This paper presents for the first time a comparison of the genetic consequences of two different types of speciation in plants of an oceanic island. Genetic data, using two different DNA methods, were obtained from more than 4,000 plants from the two major islands of the Juan Fernández Archipelago (Chile). Results show that some immigrant populations undergo major splitting events and harbor limited genetic diversity within each evolving line. In contrast, other immigrant populations establish and enlarge, but they never split, hence accumulating higher levels of genetic diversity.
Keywords: Adaptive radiation, anagenesis, cladogenesis, genetic diversity, phyletic speciation, Robinson Crusoe Islands
Abstract
Adaptive radiation is a common mode of speciation among plants endemic to oceanic islands. This pattern is one of cladogenesis, or splitting of the founder population, into diverse lineages in divergent habitats. In contrast, endemic species have also evolved primarily by simple transformations from progenitors in source regions. This is anagenesis, whereby the founding population changes genetically and morphologically over time primarily through mutation and recombination. Gene flow among populations is maintained in a homogeneous environment with no splitting events. Genetic consequences of these modes of speciation have been examined in the Juan Fernández Archipelago, which contains two principal islands of differing geological ages. This article summarizes population genetic results (nearly 4000 analyses) from examination of 15 endemic species, involving 1716 and 1870 individuals in 162 and 163 populations (with amplified fragment length polymorphisms and simple sequence repeats, respectively) in the following genera: Drimys (Winteraceae), Myrceugenia (Myrtaceae), Rhaphithamnus (Verbenaceae), Robinsonia (Asteraceae, Senecioneae) and Erigeron (Asteraceae, Astereae). The results indicate that species originating anagenetically show high levels of genetic variation within the island population and no geographic genetic partitioning. This contrasts with cladogenetic species that show less genetic diversity within and among populations. Species that have been derived anagenetically on the younger island (1–2 Ma) contain less genetic variation than those that have anagenetically speciated on the older island (4 Ma). Genetic distinctness among cladogenetically derived species on the older island is greater than among similarly derived species on the younger island. An important point is that the total genetic variation within each genus analysed is comparable, regardless of whether adaptive divergence occurs.
Introduction
Oceanic islands have long stimulated biologists to investigate patterns and processes of evolution (e.g. Darwin 1842; Wallace 1881; Whittaker and Fernández-Palacios 2007; Bramwell and Caujapé-Castells 2011). These isolated land masses, far from continental source areas, offer opportunities for determining origins of immigrants and their evolutionary history after establishment. The low probability of long-distance dispersal and successful colonization, the reduction of genetic variation in founding populations and the challenges of adaptation to new environments are all features that combine to affect processes of evolution in island archipelagos, particularly speciation.
One dimension of speciation in island plants that has received considerable attention is adaptive radiation (Carlquist 1974; Whittaker and Fernández-Palacios 2007; Rundell and Price 2009). This is a process that begins with dispersal from the original immigrant population into different habitats on the same or neighbouring island. This isolation leads to divergence of the new segregate populations, each becoming rapidly adapted to divergent habitats (Schluter 2001), such that eventually new species are recognized taxonomically. This general process of speciation is usually diagrammed (Fig. 1) as splitting events or cladogenesis (Rensch 1959). A number of dramatic species complexes have developed in oceanic islands through adaptive radiation, such as illustrated by the lobelioids (Givnish et al. 2009) and silverswords (Carlquist et al. 2003) in Hawaii, Aeonium (Liu 1989; Jorgensen and Olesen 2001) and Echium (Böhle et al. 1996) in the Canary Islands and Scalesia (Eliasson 1974) in the Gálapagos archipelago.
Figure 1.
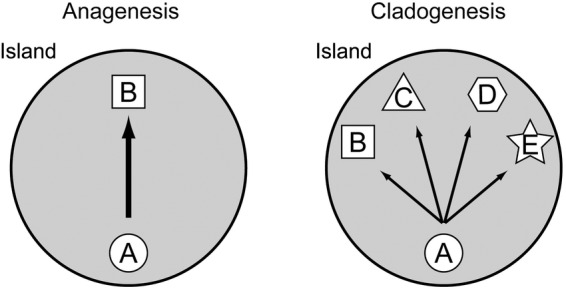
Diagram of the two principal modes of speciation in oceanic islands.
In addition to speciation via adaptive radiation (involving cladogenesis), another process, anagenesis (Fig. 1), has recently been emphasized (Stuessy et al. 1990, 2006; Whittaker et al. 2008). Some immigrant populations, especially when arriving on an island with limited ecological opportunity, proliferate in size and accumulate genetic diversity mainly through mutation and recombination. After many generations (perhaps over a million or more years), genetic changes result in different morphology that may be treated as a distinct species. This process has been labelled anagenetic speciation (Stuessy et al. 2006), being one type of progenitor-derivative speciation (Crawford 2010). It has been estimated that at least one-quarter of all endemic plant species of oceanic islands have originated via anagenesis (Stuessy et al. 2006).
Some studies have been published on the genetic consequences of cladogenesis in endemic plants of different archipelagos. Böhle et al. (1996) examined chloroplast sequence variation among endemic species of Echium (Boraginaceae) of the Canary Islands, showing very little nucleotide divergence even though the morphological variation is striking. Likewise, Baldwin (2003) examined internal transcribed spacer regions of nuclear ribosomal DNA (ITS) variation among species of the Hawaiian silverswords (Asteraceae) and again, limited sequence variation was seen. The general result from these, and other studies, is that during cladogenesis, the immigrant population becomes fragmented, with each segment containing a limited range of genetic variation in comparison with the continental progenitor population (Baldwin et al. 1998). Maximum morphological divergence occurs but with low levels of observable genetic diversity (Frankham 1997). There is some evidence (Perugganan et al. 2003) that the genetic changes responsible for the morphological adaptations involve alterations in regulatory rather than structural genes.
Results so far with anagenesis show a strikingly different pattern. Most of the investigations have been done on endemic species of Ullung Island, in which at least 88 % of the endemic species have originated anagenetically (Stuessy et al. 2006). The island is young (1.8 Ma; Kim 1985), of low elevation (<1000 m) and relatively ecologically uniform (Yim et al. 1981). Pfosser et al. (2005), using amplified fragment length polymorphisms (AFLPs), examined island and Japanese populations of Dystaenia takesimana and D. ibukiensis, respectively, and the results showed high levels of genetic variation within D. takesimana in comparison with D. ibukiensis. Similar results have been obtained in assessing the origin of Acer takesimensis and A. okomotoanum (Takayama et al. 2012, 2013a). Because there is no partitioning of the immigrant population, it survives and proliferates, during which time it accumulates genetic variation through mutation and recombination. Eventually, the level of genetic diversity may even equal (or surpass) that observed in parental source populations (Stuessy 2007).
Because the above studies have been done on different genera in different island archipelagos, it would be useful to compare the genetic consequences of both types of speciation within groups of the same island system, preferably within the same island. In this fashion, more direct comparisons can be made because the general environment is the same. Important, obviously, is to locate plant groups that have originated via both anagenesis and cladogenesis within the same archipelago. A good choice for examining the genetic consequences of anagenesis and cladogenesis in endemic plants of oceanic islands is the Juan Fernández Archipelago, Chile. Approximately 64 % of the species have originated by cladogenesis and 36 % by anagenesis (Stuessy et al. 2006). From another perspective, it is estimated that 70 % of the colonists to the islands have diverged anagenetically, in contrast to only 30 % that have diverged via adaptive radiation (Stuessy et al. 1990).
The Juan Fernández Archipelago consists of two major islands (Fig. 2): Robinson Crusoe (= Masatierra), located 667 km west of continental Chile at 33°S latitude, and Alejandro Selkirk (= Masafuera) situated 181 km further westward into the Pacific Ocean. The former is known to be ∼4 million years old and the latter 1–2 million years old (Stuessy et al. 1984). At present, these two islands are approximately the same size of 50 km2 (Stuessy 1995). The flora is small, containing 78 native and 135 endemic vascular plant species (Danton et al. 2006). From a biogeographic standpoint, this setting is particularly favourable for generating initial hypotheses, because the near island (Robinson Crusoe) is also the older one, making it highly probable as the initial site for colonization of most groups. Furthermore, the older island is hypothesized to have been much larger when formed (Stuessy et al. 1998), making it a bigger target for dispersal from the mainland.
Figure 2.
Location of the Juan Fernández Archipelago and its two major islands, Alejandro Selkirk (= Masafuera) and Robinson Crusoe (= Masatierra).
Numerous molecular markers now exist for assessing genetic variation within and among populations (Lowe et al. 2004). Amplified fragment length polymorphisms (Vos et al. 1995) have been used effectively to provide an overall evaluation of population genetic diversity (Tremetsberger et al. 2003; López-Sepúlveda et al. 2013a). These are treated as dominant markers and hence cannot be employed to determine allelic frequencies. An appropriate co-dominant and polymorphic marker that does allow allelic calculations are nuclear microsatellites or simple sequence repeats (SSRs). The challenge with this marker is to develop primers for locating sequences within the genome for comparison. Next-generation sequencing (NGS) methods are now available that allow this to be done much more easily and at reasonable cost (Takayama et al. 2011, 2013b). Numerous successful applications of SSRs have shown their efficacy to reveal genetic variation at the population level (Gleiser et al. 2008; Kikuchi et al. 2009; López-Sepúlveda et al. 2013b).
Studies using AFLPs and SSRs have already been published on a number of endemic taxa of the Juan Fernández Archipelago, representing groups that have undergone speciation via cladogenesis and anagenesis. The largest (and endemic) genus that has been investigated is Robinsonia (Asteraceae; Takayama et al. 2015), which has seven species on Robinson Crusoe Island that have originated cladogenetically and one on Alejandro Selkirk Island that has evolved anagenetically. The genus Erigeron (Asteraceae; López-Sepúlveda et al. 2015) has six species that evolved cladogenetically on the younger island, Alejandro Selkirk. These two genera were selected because Robinsonia has speciated primarily via cladogenesis on the older island, and Erigeron has done so on the younger island. Regarding anagenesis, studies have been completed on Drimys confertifolia (Winteraceae; López-Sepúlveda et al. 2014) and Rhaphithamnus venustus (Verbenaceae; P. López-Sepúlveda, K. Takayama, D. J. Crawford, J. Greimler, P. Peñailillo, M. Baeza, E. Ruiz, G. Kohl, K. Tremetsberger, A. Gatica, L. Letelier, P. Novoa, J. Novak, T. F. Stuessy, submitted for publication), which occur on both islands of the archipelago. Investigations have also been completed on Myrceugenia (Myrtaceae; López-Sepúlveda et al. 2013b), which contains one endemic species on each of the islands. The available genetic data to date, therefore, come from 15 endemic species, plus 4 close continental relatives, summing to 1870 individuals in 163 populations.
The purposes of this article are to (i) summarize published data from AFLP and SSR investigations on endemic species of the genera Drimys, Myrceugenia, Rhaphithamnus, Robinsonia and Erigeron; (ii) compare and contrast differences in genetic diversity in groups that have undergone anagenetic or cladogenetic speciation and (iii) discuss the importance of considering modes of speciation for understanding levels of genetic diversity within endemic species of oceanic archipelagos.
Methods
The data summarized here (Table 1) provide the first comprehensive genetic comparisons (with AFLPs and SSRs) in the Juan Fernández Archipelago of species that have evolved by anagenesis and cladogenesis, based on consistent samplings, laboratory methods and modes of analysis. A number of earlier studies utilizing isozymes and DNA sequences have examined genetic variation in endemic species of these islands (e.g. Crawford et al. 1998, 2001a), but these investigations were not focussed on comparing modes of speciation. Genera in the present studies were selected for their representation of anagenesis and cladogenesis and for their occurrence on the two islands of different geological ages. The samples were collected during expeditions in February 2010 and 2011 from 1870 individuals in 163 populations in 15 endemic species, hence representing 14 % of the endemic angiosperms in the archipelago. The samples provide very good geographic coverage of populations over the landscape in both islands. The term population, as used here in the sense of sampling, refers to groups of individuals that were clearly delimited spatially in the field. The number of individuals analysed per population ranged from 1 to 31. The voucher data for these samples and details of data gathering and analysis are given in the respective publications.
Table 1.
Summary of measures of genetic diversity in endemic species of the Juan Fernández Archipelago that have originated by anagenesis or cladogenesis. All average values. Data from López-Sepúlveda et al. (2013a, b, 2014), Takayama et al. (2015) and P. López-Sepúlveda, K. Takayama, D. J. Crawford, J. Greimler, P. Peñailillo, M. Baeza, E. Ruiz, G. Kohl, K. Tremetsberger, A. Gatica, L. Letelier, P. Novoa, J. Novak, T. F. Stuessy, submitted for publication. TNB, total number of bands (fragments); PPB, percentage of polymorphic bands; SDI, Shannon Diversity Index; AGDOL, average gene diversity over loci; RI, rarity index; HO, observed proportion of heterozygotes; HE, expected proportion of heterozygotes; NA, number of alleles per locus; FIS, inbreeding coefficient; AR5, allelic richness standardized by five individuals; RC, Robinson Crusoe Island; AS, Alejandro Selkirk Island.
| Species | AFLPs |
Microsatellites (SSRs) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of pops. | No. of plants | TNB | PPB | SDI | AGDOL | RI | No. of pops. | No. of plants | HO | HE | NA | FIS | AR5 | |
| Anagenesis | ||||||||||||||
| D. confertifolia (RC) | 16 | 183 | 557 | 96.5 | 125.3 | 0.26 | 1.96 | 16 | 181 | 0.48 | 0.68 | 9.00 | 0.29 | 4.12 |
| D. confertifolia (AS) | 15 | 96 | 538 | 96.5 | 114.3 | 0.23 | 2.26 | 15 | 80 | 0.35 | 0.51 | 6.38 | 0.26 | 3.24 |
| D. confertifolia (combined RC and AS) | 31 | 279 | 576 | 100 | 134.7 | 0.28 | 2.06 | 31 | 261 | 0.44 | 0.68 | 9.88 | 0.33 | 4.13 |
| M. fernandeziana (RC) | 18 | 211 | 371 | 100 | 74.6 | 0.23 | 1.76 | 18 | 231 | 0.38 | 0.49 | 10.08 | 0.19 | 3.38 |
| M. schulzei (AS) | 13 | 129 | 417 | 100 | 96.2 | 0.28 | 3.39 | 13 | 155 | 0.39 | 0.61 | 10.33 | 0.35 | 3.79 |
| R. venustus (RC) | 20 | 143 | 440 | 99.3 | 96.4 | 0.25 | 2.80 | 20 | 140 | 0.17 | 0.23 | 4.22 | 0.31 | 1.83 |
| R. venustus (AS) | 4 | 18 | 271 | 57.3 | 60.8 | 0.18 | 2.34 | 4 | 11 | 0.30 | 0.34 | 2.33 | 0.13 | 2.12 |
| R. venustus (combined RC and AS) | 24 | 161 | 443 | 100 | 98.7 | 0.26 | 2.75 | 24 | 151 | 0.18 | 0.28 | 4.56 | 0.40 | 2.04 |
| R. masafuerae (AS) | 5 | 9 | 344 | 41.4 | 84.1 | 0.15 | 2.90 | 5 | 7 | 0.36 | 0.43 | 3.50 | 0.17 | 3.08 |
| Cladogenesis | ||||||||||||||
| Robinsonia gayana (RC) | 10 | 123 | 592 | 77.2 | 111.0 | 0.16 | 2.39 | 10 | 134 | 0.34 | 0.42 | 6.30 | 0.28 | 3.04 |
| R. gracilis (RC) | 5 | 75 | 515 | 63.2 | 97.3 | 0.15 | 2.68 | 5 | 87 | 0.28 | 0.39 | 3.50 | 0.24 | 2.26 |
| R. evenia (RC) | 6 | 73 | 586 | 73.4 | 112.0 | 0.17 | 3.18 | 6 | 86 | 0.21 | 0.26 | 2.80 | 0.21 | 1.87 |
| R. saxatilis (RC) | 1 | 5 | 267 | 29.0 | 67.0 | 0.14 | 1.99 | 1 | 5 | 0.30 | 0.26 | 2.10 | −0.22 | 2.10 |
| Robinsonia (combined all RC species) | 22 | 276 | 765 | 100 | 183.7 | 0.26 | 2.77 | 22 | 312 | 0.28 | 0.66 | 8.40 | 0.61 | 3.97 |
| Robinsonia (combined all species) | 27 | 285 | 766 | 100 | 265.0 | 0.26 | 2.68 | 27 | 319 | 0.29 | 0.67 | 8.70 | 0.61 | 4.02 |
| E. fernandezianus (RC) | 13 | 240 | 403 | 90.3 | 70.7 | 0.20 | 0.58 | 13 | 271 | 0.21 | 0.29 | 4.20 | 0.31 | 2.17 |
| E. fernandezianus (AS) | 19 | 172 | 426 | 95.3 | 81.1 | 0.23 | 0.81 | 19 | 200 | 0.17 | 0.50 | 7.50 | 0.72 | 3.27 |
| E. fernandezianus (combined RC and AS) | 32 | 412 | 433 | 97.5 | 81.7 | 0.23 | 0.68 | 32 | 471 | 0.20 | 0.40 | 8.00 | 0.64 | 2.86 |
| E. ingae (AS) | 2 | 21 | 315 | 61.3 | 62.0 | 0.18 | 0.62 | 2 | 25 | 0.20 | 0.34 | 2.90 | 0.55 | 2.04 |
| E. luteoviridis (AS) | 2 | 25 | 334 | 61.5 | 60.2 | 0.18 | 0.99 | 2 | 25 | 0.05 | 0.31 | 3.10 | 0.72 | 2.19 |
| E. rupicola (AS) | 9 | 175 | 377 | 81.8 | 69.5 | 0.20 | 0.67 | 9 | 211 | 0.17 | 0.36 | 4.40 | 0.57 | 2.43 |
| E. turricola (AS) | 3 | 10 | 269 | 49.3 | 57.6 | 0.19 | 0.50 | 3 | 10 | 0.24 | 0.53 | 3.40 | 0.57 | 2.94 |
| E. stuessyi (AS) | 1 | 8 | 306 | 66.7 | 82.4 | 0.28 | 0.81 | 2 | 11 | 0.20 | 0.25 | 2.10 | 0.53 | 1.89 |
| Erigeron (combined all AS species) | 36 | 411 | 443 | 100 | 95.1 | 0.26 | 0.74 | 37 | 482 | 0.17 | 0.62 | 9.20 | 0.76 | 2.85 |
| Erigeron (combined all species) | 49 | 651 | 444 | 100 | 94.2 | 0.26 | 0.68 | 50 | 753 | 0.18 | 0.56 | 9.50 | 0.73 | 3.46 |
| Total and averages | ||||||||||||||
| Anagenesis | 91 | 789 | 419.7 | 84.4 | 93.1 | 0.23 | 2.49 | 91 | 805 | 0.35 | 0.47 | 6.55 | 0.24 | 3.08 |
| Cladogenesis | 71 | 927 | 399.1 | 68.1 | 79.2 | 0.19 | 1.38 | 72 | 1065 | 0.2 | 0.4 | 3.8 | 0.41 | 2.38 |
| Robinson Crusoe | 89 | 1053 | 466.4 | 78.6 | 94.3 | 0.19 | 2.17 | 89 | 1135 | 0.30 | 0.38 | 5.28 | 0.20 | 2.60 |
| Alejandro Serkirk | 73 | 663 | 359.7 | 71.1 | 76.8 | 0.21 | 1.53 | 74 | 735 | 0.24 | 0.42 | 4.59 | 0.46 | 2.70 |
| Anagenesis (RC) | 54 | 537 | 456.0 | 98.6 | 98.8 | 0.24 | 2.17 | 54 | 552 | 0.34 | 0.47 | 7.77 | 0.26 | 3.11 |
| Anagenesis (AS) | 37 | 252 | 392.5 | 73.8 | 88.9 | 0.21 | 2.72 | 37 | 253 | 0.35 | 0.47 | 5.64 | 0.23 | 3.06 |
| Cladogenesis (RC) | 35 | 516 | 472.6 | 66.6 | 91.6 | 0.16 | 2.16 | 35 | 583 | 0.27 | 0.32 | 3.78 | 0.17 | 2.29 |
| Cladogenesis (AS) | 36 | 411 | 337.8 | 69.3 | 68.8 | 0.21 | 0.73 | 37 | 482 | 0.17 | 0.38 | 3.90 | 0.61 | 2.46 |
Briefly, the following approaches were used for AFLPs. Four or six selective primer combinations were chosen. Numerous (24–85) primer trials were run with each genus to determine the best combination of primers for good resolution of individuals and populations. Data were obtained on an automated DNA sequencer (ABI 3130xl, Applied Biosystems, Waltham, MA, USA). Scoring was done using GeneMarker ver. 1.85 (SoftGenetics, State College, PA, USA). For analysis of AFLP data, the programs Arlequin 3.5.1.2 (Excoffier et al. 2005), FAMD ver. 1.25 (Schlüter and Harris 2006), R-Script AFLPdat (Ehrich 2006) and SPSS ver. 15.0 (SPSS; IBM, Armonk, NY, USA) were employed to determine total number of fragments (TNB), percentage of polymorphic fragments (PPB), Shannon Diversity Index (SDI), average gene diversity over loci (AGDOL) and rarity index (RI).
For SSRs, NGS methods (Takayama et al. 2011) were used to generate 6–12 loci, selected for their repeatability and scoring convenience. Polymerase chain reaction-amplified fragments were also run on the same automated sequencer and scored with GeneMarker ver. 1.85. Data analysis involved using GENEPOP 4.0 (Raymond and Rousset 1995), Micro-Checker 2.2.3 (van Oosterhout et al. 2004), FSTAT 2.9.3.2 and GENALEX 6 (Peakall and Smouse 2006). These allow analyses for observed proportion of heterozygotes (HO), expected proportion of heterozygotes (HE), number of alleles per locus (NA), inbreeding coefficient (FIS) and allelic richness standardized by five individuals (AR5).
The overall pattern of higher genetic diversities in anagenetically derived species in comparison with cladogenetically derived ones was examined by a Student's t-test (average TNB, PPB, SDI, AGDOL and RI in AFLPs, and HO, HE, NA and AR5 in SSRs) and shown in Table 2. To improve normality of HO and HE, a square-root transformation was applied. The overall patterns of higher genetic diversities in Robinson Crusoe Island (old) than Alejandro Selkirk Island (new) were also examined in the same way. The effects of two factors (speciation mode and island) and their interaction were analysed in a two-way ANOVA in R version 3.0.0 (R Core Team 2013) and shown in Table 3.
Table 2.
Summary of statistical tests based on Table 1. TNB, total number of bands (fragments); PPB, percentage of polymorphic bands; SDI, Shannon Diversity Index; AGDOL, average gene diversity over loci; RI, rarity index; HO, observed proportion of heterozygotes; HE, expected proportion of heterozygotes; NA, number of alleles per locus; AR5, allelic richness standardized by five individuals. Bold font indicates significant values (P < 0.05).
| High genetic diversity in anagenetically derived species | High genetic diversity in Robinson Crusoe Island species | |
|---|---|---|
| AFLPs | ||
| TNB | 0.351 | 0.024 |
| PPB | 0.086 | 0.235 |
| SDI | 0.101 | 0.045 |
| AGDOL | 0.050 | 0.227 |
| RI | 0.004 | 0.085 |
| SSRs | ||
| HO | 0.006 | 0.132 |
| HE | 0.061 | 0.236 |
| NA | 0.040 | 0.308 |
| AR5 | 0.038 | 0.388 |
Table 3.
Summary of two-way ANOVA based on Table 1. TNB, total number of bands (fragments); PPB, percentage of polymorphic bands; SDI, Shannon Diversity Index; AGDOL, average gene diversity over loci; RI, rarity index; HO, observed proportion of heterozygotes; HE, expected proportion of heterozygotes; NA, number of alleles per locus; AR5, allelic richness standardized by five individuals. For all F-values, the degree of freedom was 1. Bold font indicates significant values (P < 0.05).
| Factor | F-value | P-value | |
|---|---|---|---|
| AFLPs | |||
| TNB | Island | 4.78 | 0.046 |
| Speciation mode | 0.22 | 0.645 | |
| Island vs. speciation mode | 0.51 | 0.489 | |
| PPB | Island | 0.67 | 0.427 |
| Speciation mode | 2.60 | 0.129 | |
| Island vs. speciation mode | 2.05 | 0.174 | |
| SDI | Island | 3.61 | 0.078 |
| Speciation mode | 2.36 | 0.147 | |
| Island vs. speciation mode | 0.47 | 0.504 | |
| AGDOL | Island | 0.85 | 0.372 |
| Speciation mode | 4.09 | 0.063 | |
| Island vs. speciation mode | 4.67 | 0.048 | |
| RI | Island | 4.63 | 0.049 |
| Speciation mode | 13.71 | 0.002 | |
| Island vs. speciation mode | 10.53 | 0.006 | |
| SSRs | |||
| HO | Island | 2.03 | 0.176 |
| Speciation mode | 11.65 | 0.004 | |
| Island vs. speciation mode | 1.64 | 0.221 | |
| HE | Island | 0.47 | 0.502 |
| Speciation mode | 3.44 | 0.085 | |
| Island vs. speciation mode | 0.19 | 0.671 | |
| NA | Island | 0.47 | 0.502 |
| Speciation mode | 3.44 | 0.085 | |
| Island vs. speciation mode | 0.19 | 0.671 | |
| AR5 | Island | 0.10 | 0.752 |
| Speciation mode | 4.54 | 0.051 | |
| Island vs. speciation mode | 0.11 | 0.744 | |
Data from both AFLPs and microsatellites were further analysed by assessing genetic distance (Nei et al. 1983) with the NeighborNet algorithm (Bryant and Moulton 2004) implemented by SplitsTree4 ver. 4.10 (Huson and Bryant 2006) and Population 1.2.30 (Langella 1999), respectively.
For this article, to allow ease of visual comparisons of results among the species, emphasis has been placed on selected graphic presentations. SplitsTree NeighborNet was employed with the AFLP data, and the results are given in a series of graphs (Fig. 3). Neighbour-joining based on genetic distance was used for analysis of the SSRs, and simplified networks were used to show relationships among the populations (Fig. 4). For summary comparisons of genetic diversity among species, AGDOL was used with the AFLP data (Fig. 5). Not all calculated values for all original populations are presented or discussed in this review. The reader is referred to the original publications for additional methods and data.
Figure 3.
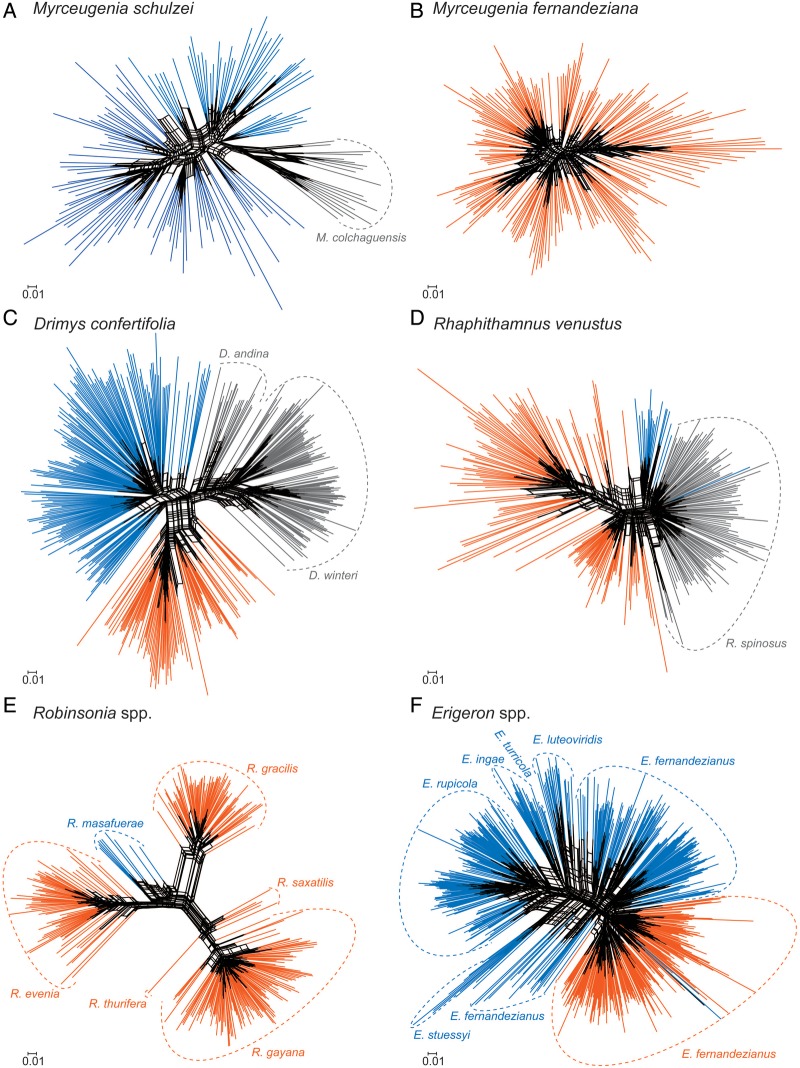
SplitsTree NeighborNet showing genetic relationships based on AFLPs among individuals in endemic species of Myrceugenia (A and B), Drimys (C), Rhaphithamnus (D), Robinsonia (E) and Erigeron (F) in the Juan Fernández Archipelago. Closely related continental relatives are also shown in A, C and D. Orange = species and populations on Robinson Crusoe Island; blue = on Alejandro Selkirk Island and black = on the or islands continent.
Figure 4.
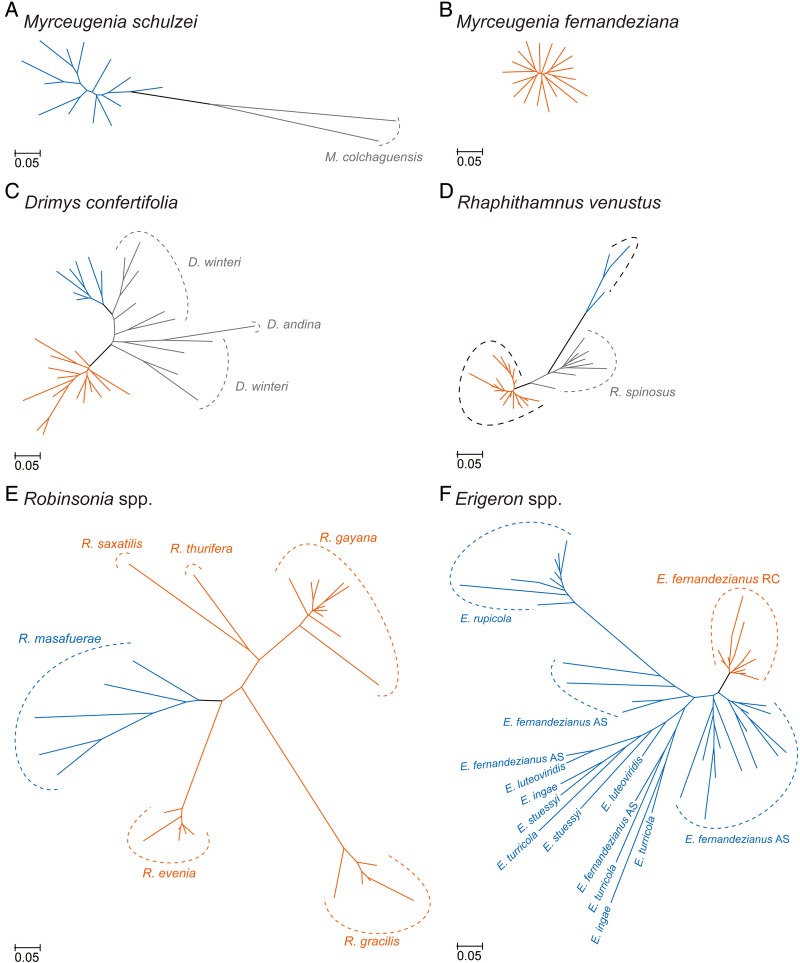
Neighbour-joining tree showing genetic relationships based on SSRs among populations in endemic species of Myrceugenia (A and B), Drimys (C), Rhaphithamnus (D), Robinsonia (E) and Erigeron (F) in the Juan Fernández Archipelago. Closely related continental relatives are also shown in A, C and D. Orange = species and populations on Robinson Crusoe Island; blue = on Alejandro Selkirk Island and black = on the continent.
Figure 5.
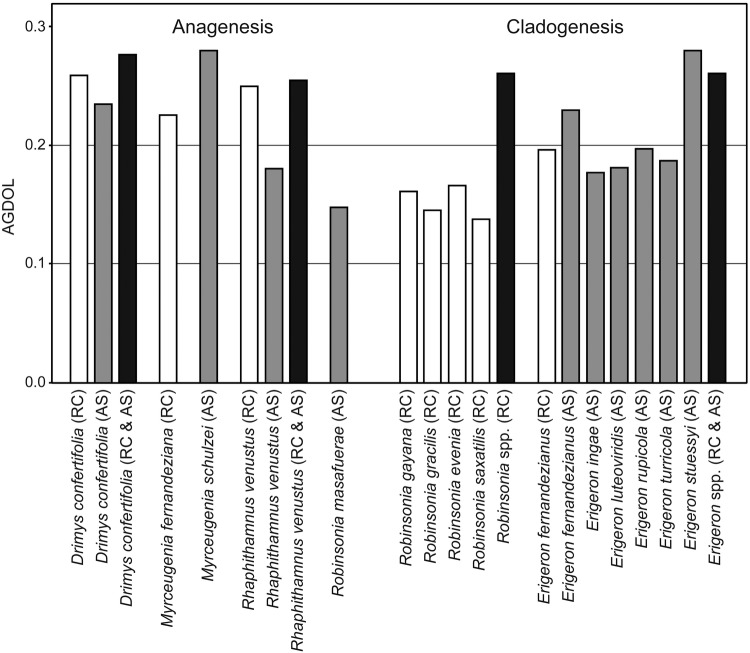
Summary of genetic diversities, AGDOL, within the endemic species of Drimys, Myrceugenia and Rhaphithamnus having originated by anagenesis, and Robinsonia and Erigeron having been derived through cladogenesis. Robinsonia masafuerae from the younger island is also an anagenetic derivative from the cladogenetic complex of Robinsonia on the older island. AS, Alejandro Selkirk Island; RC, Robinson Crusoe Island. White bar indicates an endemic species in RC, grey bar an endemic species in AS and black bar multiple species or islands combined.
Results
The results from the AFLP and SSR data analyses are given in Tables 1–4 and shown graphically in Figs 3–5. In general, the results from the two sources of genetic data are similar, with some exceptions, reinforcing confidence in the patterns seen. These data will be presented in context of the two modes of speciation, anagenesis and cladogenesis, but with attention also to the different ages of the islands. Robinson Crusoe Island is ∼4 million years old and Alejandro Selkirk 1–2 million (Stuessy et al. 1984).
Table 4.
Generalized comparison of the levels of genetic diversity obtained with AFLPs and SSRs from species that have originated via anagenesis and cladogenesis on the two islands of the Juan Fernández Archipelago. See Table 1 for the actual data. RC, Robinson Crusoe Island; AS, Alejandro Selkirk Island.
| Anagenesis |
Cladogenesis |
|||
|---|---|---|---|---|
| RC | AS | RC | AS | |
| AFLPs | ||||
| Total number of bands (TNB) | High | Medium | High | Low |
| Percentage of polymorphic bands (PPB) | High | Low | Low | Low |
| Shannon Diversity Index (SDI) | High | Medium high | Medium high | Low |
| Average gene diversity over loci (AGDOL) | High | Medium high | Low | Medium high |
| Rarity index (RI) | Medium | High | Medium | Very low |
| Microsatellites (SSRs) | ||||
| Observed proportion of heterozygotes (HO) | High | High | Medium | Low |
| Expected proportion of heterozygotes (HE) | High | High | Medium | Medium |
| Number of alleles per locus (NA) | High | Medium | Low | Low |
| Inbreeding coefficient (FIS) | Low | Low | Low | High |
| Allelic richness (AR5) | High | High | Low | Low |
Anagenesis
The results from analysis of species that have evolved anagenetically include those from Myrceugenia fernandeziana, M. schulzei, Robinsonia masafuerae, D. confertifolia and R. venustus. The first species occurs only on the older island, the second and third species only on the younger island and the last two on both islands. A number of points seem evident. First, all anagenetically derived species show considerable levels of genetic diversity (Table 1, and Figs 3 and 5), and none of them shows geographic patterns over the island landscape (López-Sepúlveda et al. 2013b, 2014, P. López-Sepúlveda, K. Takayama, D. J. Crawford, J. Greimler, P. Peñailillo, M. Baeza, E. Ruiz, G. Kohl, K. Tremetsberger, A. Gatica, L. Letelier, P. Novoa, J. Novak, T. F. Stuessy, submitted for publication). This is what might be expected from the predictions regarding anagenesis based on previous studies. Even more interesting, perhaps, is that the amount of genetic diversity differs in species on the two islands of different ages. In D. confertifolia, and R. venustus, which occur on both islands, one sees in both cases more genetic diversity (SDI) in populations on the older island than on the younger island except for estimates of SSRs in R. venustus (Table 1). The explanation of these data may relate to the time available for a genetic change to take place. Because Alejandro Selkirk Island is no more than 1–2 million years old, this must be the maximum time available for population divergence to take place. With anagenetically evolved species, all factors being equal, genetic variation increases through time, and this can be seen in the species investigated.
One case of anagenesis in the archipelago also merits comment. Robinsonia masafuerae is a species that appears to have speciated from R. evenia, with which it has been closely associated in all studies so far (Crawford et al. 1993a; Sang et al. 1995; Takayama et al. 2015). Previous investigations on ITS 1 and 2 in Robinsonia (Sang et al. 1995) have shown sequence divergence between R. evenia and R. masafuerae as only 0.0063 (two base substitutions). Although one cannot place an absolute time on this divergence, it is the lowest level among any pair of species in the genus, which correlates well with the youthful geological age of Alejandro Selkirk Island. Genetic variation in R. masafuerae is much lower from AFLP data than in R. evenia from Robinson Crusoe (Table 1 and Fig. 5), but in SSRs, the pattern reverses with the anagenetically derived species, R. masafuerae, showing more variation than any single one of the cladogenetically originated species on Robinson Crusoe (Table 1).
It is also possible to make comparisons between populations of continental progenitors with endemic island derivatives. In the case of Myrceugenia schulzei, the closest continental congener is M. colchaguensis (Landrum 1981a, b; Ruiz et al. 2004). Although the sampling of populations on the continent is limited to two populations, the amount of genetic diversity is particularly low as shown by AFLP data, although somewhat higher with SSRs (López-Sepúlveda et al. 2013b). Although M. schulzei is known only on the younger island, it did not diverge from M. fernandeziana on the older island because the two are unrelated (Murillo-Aldana et al. 2012), so much so that the latter has now been transferred to another genus (Nothomyrcia; Murillo-Aldana and Ruiz 2011). With D. confertifolia, comparisons with D. winteri and D. andina show less genetic variation in the two latter species as seen from AFLPs and SSRs (López-Sepúlveda et al. 2014). In R. venustus, which is a congener of R. spinosus (the only other known species in the genus; Moldenke 1937; Crawford et al. 1993b), the amount of genetic diversity is again greater in the population on Robinson Crusoe Island than documented on the continent, although considerably lower in the population on Alejandro Selkirk (P. López-Sepúlveda, K. Takayama, D. J. Crawford, J. Greimler, P. Peñailillo, M. Baeza, E. Ruiz, G. Kohl, K. Tremetsberger, A. Gatica, L. Letelier, P. Novoa, J. Novak, T. F. Stuessy, submitted for publication). These results support the concept that over time, considerable genetic variation can accumulate in anagenetically derived populations, so much so that the degree of variation can approximate and even surpass that in the progenitor source populations.
Cladogenesis
Two of the largest genera of the archipelago are Robinsonia with eight endemic species and Erigeron with six. Both are in Asteraceae, although unrelated and placed in different tribes (Senecioneae vs. Astereae, respectively). Robinsonia has adaptively radiated on Robinson Crusoe Island during the past 4 million years (maximum value) and Erigeron has done so on Alejandro Selkirk Island in the past 1–2 million years.
Robinsonia is the second largest genus in the archipelago. The largest is Dendroseris, also of Asteraceae but from still another tribe (Cichorieae). This latter genus is of interest as it has derived cladogenetically on the older island with three independent dispersals to the younger island and three anagenetic speciations there (Sanders et al. 1987; Pacheco et al. 1991; Sang et al. 1994). Most of these species are quite rare now, however, which precluded our being able to obtain sufficient population data for genetic evaluation. Robinsonia has eight species, but two are presumed extinct (R. berteroi and R. megacephala; Danton et al. 2006). Our studies have focussed on five species having originated cladogenetically on the older island. Comments have already been made regarding the one anagenetically derived species (R. masafuerae) on Alejandro Selkirk Island. The results from AFLP data are shown in Fig. 3 and from SSRs in Fig. 4. Most notable from the SplitsTree graph in Fig. 3 is that the different species of Robinsonia are very distinct genetically. Divergence has obviously taken place during adaptive radiation and also during a maximum time available of 4 million years. The species R. gayana, R. thurifera and R. saxatilis form an evolutionary complex, which taxonomically has been regarded as sect. Robinsonia (Skottsberg 1922, as sect. Symphyolepis; Takayama et al. 2015). Robinsonia gracilis ties with R. evenia and its close anagenetic relative R. masafuerae in sect. Eleutherolepis (Skottsberg 1922). With SSR data (Fig. 4), the species are also very distinct and genetically more cohesive, with the anagenetic species R. masafuerae showing the greatest genetic diversity. Another important point seen clearly in Figs 3 and 4 is that the range of genetic diversity within each of these cladogenetic species is limited in comparison with the anagenetically derived species discussed above.
Although Erigeron is not an endemic genus in the archipelago, six endemic species occur there having evolved via cladogenesis and adaptive radiation. The origin of this complex is unusual in that the colonist(s) presumably arrived directly to the younger island (Valdebenito et al. 1992). Amplified fragment length polymorphism and SSR data (Figs 3 and 4) reveal considerable genetic diversity within these endemic species, and each species is reasonably distinct. An exception is the Erigeron ingae complex consisting of E. ingae, E. luteoviridis and E. turricola. These species are sometimes difficult to distinguish morphologically. Solbrig (1962) and Marticorena et al. (1998), for example, placed E. turricola into synonymy with E. ingae, but Danton et al. (2006) kept them distinct. The molecular data parallel this morphological inconsistency. This may be a population complex in early stages of speciation, now undergoing divergence from within a pool of morphological and genetic variation. All of these species grow in the ‘alpine zone’ on the younger island (Skottsberg 1922), and we have not noticed any clear habitat differences among them. The species E. rupicola is confined to coastal rocks along the sea and also penetrates into the quebradas (ravines); its close relative, E. stuessyi, is also found on rocky ledges but residing inside the cool and deep ravines. Erigeron fernandezianus occurs in a broad altitudinal range (100–1200 m), and it inhabits mainly rocky areas in middle elevation plains, quebradas and ridges. This species also occurs on the older island, but it is found there in many plant communities and especially in disturbed sites. It appears, therefore, to be an example of back migration from the younger to the older island (Valdebenito et al. 1992; López-Sepúlveda et al. 2015).
Although most species of Erigeron on the younger island are distinct genetically, the degree of distinctness is much less than observed among species of Robinsonia on the older island (Figs 3 and 4). It may be that these species of Erigeron have had less time to diverge in comparison with those of Robinsonia. With the passage of time, therefore, the genetic profiles of species undergoing adaptive radiation may remain narrow due to strong directional selection in each different habitat. In both Erigeron and Robinsonia, however, the range of genetic variation seen is less than that in the anagenetically derived species.
Discussion
Comparison of anagenesis and cladogenesis
Predictions from theory (Stuessy 2007) would suggest that higher levels of genetic diversity should be found within the anagenetically derived species. This is because the founding population increases in size over time, accumulating genetic diversity mainly through mutation and recombination. One would expect no (or very little) geographic partitioning over the landscape. Likewise, due to a lack of strong selection, one would not expect to find high levels of private alleles or bands, nor a high RI. With cladogenetic speciation, on the other hand, one would expect less overall genetic diversity within each species, but with more private alleles due to strong directional selection. As for impact from the age of the islands, one would predict less total genetic diversity within anagenetically derived species on the younger island because diversity increases through time. As for the cladogenetic species, one would predict less genetic divergence (distinctness) on the younger island in comparison with species on the older island, because directional selection continues over time and refines the genetic profile of each species as it adapts to the particular ecological zone.
Results from genetic analyses of 5 anagenetic species and 10 cladogenetic species allow comparisons between the two modes of speciation and the two islands of differing ages (Tables 1–3). A number of general points can be observed (Table 4 and Fig. 5). First, in anagenetic species, the level of genetic diversity tends to be higher per species than in the cladogenetic species, especially on Robinson Crusoe Island. This can be seen in percentage of polymorphic bands, SDI, AGDOL, observed proportion of heterozygotes, expected proportion of heterozygotes, number of alleles per locus and allelic richness. Second, in the anagenetic species, the individuals on each island behave genetically as one large population, showing no genetic pattern over the landscape (López-Sepúlveda et al. 2013b, 2014; Takayama et al. 2015; P. López-Sepúlveda, K. Takayama, D. J. Crawford, J. Greimler, P. Peñailillo, M. Baeza, E. Ruiz, G. Kohl, K. Tremetsberger, A. Gatica, L. Letelier, P. Novoa, J. Novak, T. F. Stuessy, submitted for publication). This is true on both islands of differing ages. This suggests that this pattern can develop easily within 1–2 million years and that it can persist for up to 4 million. This is consistent with the results reported for Ullung Island, Korea, which is known to be 1.8 million years old (Pfosser et al. 2005; Takayama et al. 2012, 2013a). Third, the ability of an immigrant population to radiate adaptively has much to do with the properties of the colonists (and progenitors) and less with differences of habitat. Some colonists remain as a single larger population and are not responsive to adaptive change in different ecological zones, whereas others disperse well to micro-zones and quickly become modified morphologically and genetically. Fourth, perhaps most importantly, the total amount of genetic diversity within an anagenetically derived species in comparison with an entire adaptively radiating lineage is approximately the same (Fig. 5).
Genetics of speciation in endemic plants of oceanic islands
A number of previous studies have assessed levels of genetic variation within and among populations of endemic species of the Juan Fernández Archipelago with other markers such as isozymes, random amplified polymorphic DNA (RAPDs) and inter simple sequence repeats (ISSRs). Isozymes have been analysed in Dendroseris (Crawford et al. 1987), Chenopodium sanctae-clarae (Crawford et al. 1988), Wahlenbergia (Crawford et al. 1990), Robinsonia (Crawford et al. 1992), Lactoris (Crawford et al. 1994) and Myrceugenia (Jensen et al. 2002). RAPDs have been investigated in Dendroseris (Esselman et al. 2000) and Lactoris (Brauner et al. 1992), and ISSRs also in Lactoris (Crawford et al. 2001b).
Crawford et al. (2001a) summarized the results from isozyme studies on 29 endemic species of the Juan Fernández Archipelago, and this represents the best set of observations to compare with the AFLP and SSR data summarized here. The most conspicuous result is that the mean genetic diversities at the species level are low (Hes = 0.065). Higher levels of diversity were seen in larger populations or in many small populations and also in outcrossing species in contrast to selfers. Of relevance for comparisons to the present study, isozymes have been analysed from four species of Robinsonia and in M. fernandeziana, E. fernandezianus and R. venustus. It is difficult to compare the results of the isozymes because they provide less detailed genetic information than from AFLPs and SSRs. Isozyme studies on the endemic Lactoris fernandezianus, for example Crawford et al. (1994), revealed virtually no variation, but ISSRs showed considerable variation within and among populations (Crawford et al. 2001b). Studies on isozymes (Crawford et al. 1987) and RAPDs (Esselman et al. 2000) from Dendroseris showed greater resolution of relationships from the latter. The isozyme data for the four cladogenetically derived species of Robinsonia show higher levels of genetic variation than in the anagenetic R. venustus (Crawford et al. 1993b) and Myrceugenia (Jensen et al. 2002), which would be in contrast to the trends documented here. It is important, therefore, that for questions involving population genetics in endemic plants of oceanic islands, rapidly evolving markers need to be used.
The employment of AFLPs and SSRs in the present study from 15 species of the Juan Fernández Archipelago, therefore, does provide detailed genetic data at the population level for purposes of comparing consequences of different modes of speciation. A general review has recently been published on the general topic of interpretation of genetic variation within endemic species of oceanic islands (Stuessy et al. 2014), and the present data corroborate ideas summarized there. Clearly, the alternative modes of speciation, anagenesis and cladogenesis result in different genetic consequences. Interpretation of the evolutionary significance of levels of genetic diversity, therefore, must be done in context of type of speciation. As can be seen in the results of adaptive radiation in Erigeron and Robinsonia, on the young and older islands, respectively, the geological age of the island also matters, as this provides the time dimension in which the evolutionary processes unfold.
Another very significant impact on levels of genetic variation in populations of endemic plants of oceanic islands is that from human activity. Because oceanic islands often have agreeable climates and attractive beaches, people have come to live, play and build homes and apartments, all of which have caused pressures on the native vegetation. In the Juan Fernández Archipelago, for example, people have been living continuously on Robinson Crusoe Island for >300 years (Woodward 1969; Wester 1991). It is not impossible that the species of Robinsonia on the older island have suffered some genetic loss due to human activity. Although these species occur either on high ridges or in deep forests, far removed from most persons who live at sea level in the village (San Juan Bautista), incursions into the native forest must have taken place and some plants destroyed. It is known that two species of Robinsonia, both on Robinson Crusoe Island, are now extinct (R. berteroi and R. megacephala; Danton and Perrier 2005; Danton et al. 2006). Assessing the level of human impact on the vegetation of an oceanic island, therefore, is challenging. At least in the Juan Fernández Archipelago, there were no aboriginal peoples, and human activity could only have begun with discovery by Europeans (Juan Fernández; Medina 1974) at the end of the 16th century. Since that time, however, considerable negative impact from human activity has been documented in the archipelago (Wester 1991; Matthei et al. 1993; Stuessy et al. 1997; Swenson et al. 1997; Cuevas and Leersum 2001; Greimler et al. 2002; Dirnböck et al. 2003; Cuevas et al. 2004; Ricci 2006; Vargas et al. 2011), especially from introduced animals, such as rats, rabbits and goats (e.g. Camus et al. 2008). These combined activities have surely had some impact on the levels of genetic variation within and among populations.
Sources of Funding
This work was supported by an FWF (Austrian Science Fund) grant (P21723-B16) to T.F.S. and a Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad (grant 526) to Ko.T.
Contributions by the Authors
Ko.T. conceived the idea behind the article; all authors participated in the field work except G.K. and Ka.T.; J.N., P.L.-S., G.K. and Ko.T. completed the laboratory work; J.N. coordinated the NGS data acquisition; T.F.S. and Ko.T wrote the initial draft and all authors contributed to subsequent drafts and offered comments for improvement.
Conflict of Interest Statement
None declared.
Acknowledgements
We very much appreciate the generous logistic and facility support of Sr Iván Leiva, Chief of the Robinson Crusoe Islands national park, administered by the Corporación Nacional Forestal (CONAF); the help and cooperation in fieldwork from the CONAF guides, especially Jorge Angulo, Danilo Arredondo, Danilo Arredondo, Jr, Oscar Chamorro, Michael González, Bernardo López, Eduardo Paredes, Ramon Schiller and Manuel Tobar; and the Armada de Chile for logistic support in transporting supplies from the continent to the islands. The results presented in this paper form part of an Open Partnership Joint Project of the JSPS Bilateral Joint Research program.
Literature Cited
- Baldwin BG. 2003. A phylogenetic perspective on the origin and evolution of Madiinae. In: Carlquist S, Baldwin BG, Carr GD, eds. Tarweeds & silverswords: evolution of the Madiinae (Asteraceae). St. Louis: Missouri Botanical Garden Press, 193–228. [Google Scholar]
- Baldwin BG, Crawford DJ, Francisco-Ortega J, Kim S-C, Sang T, Stuessy TF. 1998. Molecular phylogenetic insights on the origin and evolution of oceanic island plants. In: Soltis DE, Soltis PS, Doyle JJ, eds. Molecular systematics of plants II: DNA sequencing. Boston: Kluwer, 410–441. [Google Scholar]
- Böhle U-R, Hilger HH, Martin WF. 1996. Island colonization and evolution of the insular woody habit in Echium L. (Boraginaceae). Proceedings of the National Academy of Sciences of the USA 93:11740–11745. 10.1073/pnas.93.21.11740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramwell D, Caujapé-Castells J. 2011. The biology of island floras. Cambridge: Cambridge University Press. [Google Scholar]
- Brauner S, Crawford DJ, Stuessy TF. 1992. Ribosomal DNA and RAPD variation in the rare plant family Lactoridaceae. American Journal of Botany 79:1436–1439. 10.1093/molbev/msh018 [DOI] [Google Scholar]
- Bryant D, Moulton V. 2004. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Molecular Biology and Evolution 21:255–265. 10.1093/molbev/msh018 [DOI] [PubMed] [Google Scholar]
- Camus P, Castro S, Jaksic F. 2008. El conejo europeo en Chile: Historia de una invasión histórica. Historia 41:305–339. [Google Scholar]
- Carlquist S. 1974. Island biology. New York: Columbia University Press. [Google Scholar]
- Carlquist S, Baldwin BG, Carr GD, eds. 2003. Tarweeds & silverswords: evolution of the Madiinae (Asteraceae). St. Louis: Missouri Botanical Garden. [Google Scholar]
- Crawford DJ. 2010. Progenitor-derivative species pairs and plant speciation. Taxon 59:1413–1423. [Google Scholar]
- Crawford DJ, Stuessy TF, Silva M. 1987. Allozyme divergence and the evolution of Dendroseris (Compositae: Lactuceae) on the Juan Fernandez Islands. Systematic Botany 12:435–443. 10.2307/2419268 [DOI] [Google Scholar]
- Crawford DJ, Stuessy TF, Silva M. 1988. Allozyme variation in Chenopodium sanctae-clarae, an endemic species of the Juan Fernandez Islands, Chile. Biochemical Systematics and Ecology 16:279–284. 10.1016/0305-1978(88)90008-7 [DOI] [Google Scholar]
- Crawford DJ, Stuessy TF, Lammers TG, Silva M, Pacheco P. 1990. Allozyme variation and evolutionary relationships among three species of Wahlenbergia (Campanulaceae) in the Juan Fernandez Islands. Botanical Gazette 151:119–124. 10.1086/337811 [DOI] [Google Scholar]
- Crawford DJ, Stuessy TF, Haines DW, Cosner MB, Silva M, Lopez P. 1992. Allozyme diversity within and divergence among four species of Robinsonia (Asteraceae: Senecioneae), a genus endemic to the Juan Fernandez Islands, Chile. American Journal of Botany 79:962–966. 10.2307/2445008 [DOI] [Google Scholar]
- Crawford DJ, Stuessy TF, Cosner MB, Haines DW, Silva M. 1993a. Ribosomal and chloroplast DNA restriction site mutations and the radiation of Robinsonia (Asteraceae: Senecioneae) on the Juan Fernandez Islands. Plant Systematics and Evolution 184:233–239. 10.1007/BF00937437 [DOI] [Google Scholar]
- Crawford DJ, Stuessy TF, Rodriguez R, Rondinelli M. 1993b. Genetic diversity in Rhaphithamnus venustus (Verbenaceae), a species endemic to the Juan Fernandez Islands. Bulletin of the Torrey Botanical Club 120:23–28. 10.2307/2996659 [DOI] [Google Scholar]
- Crawford DJ, Stuessy TF, Cosner MB, Haines DW, Wiens D, Peñaillo P. 1994. Lactoris fernandeziana (Lactondaceae) on the Juan Fernandez Islands: allozyme uniformity and field observations. Conservation Biology 8:277–280. 10.1046/j.1523-1739.1994.08010277.x [DOI] [Google Scholar]
- Crawford DJ, Sang T, Stuessy TF, Kim S-C, Silva M. 1998. Dendroseris (Asteraceae: Lactuceae) and Robinsonia (Asteraceae: Senecioneae) on the Juan Fernandez Islands: similarities and differences in biology and phylogeny. In: Stuessy TF, Ono M, eds. Evolution and speciation of island plants. Cambridge: Cambridge University Press, 97–119. [Google Scholar]
- Crawford DJ, Ruiz E, Stuessy TF, Tepe E, Aqueveque P, González F, Jensen RJ, Anderson GJ, Bernardello G, Baeza CM, Swenson U, Silva M. 2001a. Allozyme diversity in endemic flowering plant species of the Juan Fernandez Archipelago, Chile: ecological and historical factors with implications for conservation. American Journal of Botany 88:2195–2203. 10.2307/3558381 [DOI] [PubMed] [Google Scholar]
- Crawford DJ, Tago-Nakazawa M, Stuessy TF, Anderson GJ, Bernardello G, Ruiz E, Jensen RJ, Baeza C, Wolfe AD, Silva M. 2001b. Intersimple sequence repeat (ISSR) variation in Lactoris fernandeziana (Lactoridaceae), a rare endemic of the Juan Fernández Archipelago, Chile. Plant Species Biology 16:185–192. 10.1046/j.1442-1984.2001.00065.x [DOI] [Google Scholar]
- Cuevas JG, van Leersum G. 2001. Project “Conservation, restoration, and development of the Juan Fernández Islands, Chile”. Revista Chilena de Historia Natural 74:899–910. 10.4067/S0716-078X2001000400016 [DOI] [Google Scholar]
- Cuevas JG, Marticorena A, Cavieres LA. 2004. New additions to the introduced flora of the Juan Fernández Islands: origin, distribution, life history traits, and potential of invasion. Revista Chilena de Historia Natural 77:523–538. 10.4067/S0716-078X2004000300011 [DOI] [Google Scholar]
- Danton P, Perrier C. 2005. Note sur la disparition d'une espèce emblématique, Robinsonia berteroi (DC.) Sanders, Stuessy & Martic. (Asteraceae) dans l'ile Robinson Crusoe, archipel Juan Fernández (Chili). Journal de Botanique de la Société Botanique de France 31:1–6. [Google Scholar]
- Danton P, Perrier C, Martinez Reyes G. 2006. Nouveau catalogue de la flore vasculaire de l'archipel Juan Fernández (Chili). Nuevo catálogo de la flora vascular del Archipiélago Juan Fernández (Chile). Acta Botanica Gallica 153:399–587. 10.1080/12538078.2006.10515559 [DOI] [Google Scholar]
- Darwin C. 1842. The structure and distribution of coral reefs. London: Smith, Elder and Co. [Google Scholar]
- Dirnböck T, Greimler J, López P, Stuessy TF. 2003. Predicting future threats to the native vegetation of Robinson Crusoe Island, Juan Fernandez Archipelago, Chile. Conservation Biology 17:1650–1659. 10.1111/j.1523-1739.2003.00173.x [DOI] [Google Scholar]
- Ehrich D. 2006. AFLPdat: a collection of R functions for convenient handling of AFLP data. Molecular Ecology Notes 6:603–604. 10.1111/j.1471-8286.2006.01380.x [DOI] [Google Scholar]
- Eliasson U. 1974. Studies in Galapágos Plants XIV. The genus Scalesia Arn. Opera Botanica 36:1–117. [Google Scholar]
- Esselman EJ, Crawford DJ, Brauner S, Stuessy TF, Anderson GJ, Silva M. 2000. RAPD marker diversity within and divergence among species of Dendroseris (Asteraceae: Lactuceae). American Journal of Botany 87:591–596. 10.2307/2656603 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Frankham R. 1997. Do island populations have less genetic variation than mainland populations? Heredity 78:311–327. 10.1038/hdy.1997.46 [DOI] [PubMed] [Google Scholar]
- Givnish TJ, Millam KC, Mast AR, Paterson TB, Theim TJ, Hipp AL, Henss JM, Smith JF, Wood KR, Sytsma KJ. 2009. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proceedings of the Royal Society B Biological Sciences 276:407–416. 10.1098/rspb.2008.1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleiser G, Verdú M, Segarra-Moragues JG, González-Martínez SC, Pannell JR. 2008. Disassortative mating, sexual specialization, and the evolution of gender dimorphism in heterodichogamous Acer opalus. Evolution 62:1676–1688. 10.1111/j.1558-5646.2008.00394.x [DOI] [PubMed] [Google Scholar]
- Greimler J, Stuessy TF, Swenson U, Baeza CM, Matthei O. 2002. Plant invasions on an oceanic archipelago. Biological Invasions 4:73–85. 10.1023/A:1020565510507 [DOI] [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23:254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Jensen RJ, Schwoyer M, Crawford DJ, Stuessy TF, Anderson GJ, Baeza CM, Silva M, Ruiz E. 2002. Patterns of morphological and genetic variation among populations of Myrceugenia fernandeziana (Myrtaceae) on Masatierra Island: implications for conservation. Systematic Botany 27:534–547. [Google Scholar]
- Jorgensen TH, Olesen JM. 2001. Adaptive radiation of island plants: evidence from Aeonium (Crassulaceae) of the Canary Islands. Perspectives in Plant Ecology, Evolution and Systematics 4:29–42. 10.1078/1433-8319-00013 [DOI] [Google Scholar]
- Kikuchi S, Shibata M, Tanaka H, Yoshimaru H, Niiyama K. 2009. Analysis of the disassortative mating pattern in a heterodichogamous plant, Acer mono Maxim. using microsatellite markers. Plant Ecology 204:43–54. 10.1007/s11258-008-9564-1 [DOI] [Google Scholar]
- Kim YK. 1985. Petrology of Ulreung volcanic island, Korea—Part 1. Geology. Journal of the Japanese Association of Mineralogists, Petrologists and Economic Geologists 80:128–135. 10.2465/ganko1941.80.128 [DOI] [Google Scholar]
- Landrum L. 1981a. A monograph of the genus Myrceugenia (Myrtaceae). Bronx, NY: New York Botanical Garden Press. [Google Scholar]
- Landrum LR. 1981b. The phylogeny and geography of Myrceugenia (Myrtaceae). Brittonia 33:105–129. 10.2307/2806583 [DOI] [Google Scholar]
- Langella O. 1999. Populations, 1.2.30. http://www.bioinformatics.org/~tryphon/populations/ (15 October 2014).
- Liu H-Y. 1989. Systematics of Aeonium (Crassulaceae). Special Publications of the National Museum of Natural Science (Taichung), No. 3, 1–102. [Google Scholar]
- López-Sepúlveda P, Tremetsberger K, Ortiz MA, Baeza CM, Peñailillo P, Stuessy TF. 2013a. Radiation of the Hypochaeris apargioides complex (Asteraceae: Cichorieae) of southern South America. Taxon 62:550–564. 10.12705/623.14 [DOI] [Google Scholar]
- López-Sepúlveda P, Takayama K, Greimler J, Peñailillo P, Crawford DJ, Baeza M, Ruiz E, Kohl G, Tremetsberger K, Gatica A, Letelier L, Novoa P, Novak J, Stuessy TF. 2013b. Genetic variation (AFLPs and nuclear microsatellites) in two anagenetically derived endemic species of Myrceugenia (Myrtaceae) on the Juan Fernández Islands, Chile. American Journal of Botany 100:722–734. 10.3732/ajb.1200541 [DOI] [PubMed] [Google Scholar]
- López-Sepúlveda P, Takayama K, Greimler J, Crawford DJ, Peñailillo P, Baeza M, Ruiz E, Kohl G, Tremetsberger K, Gatica A, Letelier L, Novoa P, Novak J, Stuessy TF. 2014. Progressive migration and anagenesis in Drimys confertifolia of the Juan Fernández Archipelago, Chile. Journal of Plant Research 128:73–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Sepúlveda P, Takayama K, Crawford DJ, Greimler J, Peñailillo P, Baeza M, Ruiz E, Kohl G, Tremetsberger K, Gatica A, Letelier L, Novoa P, Novak J, Stuessy TF. 2015. Speciation and biogeography of Erigeron (Asteraceae) endemic to the Juan Fernández Archipelago, Chile, based on AFLPs and SSRs. Systematic Botany, 40: 10.1600/036364415X689311. [DOI] [Google Scholar]
- Lowe A, Harris S, Ashton P. 2004. Ecological genetics: design, analysis, and application. Oxford: Blackwell. [Google Scholar]
- Marticorena C, Stuessy TF, Baeza M. 1998. Catálogo de la flora vascular del Archipiélago de Juan Fernández, Chile. Gayana Botánica 55:187–211. [Google Scholar]
- Matthei O, Marticorena C, Stuessy TF. 1993. La flora adventicia del archipiélago de Juan Fernández. Gayana Botánica 50:69–102. [Google Scholar]
- Medina JT. 1974. El piloto Juan Fernández, descubridor de las islas que llevan su nombre, y Juan Jufre, armador de la expedición que hizo en busca de otras en el Mar del Sur. Santiago: Editora Nacional Gabriela Mistral. [Google Scholar]
- Moldenke HN. 1937. A monograph of the genus Rhaphithamnus. Repertorium Novarum Specierum Regni Vegetabilis 42:62–82. 10.1002/fedr.19370420111 [DOI] [Google Scholar]
- Murillo-Aldana J, Ruiz E. 2011. Revalidación de Nothomyrcia (Myrtaceae), un género endémico del Archipiélago de Juan Fernández. Gayana Botánica 68:129–134. 10.4067/S0717-66432011000200002 [DOI] [Google Scholar]
- Murillo-Aldana J, Ruiz-P E, Landrum LR, Stuessy TF, Barfuss MHJ. 2012. Phylogenetic relationships in Myrceugenia (Myrtaceae) based on plastid and nuclear DNA sequences. Molecular Phylogenetics and Evolution 62:764–776. 10.1016/j.ympev.2011.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Tajima F, Tateno Y. 1983. Accuracy of estimated phylogenetic trees from molecular data II. Gene frequency data. Journal of Molecular Evolution 19:153–170. 10.1007/BF02300753 [DOI] [PubMed] [Google Scholar]
- Pacheco P, Crawford DJ, Stuessy TF, Silva OM. 1991. Flavonoid evolution in Dendroseris (Compositae, Lactuceae) from the Juan Fernandez Islands, Chile. American Journal of Botany 78:534–543. 10.2307/2445263 [DOI] [Google Scholar]
- Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6:288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perugganan MD, Remington DL, Robichaux RH. 2003. Molecular evolution of regulatory genes in the silversword alliance. In: Carlquist S, Baldwin BG, Carr GD, eds. Tarweeds & silverswords: evolution of the Madiinae (Asteraceae). St. Louis: Missouri Botanical Garden Press, 171–182. [Google Scholar]
- Pfosser M, Jakubowsky G, Schlüter PM, Fer T, Kato H, Stuessy TF, Sun B-Y. 2005. Evolution of Dystaenia takesimana (Apiaceae), endemic to Ullung Island, Korea. Plant Systematics and Evolution 256:159–170. 10.1007/s00606-005-0374-9 [DOI] [Google Scholar]
- Raymond M, Rousset F. 1995. GENEPOP (version 1.2): population genetics software for exact test and ecumenicism. Journal of Heredity 86:248–249. [Google Scholar]
- R Core Team. 2013. A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rensch B. 1959. Evolution above the species level. New York: Columbia University Press. [Google Scholar]
- Ricci M. 2006. Conservation status and ex situ cultivation efforts of endemic flora of the Juan Fernández Archipelago. Biodiversity and Conservation 15:3111–3130. 10.1007/s10531-005-5404-y [DOI] [Google Scholar]
- Ruiz E, Crawford DJ, Stuessy TF, González F, Samuel R, Becerra J, Silva M. 2004. Phylogenetic relationships and genetic divergence among endemic species of Berberis, Gunnera, Myrceugenia and Sophora of the Juan Fernández Islands (Chile) and their continental progenitors based on isozymes and nrITS sequences. Taxon 53:321–332. 10.2307/4135611 [DOI] [Google Scholar]
- Rundell RJ, Price TD. 2009. Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends in Ecology and Evolution 24:394–399. 10.1016/j.tree.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Sanders RW, Stuessy TF, Marticorena C, Silva M. 1987. Phytogeography and evolution of Dendroseris and Robinsonia, tree-Compositae of the Juan Fernandez Islands. Opera Botanica 92:195–215. [Google Scholar]
- Sang T, Crawford DJ, Kim S-C, Stuessy TF. 1994. Radiation of the endemic genus Dendroseris (Asteraceae) on the Juan Fernandez Islands: evidence from sequences of the ITS regions of nuclear ribosomal DNA. American Journal of Botany 81:1494–1501. 10.2307/2445322 [DOI] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF, Silva M. 1995. ITS sequences and the phylogeny of the genus Robinsonia (Asteraceae). Systematic Botany 20:55–64. 10.2307/2419632 [DOI] [Google Scholar]
- Schluter D. 2001. Ecology and the origin of species. Trends in Ecology and Evolution 16:372–380. 10.1016/S0169-5347(01)02198-X [DOI] [PubMed] [Google Scholar]
- Schlüter PM, Harris SA. 2006. Analysis of multilocus fingerprinting data sets containing missing data. Molecular Ecology Notes 6:569–572. 10.1111/j.1471-8286.2006.01225.x [DOI] [Google Scholar]
- Skottsberg C. 1922. The phanerogams of the Juan Fernandez Islands. In: Skottsberg C, ed. The natural history of Juan Fernandez and Easter Island, Vol. 2 Uppsala: Almqvist & Wiksells, 95–240. [Google Scholar]
- Solbrig OT. 1962. The South American species of Erigeron. Contributions from the Gray Herbarium of Harvard University 191:3–79. [Google Scholar]
- Stuessy TF. 1995. Juan Fernández Islands. In: Davis SD, Heywood VH, Hamilton AC, eds. Centres of plant diversity: a guide and strategy of their conservation. Cambridge: IUCN Publications Unit, 565–568. [Google Scholar]
- Stuessy TF. 2007. Evolution of specific and genetic diversity during ontogeny of island floras: the importance of understanding process for interpreting island biogeographic patterns. In: Ebach MC, Tangney RS, eds. Biogeography in a changing world. Boca Raton: CRC Press, 117–133. [Google Scholar]
- Stuessy TF, Foland KA, Sutter JF, Sanders RW, Silva M. 1984. Botanical and geological significance of potassium-argon dates from the Juan Fernandez Islands. Science 225:49–51. 10.1126/science.225.4657.49 [DOI] [PubMed] [Google Scholar]
- Stuessy TF, Crawford DJ, Marticorena C. 1990. Patterns of phylogeny in the endemic vascular flora of the Juan Fernandez Islands, Chile. Systematic Botany 15:338–346. 10.2307/2419187 [DOI] [Google Scholar]
- Stuessy TF, Swenson U, Marticorena C, Mathei O, Crawford DJ. 1997. Loss of plant diversity and extinction on Robinson Crusoe Island, Chile. In: Peng C-I, ed. Rare, threatened, and endangered floras of Asia and the Pacific Rim. Taipei: Academia Sinica; (Monograph Series No. 16), 147–257. [Google Scholar]
- Stuessy TF, Crawford DJ, Marticorena C, Rodriguez R. 1998. Island biogeography of angiosperms of the Juan Fernandez archipelago. In: Stuessy TF, Ono M, eds. Evolution and speciation of island plants. Cambridge: Cambridge University Press, 121–138. [Google Scholar]
- Stuessy TF, Jakubowsky G, Gómez RS, Pfosser M, Schluter PM, Fer T, Sun B-Y, Kato H. 2006. Anagenetic evolution in island plants. Journal of Biogeography 33:1259–1265. 10.1111/j.1365-2699.2006.01504.x [DOI] [Google Scholar]
- Stuessy TF, Takayama K, López-Sepúlveda P, Crawford DJ. 2014. Interpretation of patterns of genetic variation in endemic plant species of oceanic islands. Botanical Journal of the Linnean Society 174:276–288. 10.1111/boj.12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson U, Stuessy TF, Baeza M, Crawford DJ. 1997. New and historical plant introductions, and potential pests in the Juan Fernández Islands, Chile. Pacific Science 51:233–253. [Google Scholar]
- Takayama K, López P, König C, Kohl G, Novak J, Stuessy TF. 2011. A simple and cost-effective approach for microsatellite isolation in non-model plant species using small-scale 454 pyrosequencing. Taxon 60:1442–1449. [Google Scholar]
- Takayama K, Sun B-Y, Stuessy TF. 2012. Genetic consequences of anagenetic speciation in Acer okamotoanum (Sapindaceae) on Ullung Island, Korea. Annals of Botany 109:321–330. 10.1093/aob/mcr280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, Sun B-Y, Stuessy TF. 2013a. Anagenetic speciation in Ullung Island, Korea: genetic diversity and structure in the island endemic species, Acer takesimense (Sapindaceae). Journal of Plant Research 126:323–333. 10.1007/s10265-012-0529-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, López-Sepúlveda P, Kohl G, Novak J, Stuessy TF. 2013b. Development of microsatellite markers in Robinsonia (Asteraceae) an endemic genus of the Juan Fernández Archipelago, Chile. Conservation Genetics Resources 5:63–67. 10.1007/s12686-012-9734-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, López-Sepúlveda P, Greimler J, Crawford DJ, Peñailillo P, Baeza M, Ruiz E, Kohl G, Tremetsberger K, Gatica A, Letelier L, Novoa P, Novak J, Stuessy TF. 2015. Relationships and genetic consequences of contrasting modes of speciation among endemic species of Robinsonia (Asteraceae, Senecioneae) of the Juan Fernández Archipelago, Chile, based on AFLPs and SSRs. New Phytologist 205:415–428. 10.1111/nph.13000 [DOI] [PubMed] [Google Scholar]
- Tremetsberger K, Stuessy TF, Guo Y-P, Baeza CM, Weiss H, Samuel RM. 2003. Amplified fragment length polymorphism (AFLP) variation within and among populations of Hypochaeris acaulis (Asteraceae) of Andean southern South America. Taxon 52:237–245. 10.2307/3647392 [DOI] [Google Scholar]
- Valdebenito H, Stuessy TF, Crawford DJ, Silva M. 1992. Evolution of Erigeron (Compositae) in the Juan Fernandez Islands, Chile. Systematic Botany 17:470–480. 10.2307/2419485 [DOI] [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4:535–538. 10.1111/j.1471-8286.2004.00684.x [DOI] [Google Scholar]
- Vargas R, Reif A, Faúndez MJ. 2011. The forests of Robinson Crusoe Island, Chile: an endemism hotspot in danger. Bosque 32:155–164. 10.4067/S0717-92002011000200006 [DOI] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Friters A, Pot J, Paleman J, Kuiper M, Zabeau M. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23:4407–4414. 10.1093/nar/23.21.4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AR. 1881. Island life. London: Macmillan and Co. [Google Scholar]
- Wester L. 1991. Invasions and extinctions on Masatierra (Juan Fernández Islands): a review of early historical evidence. Journal of Historical Geography 17:18–34. 10.1016/0305-7488(91)90003-E [DOI] [Google Scholar]
- Whittaker RJ, Fernández-Palacios JM. 2007. Island biogeography, 2nd edn Oxford: Oxford University Press. [Google Scholar]
- Whittaker RJ, Triantis KA, Ladle RJ. 2008. A general dynamic theory of oceanic island biogeography. Journal of Biogeography 35:977–994. 10.1111/j.1365-2699.2008.01892.x [DOI] [Google Scholar]
- Woodward RL. 1969. Robinson Crusoe’s Island: a history of the Juan Fernandez Islands. Chapel Hill: University of North Carolina Press. [Google Scholar]
- Yim Y-J, Lee E-B, Kim S-H. 1981. Vegetation of Ulreung and Dogdo Islands. A Report on the Scientific Survey of the Ulreung and Dogdo Islands Seoul: The Korean Association for Conservation of Nature. [Google Scholar]