4 | 28.4 | 21.3 |
| MeJA Isomer 2 | 0.002 (8.9) | 9.8 | 91.9 | 3.3 | 13.3 | 16.3 |
| MeIAA | 0.002 (10.6) | 8.8 | 99.1 | 4.0 | 7.3 | 18.9 |
| Linoleic Acid, Me Ester | 0.002 (6.8) | 6.8 | 55.7 | 4.9 | 8.9 | 17.4 |
| Linolenic Acid, Me Ester | 0.002 (6.8) | 7.7 | 86.1 | 10.5 | 8.7 | 15.5 |
| MeABA Isomer 1 | 0.005 (18.0) | 8.6 | 102.1 | 3.9 | 6.6 | 17.9 |
| MeABA Isomer 2 | 0.02 (71.9) | 3.2 | 109.8 | 3.6 | 9.5 | 24.6 |
| 13-epi-OPDA, Me Ester Isomer 1 | 0.01 (32.6) | 9.8 | 55.6 | 15.7 | 10.7 | 15.3 |
| 13-epi-OPDA, Me Ester Isomer 2 | 0.01 (32.6) | 4.1 | 59.4 | 14.8 | 14.4 | 18.2 |
The intra-/inter-day reproducibility was determined within the M. truncatula metabolite matrix
LOQ was determined by an RSD (%) ≤10 (n = 6). The higher LOQ of the minor derivatisation product of SA was due to only representing a very small part of the total SA and has therefore been omitted
Instrument capabilities are reflected in the detection limits achieved by analysis of pure standards, whereas plant matrices present a complex and dynamic system for GC–MS analysis. Consequently, the detection limit of pure standards will typically be lower than can be achieved within plant matrix. Muller et al. (2002) synthesised five isotopically labelled phytohormones (SA, JA, IAA, ABA and OPDA) and determined their respective detection limits as standards, and compared these values to when the labelled phytohormones were spiked into an extract from Arabidopsis thaliana. The lowest concentration that meets detection limit criteria of minimum S/N for these standards within plant matrix was 11.5–15 times greater when compared to pure standards (Muller et al. 2002).
Recovery
As several of the phytohormones are known to be present at basal levels within plant tissues, recovery was calculated by the analysis of six replicates of unchallenged M. truncatula foliar tissue extract, against a further six replicates spiked with 100 µL of a 5 ng µL−1 solution of the metabolite standards. A recovery of 75 % or better was achieved for nine of the 11 metabolites (Table 3). This route of analysis showed a high degree of reproducibility as all analytes (except linolenic acid, methyl ester and 13-epi-OPDA, methyl ester) showed %RSD’s below 10. The high %RSD of linolenic acid was likely an artefact of the high endogenous level of this compound within M. truncatula tissue, and therefore working closer to the upper limit of quantitation of this metabolite.
Intra- and inter-day reproducibility
Metabolomics analyses produce multi-dimensional datasets, combining numerous treatment groups, biological replication, quality controls, and often longitudinal data. Batch variation adds further dimension to particularly large studies, introduced through extraction batches and preventative instrument maintenance. Previous work demonstrated the excellent stability of MCF derivatives of 26 amino and non-amino organic acids when stored at room temperature for 72 h (RSD <10 %) (Villas-Boas et al. 2011). This method however presents a high volume injection of a plant matrix into the GC inlet, potentially making it particularly susceptible to intra- and inter-day variation. In this work, we therefore sought to examine the intra and inter-day reproducibility of the MCF preparation and GC–MS analysis of the 11 phytohormones. The intraday reproducibility was calculated from the 16 spiked foliar preparations, analysed among a sample sequence of 50 injections, with the reported RSDs capturing the entire extraction and derivatisation, through to analytical variation. The inter-day measurements represent the reproducibility of the entire method repeated over three consecutive days, whilst also capturing the variability introduced through daily preventative instrument maintenance (Table 3). These presented methods achieved a RSD of less than 15 % for ten of the 11 phytohormones, with only the minor isomer of JA exceeding this value for intraday reproducibility. The inter-day reproducibility was below 20 % RSD for eight of the 11 phytohormones. Though still within an acceptable RSD, the remaining three phytohormones, SA, JA and ABA, which reported a higher RSD, likely suffered in reproducibility due to representation by multiple isomers or derivatisation products, and the consequential division of intensity amongst multiple peaks. The added robustness of the intraday analyses over the inter-day also demonstrate the particular reliability of this method to phytohormone preparation and derivatisation, more specifically.
Concluding remarks
The complex interplay between various phytohormones is crucial for many plant processes including response to biotic and abiotic stress. Therefore it is important to be able to easily and accurately measure changes in these compounds under various conditions where sample material may be limited. Here we have presented a very quick and robust method for the analysis of 11 metabolites; phytohormones or their biochemical pathway intermediates by GC–MS, and demonstrated application using the foliar tissue of M. truncatula, requiring only 100 mg of plant tissue. The method presents a high degree of reproducibility in the isolation of these target metabolites, and calibration linearity in the range of four orders of magnitude. This protocol is particularly high through-put in terms of sample preparation and instrumentation, requires no specialised equipment for phytohormone isolation and preparation and therefore samples can be generated in a wide range of laboratories and enables analysis of large, highly replicated experiments in a short turnaround time.
Metabolites with higher volatility including many phytohormones, require particular consideration during preparative processes, but are an important component of the plant metabolome. The application of targeted methods such as these towards achieving wider metabolome coverage will benefit non-targeted metabolomics data sets. The plant science and metabolomics community would also be well served by the application of these methods to other species and tissue types and even the extension to non-targeted data acquisition methods, such as those validated by Villas-Bôas et al. (2003) and Smart et al. (2010) for a more comprehensive picture of the plant metabolome.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (DOCX 24 kb). Supplementary Fig. 1. The preparative workflow of the described methods of pytohormone isolation and derivatisation
Supplementary material 2 (DOCX 66 kb). Supplementary Fig. 2 Chemical structures of the 11 phytohormone MCF-derivatives
Supplementary material 3 (DOCX 18 kb). Supplementary Table 1 Preparation of the calibration standard curves. All preparations maintained the described 200:167:34 (1 % NaOH:methanol:pyridine) ratio
Acknowledgments
We thank Jonathon Anderson and Louise Thatcher for helpful comments on the manuscript.
Funding
Funds for the research for Lars G. Kamphuis and Karam B. Singh were supported through CSIRO, including a CSIRO Office of Chief Executive Post-Doctoral Fellowship. Metabolomics Australia is a Bioplatforms Australia (BPA) funded initiative. Joel P. A. Gummer and Catherine Rawlinson were supported through BPA. Murdoch University and the SSML supported development costs.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Ethical statement
No ethical requirements were needed.
Footnotes
Catherine Rawlinson and Lars G. Kamphuis share first authorship.
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-015-0837-0) contains supplementary material, which is available to authorized users.
References
- Anderson JP, Lichtenzveig J, Gleason C, Oliver RP, Singh KB. The B-3 ethylene response factor MtERF1-1 mediates resistance to a subset of root pathogens in Medicago truncatula without adversely affecting symbiosis with rhizobia. Plant Physiology. 2010;154:861–873. doi: 10.1104/pp.110.163949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Singh KB. Interactions of Arabidopsis and M. truncatula with the same pathogens differ in dependence on ethylene and ethylene response factors. Plant Signaling and Behavior. 2011;6:551–552. doi: 10.4161/psb.6.4.14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkawi LS, Tam Y-Y, Tillman JA, Normanly J, Cohen JD. A high-throughput method for the quantitative analysis of auxins. Nature Protocols. 2010;5:1609–1618. doi: 10.1038/nprot.2010.118. [DOI] [PubMed] [Google Scholar]
- Birkemeyer C, Kolasa A, Kopka J. Comprehensive chemical derivatization for gas chromatography-mass spectrometry-based multi-targeted profiling of the major phytohormones. Journal of Chromatography A. 2003;993:89–102. doi: 10.1016/S0021-9673(03)00356-X. [DOI] [PubMed] [Google Scholar]
- Boiero L, Perrig D, Masciarelli O, Penna C, Cassán F, Luna V. Phytohormone production by three strains of Bradyrhizobium japonicum and possible physiological and technological implications. Applied Microbiology and Biotechnology. 2007;74:874–880. doi: 10.1007/s00253-006-0731-9. [DOI] [PubMed] [Google Scholar]
- Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang P, Karp PD. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome Databases. Nucleic Acids Research. 2014;42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M-L, Fu X-M, Liu J-Q, Ye T-T, Hou S-Y, Huang Y-Q, Yuan B-F, Wu Y, Feng Y-Q. Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC–ESI-Q-TOF-MS analysis. Journal of Chromatography B. 2012;905:67–74. doi: 10.1016/j.jchromb.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Chiwocha SDS, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross ARS, Kermode AR. A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. The Plant Journal. 2003;35:405–417. doi: 10.1046/j.1365-313X.2003.01800.x. [DOI] [PubMed] [Google Scholar]
- Durgbanshi A, Arbona V, Pozo O, Miersch O, Sancho JV, Gómez-Cadenas A. Simultaneous determination of multiple phytohormones in plant extracts by liquid chromatography–electrospray tandem mass spectrometry. Journal of Agricultural and Food Chemistry. 2005;53:8437–8442. doi: 10.1021/jf050884b. [DOI] [PubMed] [Google Scholar]
- Engelberth J, Schmelz EA, Alborn HT, Cardoza YJ, Huang J, Tumlinson JH. Simultaneous quantification of jasmonic acid and salicylic acid in plants by vapor phase extraction and gas chromatography-chemical ionization-mass spectrometry. Analytical Biochemistry. 2003;312:242–250. doi: 10.1016/S0003-2697(02)00466-9. [DOI] [PubMed] [Google Scholar]
- Gough C, Jacquet C. Nod factor perception protein carries weight in biotic interactions. Trends in Plant Science. 2013;18:566–574. doi: 10.1016/j.tplants.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Hamon C, Baranger A, Miteul H, Lecointe R, Le Goff I, Deniot G, Onfroy C, Moussart A, Prosperi JM, Tivoli B, Delourme R, Pilet-Nayel ML. A complex genetic network involving a broad-spectrum locus and strain-specific loci controls resistance to different pathotypes of Aphanomyces euteiches in Medicago truncatula. Theoretical and Applied Genetics. 2010;120:955–970. doi: 10.1007/s00122-009-1224-x. [DOI] [PubMed] [Google Scholar]
- Hilou A, Zhang H, Franken P, Hause B. Do jasmonates play a role in arbuscular mycorrhiza-induced local bioprotection of Medicago truncatula against root rot disease caused by Aphanomyces euteiches? Mycorrhiza. 2014;24:45–54. doi: 10.1007/s00572-013-0513-z. [DOI] [PubMed] [Google Scholar]
- Husek P. Chloroformates in gas chromatography as general purpose derivatizing agents. Journal of Chromatography B. 1998;717:57–91. doi: 10.1016/S0378-4347(98)00136-4. [DOI] [PubMed] [Google Scholar]
- Kamphuis LG, Lichtenzveig J, Oliver RP, Ellwood SR. Two alternative recessive quantitative trait loci influence resistance to spring black stem and leaf spot in Medicago truncatula. BMC Plant Biology. 2008;8:30. doi: 10.1186/1471-2229-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis LG, Lichtenzveig J, Peng K, Guo S, Klingler JP, Siddique K, Gao LL, Singh KB. Physiology and genetics of resistance to spotted alfalfa aphid (Therioaphis trifolii) in Medicago truncatula. Journal of Experimental Botany. 2013;64:5157–5172. doi: 10.1093/jxb/ert305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis LG, Williams AH, Küster H, Trengove RD, Singh KB, Oliver RP, Ellwood SR. Phoma medicaginis stimulates the induction of the octadecanoid and phenylpropanoid pathways in Medicago truncatula. Molecular Plant Pathology. 2012;13:593–603. doi: 10.1111/j.1364-3703.2011.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis LG, Zulak K, Gao L-L, Anderson JP, Singh KB. Plant–aphid interactions with a focus on legumes. Functional Plant Biology. 2013;40:1271–1284. doi: 10.1071/FP13090. [DOI] [PubMed] [Google Scholar]
- Krajinski F, Frenzel A. Towards the elucidation of AM-specific transcription in Medicago truncatula. Phytochemistry. 2007;68:75–81. doi: 10.1016/j.phytochem.2006.09.035. [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Current Opinion in Plant Biology. 2002;5:325–331. doi: 10.1016/S1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Li D, Zhang Y, Hu X, Shen X, Ma L, Su Z, Wang T, Dong J. Transcriptional profiling of Medicago truncatula under salt stress identified a novel CBF transcription factor MtCBF4 that plays an important role in abiotic stress responses. BMC Plant Biology. 2011;11:109. doi: 10.1186/1471-2229-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Duchting P, Weiler EW. A multiplex GC-MS/MS technique for the sensitive and quantitative single-run analysis of acidic phytohormones and related compounds, and its application to Arabidopsis thaliana. Planta. 2002;216:44–56. doi: 10.1007/s00425-002-0866-6. [DOI] [PubMed] [Google Scholar]
- Müller M, Munné-Bosch S. Rapid and sensitive hormonal profiling of complex plant samples by liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Plant Methods. 2011;7:37. doi: 10.1186/1746-4811-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X. Simultaneous quantification of major phytohormones and related compounds in crude plant extracts by liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry. 2008;69:1773–1781. doi: 10.1016/j.phytochem.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Perrine FM, Rolfe BG, Hynes MF, Hocart CH. Gas chromatography-mass spectrometry analysis of indoleacetic acid and tryptophan following aqueous chloroformate derivatisation of Rhizobium exudates. Plant Physiology and Biochemistry. 2004;42:723–729. doi: 10.1016/j.plaphy.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Pilet PE. Action of gibberellic acid on auxin transport. Nature. 1965;208:1344–1345. doi: 10.1038/2081344b0. [DOI] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annual Review in Phytopathology. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Schmelz, E.A., Engelberth, J., Alborn, H.T., O’Donnell, P., Sammons, M., Toshima, H. & Tumlinson, J.H. (2003). Simultaneous analysis of phytohormones, phytotoxins, and volatile organic compounds in plants. In Proceedings of the National Acadamy of Science, USA (Vol. 100, pp. 10552–10557) [DOI] [PMC free article] [PubMed]
- Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT. The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. The Plant Journal. 2004;39:790–808. doi: 10.1111/j.1365-313X.2004.02168.x. [DOI] [PubMed] [Google Scholar]
- Smart KF, Aggio RB, Van Houtte JR, Villas-Bôas SG. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nature Protocols. 2010;5:1709–1729. doi: 10.1038/nprot.2010.108. [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J. Auxin and plant-microbe interactions. Cold Spring Harbor Perspectives in Biology. 2011;3:a001438. doi: 10.1101/cshperspect.a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J, Lalanne D, Pelletier S, Torres-Jerez I, Righetti K, Bandyopadhyay K, Leprince O, Chatelain E, Vu BL, Gouzy J, Gamas P, Udvardi MK, Buitink J. A regulatory network-based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiology. 2013;163:757–774. doi: 10.1104/pp.113.222380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villas-Bôas SG, Delicado DG, Åkesson M, Nielsen J. Simultaneous analysis of amino and nonamino organic acids as methyl chloroformate derivatives using gas chromatography-mass spectrometry. Analytical Biochemistry. 2003;322:134–138. doi: 10.1016/j.ab.2003.07.018. [DOI] [PubMed] [Google Scholar]
- Villas-Boas SG, Smart KF, Sivakumaran S, Lane GA. Alkylation or silylation for analysis of amino and non-amino organic acids by GC–MS? Metabolites. 2011;1:3–20. doi: 10.3390/metabo1010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Chen L, Zhao M, Tian Q, Zhang WH. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics. 2011;12:367. doi: 10.1186/1471-2164-12-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (DOCX 24 kb). Supplementary Fig. 1. The preparative workflow of the described methods of pytohormone isolation and derivatisation
Supplementary material 2 (DOCX 66 kb). Supplementary Fig. 2 Chemical structures of the 11 phytohormone MCF-derivatives
Supplementary material 3 (DOCX 18 kb). Supplementary Table 1 Preparation of the calibration standard curves. All preparations maintained the described 200:167:34 (1 % NaOH:methanol:pyridine) ratio