Abstract
In plants, LRR-RLKs play central roles in regulating perception of extracellular signals and initiation of cellular responses under various environmental challenges. Arabidopsis SERK genes, including SERK1 to SERK5, constitute a LRR-RLK sub-family. SERK1, SERK2, SERK3/BAK1, and SERK4/BKK1 have been well characterized to function as crucial regulators in multiple physiological processes such as brassinosteroid signaling, cell death control, pathogenesis, and pollen development. Despite extremely high sequence identity with BKK1, SERK5 is reported to have no functional overlapping with BKK1, which is previously identified to regulate BR and cell death control pathways, probably due to a natural mutation in a highly conserved RD motif in the kinase domain of SERK5 in Col-0 ecotype. Through a gene sequencing analysis in several Arabidopsis accessions, we are able to identify SERK5 in Landsberg erecta (Ler) genome encoding a LRR-RLK with an intact RD motif. Overexpression of SERK5-Ler partially suppresses the BR defective phenotypes of bri1-5 and bak1-3 bkk1-1, indicating SERK5-Ler functions as a positive regulator in BR signaling. Furthermore, the interaction between SERK5-Ler and BRI1 is confirmed by yeast two-hybrid and BiFC assays, and the genetic result showing that elevated expression of a kinase-dead form of SERK5-Ler causes a dominant-negative phenotype in bri1-5. In addition, overexpression of SERK5-Ler is capable of delaying, not completely suppressing, the cell death phenotype of bak1-3 bkk1-1. In this study, we first reveal that SERK5-Ler is a biologically functional component in mediating multiple signaling pathways.
Keywords: Arabidopsis, brassinosteroid, cell death, LRR-RLK, natural variation, RD kinase, SERK, signal transduction
Introduction
During evolution, plants have developed multiple strategies to sense surrounding cues in order to adapt to the environment. Perception of extracellular stimuli and initiation of intracellular signaling cascades serve as key processes for plant cells to make different responses under normal and various stressed conditions. Due to their unique structural properties, receptor-like protein kinases (RLKs) in plants play essential roles in cell-cell and cell-environment communications (Li, 2010). RLKs contain an extracellular domain and a cytosolic kinase domain, connected via a single-pass transmembrane helix. The ectodomains of RLKs perceive the apoplastic signals including phytohormones and small peptides followed by the activation of cytosolic kinase domain triggering protein phosphorylation cascades as responses. In Arabidopsis, there are more than 610 RLKs, including about 135 receptor-like cytoplasmic kinases (RLCKs), which lack the ectodomains (Shiu and Bleecker, 2001a). As the largest RLK subfamily, leucine-rich repeat RLK (LRR-RLK) family contain at least 223 members, featured by 1–32 copies of leucine-rich repeats in their extracellular domains (Shiu and Bleecker, 2001a,b; Lehti-Shiu et al., 2009). LRR-RLKs are grouped into 13 subfamilies (LRR I to XIII), based on their protein sequence similarity in their kinase domains (Shiu and Bleecker, 2001a). In the kinase subdomain VIb of a number of LRR-RLKs, an Arg (R) residue resides preceding a highly conserved Asp (D) residue, referred to an RD motif, followed by the activation loop. Arg (R) residue is sometimes replaced by uncharged residues such as Leu (L), Gly (G), Cys (C), or Phe (F) in some LRR-RLKs (Krupa et al., 2004). Thus, LRR-RLKs can also be categorized into RD kinases and non-RD kinases according to the presence or absence of an RD motif in their kinase domains. For most RD LRR-RLKs, the phosphorylation in the activation loop is crucial for triggering kinase activity which usually displays high (auto- and trans-) phosphorylation ability. Non-RD LRR-RLKs do not require phosphorylating the activation loop and usually exhibit lower kinase activities (Johnson et al., 1996; Nolen et al., 2004; Schwessinger et al., 2011). Distinct molecular mechanisms contribute to non-RD kinase activation, such as keeping constitutively active, release of C-terminus to remove auto-inhibition, or Tyr (Y) phosphorylation in kinase domain (Kobe et al., 1996; Mayans et al., 1998; Nolen et al., 2001).
One well-established model for paired LRR-RLK receptors functioning in signal sensing and transduction includes brassinosteroid (BR) receptor BRASSINOSTEROID INSENSITIVE 1 (BRI1) (Li and Chory, 1997) and its co-receptor BRI1-ASSOCIATED KINASE 1 (BAK1) (Li et al., 2002; Nam and Li, 2002). Genetics, biochemistry, and protein structure analyses have demonstrated that BRI1 is an indispensable bona fide receptor of brassinolide (BL), the most biologically active form of BR (He et al., 2000; Wang et al., 2001; Kinoshita et al., 2005; Hothorn et al., 2011; She et al., 2011). BAK1 was identified as an interacting protein of BRI1 (Li et al., 2002; Nam and Li, 2002), acting as a co-receptor of BRI1 (Li, 2010). Recent genetic and protein structure results support the irreplaceable role of BAK1 and its homologs in BR signaling via a direct association with a BL molecule (Gou et al., 2012; Santiago et al., 2013; Sun et al., 2013b). Upon BR perception, the BRI1-BAK1 complex initiates downstream signaling cascades and ultimately finely tunes the transcriptional programs to regulate various aspects of plant development, including cell expansion and division, vascular differentiation, cell proliferation, male fertility, senescence, induction of flowering, and stress responses (Clouse and Sasse, 1998; He et al., 2001; Nakaya et al., 2002; Symons et al., 2006; Divi and Krishna, 2009; Ye et al., 2010; Belkhadir and Jaillais, 2015).
BAK1 belongs to SOMATIC-EMBRYOGENESIS RECEPTOR-LIKE KINASE (SERK) sub-family with five members (SERK1 to SERK5) (Hecht et al., 2001). BAK1 is also known as SERK3, and SERK4, the closest homolog of BAK1, was named as BAK1-Like 1 (BKK1) (He et al., 2007). In Arabidopsis, SERKs participate in multiple signaling pathways in which they are often functionally redundant (He et al., 2007; Albrecht et al., 2008; Roux et al., 2011; Schwessinger et al., 2011; Du et al., 2012; Gou et al., 2012). In addition to BAK1, SERK1/2 and BKK1 were also discovered to interact with BRI1, functioning as positive regulators of BR responses (Karlova et al., 2006; He et al., 2007; Albrecht et al., 2008; Gou et al., 2012; Santiago et al., 2013).
Intriguingly, our previous studies revealed a novel function of BAK1 and BKK1 in a BR-independent cell death control pathway (He et al., 2007). bak1-4 bkk1-1, a null double mutant, shows a spontaneous cell death phenotype within a week after germination even on the sterilized medium (He et al., 2007). BAK1 was also found to associate with FLAGELLIN SENSITIVE 2 (FLS2), Elongation Factor-Tu Receptor 1 (EFR1) and BAK1-interacting receptor-like kinase 1 (BIR1), respectively, to regulate innate immunity (Chinchilla et al., 2007; Heese et al., 2007; Gao et al., 2009; Schulze et al., 2010; Sun et al., 2013a). BAK1, therefore, has been proposed to be an adaptor protein required for various ligand-binding RLKs to perceive diverse signal molecules and to initiate corresponding downstream signaling pathways (He et al., 2013).
BKK1 is the closed paralog of BAK1 and plays a redundant role with BAK1 at least in BR and cell death control pathways. The closest paralog of BKK1 (At2G13790) is SERK5 (At2G13800) and they exist as a pair of tandem-repeated genes on chromosome 2 (He et al., 2007). SERK5 shares 85% amino acids identity with BKK1, suggesting BKK1 and SERK5 are very likely functionally redundant. However, no reports to date have demonstrated SERK5 is involved in regulating BR signaling or cell death control (He et al., 2007; Albrecht et al., 2008; Jeong et al., 2010; Roux et al., 2011; Gou et al., 2012). Our previous studies showed that overexpression of BKK1, not SERK5, is able to partially suppress bri1-5, a weak BRI1 mutant allele (He et al., 2007; Gou et al., 2012). By using coimmunoprecipitation and mass spectrometry analyses, it has been shown that EFR and FLS2 form heteromerization, in a ligand-induced manner, with SERK1, SERK2, BAK1 and BKK1 but not SERK5 (Roux et al., 2011).
Detailed sequencing analysis indicates that SERK5 in Col-0 ecotype, which was used in previous studies, bears an amino acid substitution of Leu (L) for Arg (R) within the RD motif in the kinase domain, and this substitution might abolish the function of Col-0 SERK5 in the BR signaling pathway (He et al., 2007). It remains to be determined that whether SERK5 containing an intact RD motif exists in other Arabidopsis ecotypes and whether it is functionally active.
In this study, we aimed to elucidate the biological functions of SERK5. To this end, we analyzed SERK5 sequences in five Arabidopsis accessions. Interesting enough, SERK5 in the genome of Landsberg erecta (Ler) ecotype was identified to encode a LRR-RLK with a normal RD motif. By using genetic, biochemistry and molecular biology approaches, SERK5-Ler has been analyzed in BR signaling as a functional regulator. Overexpression of SERK5-Ler was capable of delaying cell death phenotype of bak1-3 bkk1-1, a weak double mutant, suggesting SERK5-Ler also plays a role in cell death control.
To date, little is yet known about the functional roles of SERK5 in Arabidopsis. Our results first unfold SERK5 in an Arabidopsis accession as a biologically functional mediator in regulating BR and cell death pathways, similar to its closest paralog, BKK1.
Materials and methods
Materials and plant growth conditions
Arabidopsis thaliana ecotypes Landsberg erecta (Ler), Columbia-0 (Col-0), Wassilewskija-2 (Ws-2), Lanark-0 (Lan-0) and C24 were used in this study. Plants were grown at 22°C under 16 h light/8 h dark, unless otherwise specified. For BR sensitivity assays, seedlings were grown on 1/2 MS medium supplemented with 1% sucrose, 0.8% agar and various concentrations of 24-epiBL (Sigma, St. Louis, MO).
Gene cloning and Arabidopsis transformation
Full-length cDNAs and genomic DNA fragments of SERK5 in five ecotypes were cloned into expression binary vector pB35GWF by a gateway strategy as described in previous study (Gou et al., 2012). Site-directed mutagenesis was performed according to the manual of the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). All of the cloned DNA sequences were confirmed by sequencing analysis. The cloned genes were transformed into Arabidopsis plants by a floral dip method (Clough and Bent, 1998). T1 seedlings were screened using 0.1% (v/v) basta in the soil. All the primers used for cloning are listed in Table S1.
Determination of plant fresh weight and leaf area
Thirty five-day-old seedlings grown in the greenhouse were weighted and the data were analyzed by SPSS 15.0 software (http://www-01.ibm.com/software/analytics/spss/). For leaf area measurement, fully expanded leaves of 30-day-old seedlings grown in the greenhouse were striped and imaged. The areas of the leaves were determined using Image-Pro Plus software (http://www.mediacy.com/index.aspx?page=IPP). All measurements were repeated at least three times. About 30–60 seedlings were measured each time. All data were presented as the mean ± standard deviation (SD). Student's t-test was used to show statistical differences.
Root and hypocotyl growth analyses
The seeds planted on 1/2 MS medium were vernalizated at 4°C for 2 days. Seedlings were grown vertically in continuous light or dark at 22°C. The root lengths were measured 8 days after germination in the light and the hypocotyl lengths were measured 8 days after germination in the dark.
Gene expression analyses
Total RNA was extracted from the seedlings according to the manual of the RNAprep Pure Plant Kit [TIANGEN BIOTECH (BEIJING) CO., LTD]. For semi-quantitative RT-PCR to confirm overexpression of SERKs in the transgenic plants, RNA was isolated from the leaves of 4-week-old plants grown in the soil. For BR sensitivity test, total RNA was isolated from 10-day-old seedlings grown on 1/2 MS medium treated with or without 1 μM 24-epiBL for 3 h. For the analysis of expression levels of defense- and senescence-related genes, total RNA was isolated from the 13-day-old seedlings grown on 1/2 MS medium. Two micrograms of total RNA was reversely transcribed in a 20 μl volume using Superscript III reverse transcriptase (Invitrogen). The transcripts of specific genes were quantitatively analyzed with SYBR Premix ExTaq kit (TaKaRa). ACTIN2 was used as an internal reference. The primers used for semi-quantitative RT-PCR and qRT-PCR are listed in Table S1.
Protein extraction and western analysis
Total protein was extracted from 3-week-old plants as described by Wang et al. (2005). Protein samples were separated on 10% (w/v) SDS-PAGE gel. Western blotting using α-FLAG antibodies (Sigma, St. Louis, MO) were performed as previously described (Li et al., 2002).
Trypan-blue staining
The leaves from 10 to16-day-old plants were incubated in trypan-blue staining solution (8 ml of 95% ethanol, 1 ml of ddH2O, 1 ml of lactic acid, 1 ml of glycerol, 1 ml of phenol, 0.2 mg trypan-blue) in 1.5 ml tubes and boiled in a water bath for 2–3 min. After cooling to room temperature, the solution was replaced with chloral hydrate (2.5 g/ml). The chloral hydrate solution was replaced a few times until the leaves were fully destained.
Mating-based split ubiquitin system
The mating-based split ubiquitin system (mbSUS) method (Obrdlik et al., 2004) was utilized to test the interaction between BRI1 and SERKs. The full-length BRI1 was introduced into pMetYCgate vector (bait vector) by gateway cloning technology and transformed into the yeast THY.AP4 (Cubs) strain. The full-length SERK5-Ler, SERK5, and BKK1 were introduced into pX-NubWTgate vector (prey vector) by gateway cloning technology and transformed into the yeast THY.AP5 (Nubs) strain. BRI1 was also introduced into pX-NubWTgate vector, wherein SERK5-Ler, SERK5 and BKK1 were introduced into pMetYCgate vector, followed by being transformed into the yeast THY.AP5 (Nubs) strain and THY.AP4 (Cubs) strain, respectively. Yeast-diploid colonies were obtained after mating and selected on the selection medium. The protein-protein interaction was tested by growing yeast on the synthetic minimal medium lacking adenine, histidine, leucine, and tryptophan (-AHLT) and containing 0, 75, 150, and 400 μM methionine. Interactions were also verified using β-galactosidase assays as previously described (Obrdlik et al., 2004). The strength of interaction was quantified by measuring the Integrated Optical Density (IOD) of the growing yeast using Image-Pro Plus software (http://www.mediacy.com/index.aspx?page=IPP).
Bimolecular fluorescence complementation (BIFC) assay
The full-length cDNA of BRI1 was cloned into vector pEG-GWR-nYFP (BRI1-nYFP), and the full-length cDNAs of SERK5-Ler, BKK1 and SERK5 were cloned into vector pEG-GWR-cYFP (SERK5-Ler-cYFP, SERK5-cYFP, and BKK1-cYFP). The above constructs were transformed into Agrobacterium tumefaciens strain GV3101 using electroporation. The transformed GV3101 cells were cultured overnight in LB medium containing 20 μM acetosyringone. Re-suspended the cultures to OD600 nm = 0.5 with injection buffer (liquid MS medium containing 150 μM acetosyringone, 10 mM MgCl2, 10 mM MES, pH 5.7). BRI-nYFP and SERK5-Ler/SERK5/BKK1-cYFP cultures were mixed by equal volumes. The mixtures were incubated for at least 2 h at room temperature, and were co-injected into the leaves of 3-week-old Nicotiana benthamiana. After 72 h, YFP fluorescence emitted from the epidermal cells of the injected leaves was observed with confocal laser scanning microscopy (Olympus FV1000-IX81).
Results
Landsberg erecta (Ler) SERK5 is an RD LRR-RLK
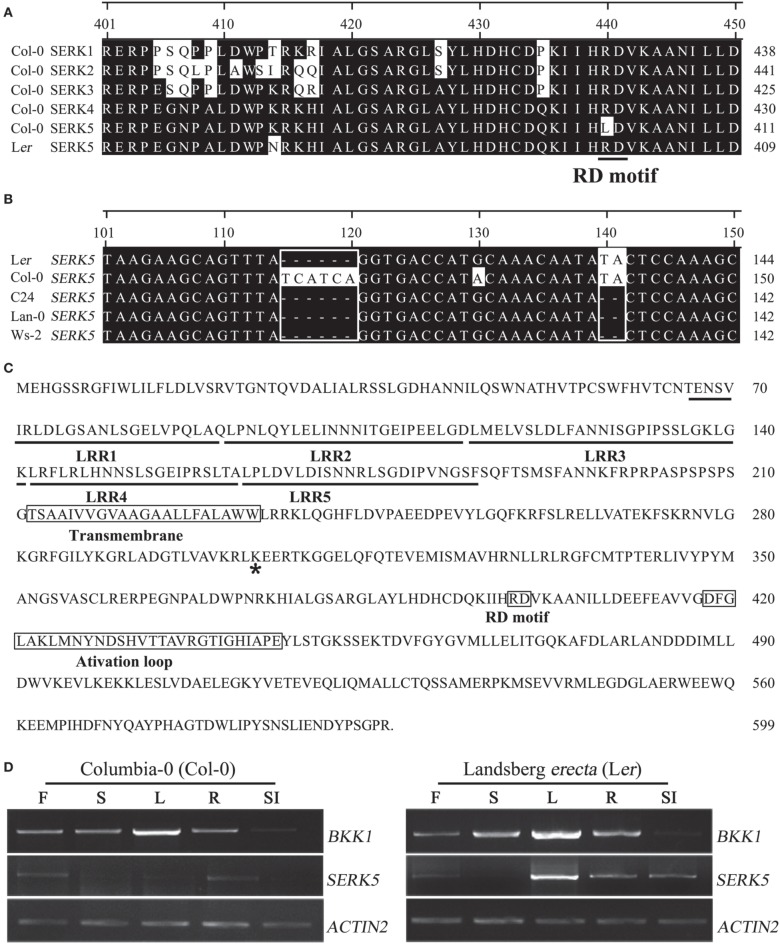
Gain-of-function and loss-of-function genetic analyses have demonstrated that SERK1, SERK2, SERK3/BAK1, and SERK4/BKK1, but not SERK5, in Col-0 ecotype regulate BR signaling (Karlova et al., 2006; He et al., 2007; Albrecht et al., 2008; Gou et al., 2012). Considering that BKK1 and SERK5 share extremely high protein sequence identity (85%), it is puzzling that SERK5 exhibits no functional redundancy with BKK1. This may be explicated by detailed sequence analysis showing that SERK5 in Col-0 contains a substitution of R by L in RD motif, which is different from BKK1, a canonical RD kinase (He et al., 2007). We thus speculated that the eradication of RD motif in SERK5 may abolish its biological functions in Col-0 ecotype. We started analyzing SERK5 sequences in different Arabidopsis accessions. SERK5 cDNAs from Landsberg erecta (Ler), Wassilewskija-2 (Ws-2), Columbia-0 (Col-0), Lanark-0 (Lan-0), and C24 ecotypes were cloned using a gateway strategy. Sequence alignment results indicated that SERK5 in Ler, Ws-2, Lan-0, or C24, but not in Col-0, contains an RD motif (Figure 1A). Unexpectedly, the cDNA sequences of SERK5 from C24, Lan-0 and Ws-2 contains a two-nucleotide base (TA) deletion (Figure 1B). As a result, the SERK5 in C24, Lan-0 and Ws-2 is not able to be properly translated, due to a shifted open reading frame. Furthermore, SERK5 in ecotype C24, Lan-0 and Ws-2 and Ler has a six-nucleotide deletion, leading to absence of two Ser (S) residues (Figure 1B). In order to further verify the SERK5 cDNAs, genomic DNA fragments of SERK5 in five ecotypes were cloned. Sequencing results confirmed that only SERK5 genomic DNA sequence in Ler has no TA deletion and maintains a typical RD motif (data not shown). The full-length CDS of SERK5 in Ler is 1800 bp, encoding a protein of 599 amino acids (Figure 1C), showing 98% and 85% similarity to SERK5 and BKK1 in Col-0, respectively. Given that SERK5 in Ler is truly an RD LRR-RLK, it is still essential to show SERK5 is indeed expressed in Ler ecotype if one claims SERK5 in Ler is physiologically active. By using RT-PCR assay with same cycles (30 cycles), we analyzed the expression patterns of SERK5 in various tissues in both Col-0 and Ler backgrounds, starting with equally amount of total RNA. Our results suggested that the expression levels of SERK5 are much lower in Col-0 than those in Ler (Figure 1D). By contrast, BKK1 was highly expressed in almost all tissues and exhibited the same expression patterns in Col-0 and Ler. Interestingly, compared to SERK5 in Col-0, the transcripts of SERK5 in Ler were significantly higher, especially in rosette leaves where BKK1 also showed the highest expression level (Figure 1D). These results indicated that SERK5 is substantially expressed in Ler with markedly higher expression levels than in Col-0 in which SERK5 is inactive. Taken together, SERK5 in Ler ecotype was identified as a typical RD LRR-RLK, which is subsequently named as SERK5-Ler. In this article, all SERKs refer to those in Col-0 ecotype unless being specifically designated.
Figure 1.
SERK5 in Landsberg erecta (Ler) ecotype encodes a LRR-RLK with a normal RD motif. (A) Amino acid sequence alignment (position 401–450) of SERK1-5 in Col-0 ecotype and SERK5 in Ler ecotype. SERK5-Ler contains a normal RD motif. R is substituted by L at position 401 in SERK5-Col. (B) cDNA sequence alignment (position 101–150) of SERK5 in Ler, Col-0, C24, Lan-0, and Ws-2. SERK5 in Col-0 contains extra six nucleotides in alignment position 115–120. SERK5 in C24, Lan-0, and Ws-2 contains two-nucleotide (TA) deletion at alignment position 140–141. (C) Full protein sequence of SERK5 in Ler. Asterisk mark indicates the conserved lysine (K) in ATP binding domain. (D) Expression levels of BKK1 and SERK5 in Col-0 (left) and Landsberg erecta (Ler) (right) ecotype are shown in all plant tissue. Semi-quantitative RT-PCR was carried out with RNA isolated from the 4 weeks old Col-0 and Ler ecotype Arabidopsis grown in soil. The PCR amplification cycles for BKK1, SERK5 and quantitative control ACTIN2, are 30, 30, and 19, respectively. F, flowers; S, stems; L, rosette leaves; R, roots; SI, siliques.
Overexpressing SERK5-Ler can partially suppress bri1-5 phenotype
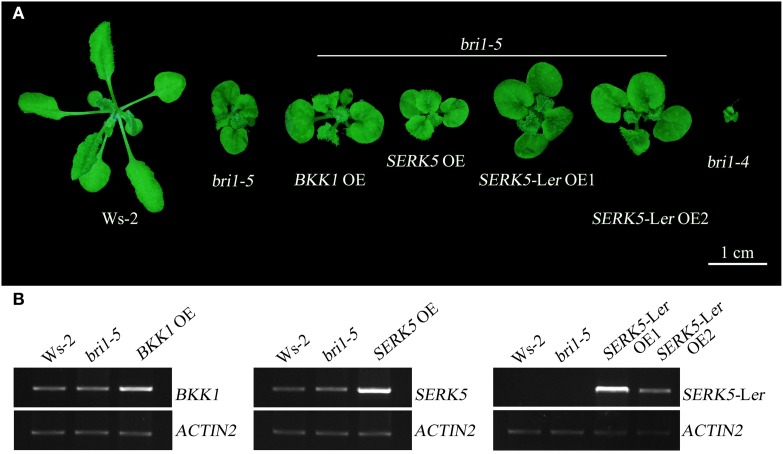
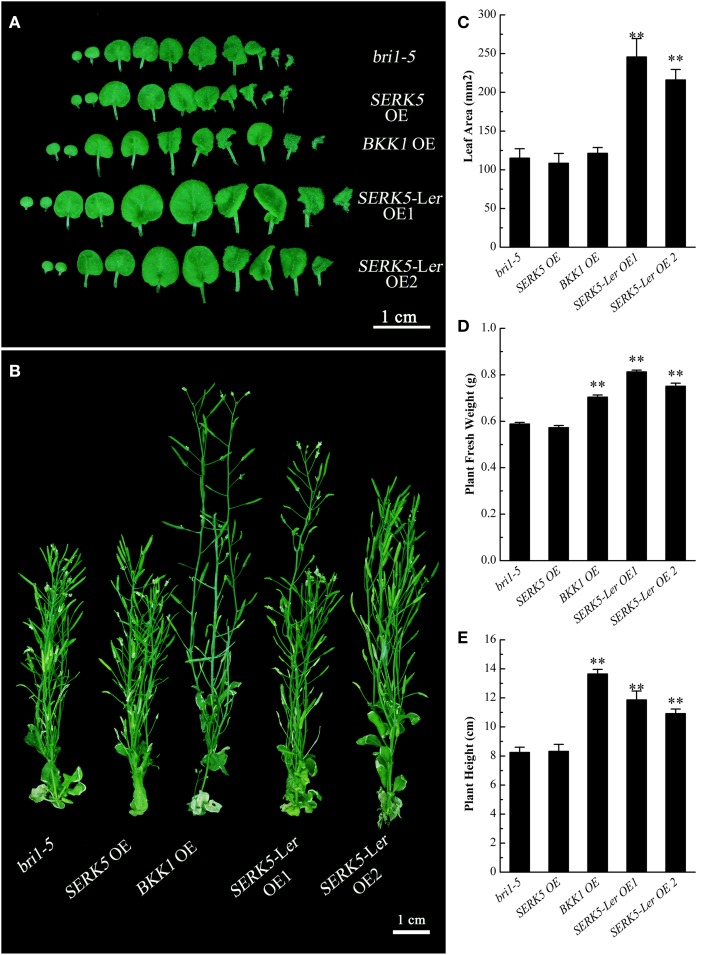
We have shown in the earlier reports that overexpression of SERK1 to SERK4 can partially suppress the BR defective phenotype of bri1-5 (He et al., 2007; Gou et al., 2012). We speculated that SERK5-Ler, possessing a normal RD motif, may share similar biological functions with other SERK members in BR signaling. To test this hypothesis, we used the CaMV35S promoter to drive SERK5-Ler in bri1-5. Two overexpression transgenic lines, named as SERK5-Ler OE1 and SERK5-Ler OE2, were confirmed by RT-PCR and western-blotting (Figure 2B, Figure S1A). Indeed, elevated expression of SERK5-Ler or BKK1, but not SERK5, was able to partially suppress bri1-5. Overexpressing SERK5-Ler dramatically enlarged the leaf size of bri1-5, which was not observed in bri1-5 lines overexpressing BKK1 (Figures 3A,C). Overexpression of BKK1 was capable of suppressing the phenotypes of shortened petioles in bri1-5, while enhanced expression of SERK5-Ler failed to do so (Figure S2A). This result indicated that BKK1 and SERK5-Ler may play distinct roles in BR signaling during different developmental stages or in diverse plant tissues. Overexpression of either BKK1 or SERK5-Ler in bri-5 also noticeably increased the plant height compared to bri-5 background at 55 days (Figures 3B,E). Overexpression of SERK5-Ler or BKK1 also increased bri1-5 fresh weight (Figure 3D).
Figure 2.
Overexpressing SERK5-Ler can partially suppress bri1-5 phenotype. (A) Overexpression of SERK5-Ler and BKK1, but not SERK5, can partially suppresses the phenotypes of bri1-5, a weak BRI1 mutant. bri1-4 is a null BRI1 mutant allele. (B) RT-PCR analyses to confirm the elevated expression of the transgenes in bri1-5. The PCR amplification cycles for BKK1, SERK5, SERK5-Ler and quantitative control ACTIN2, are 28, 28, 27, and 19, respectively.
Figure 3.
Detailed phenotypic analyses of bri1-5 overexpressing SERKs. (A) Overexpression of SERK5-Ler, but not BKK1 and SERK5, enlarges the leaf size of bri1-5. (B) Overexpression of SERK5-Ler and BKK1, but not and SERK5, increases the plant height of bri1-5. (C) Statistic analyses of leaf areas of the plants presented in (A). (D) Statistic analyses of plant fresh weight of 35-day-old seedlings. (E) Statistic analyses of plant heights presented in (B). The data are shown as means ± standard deviation (SD) (n = 20). Student's t-test indicated the differences are statistically significant (**P < 0.01). Experiments were repeated three times with similar results.
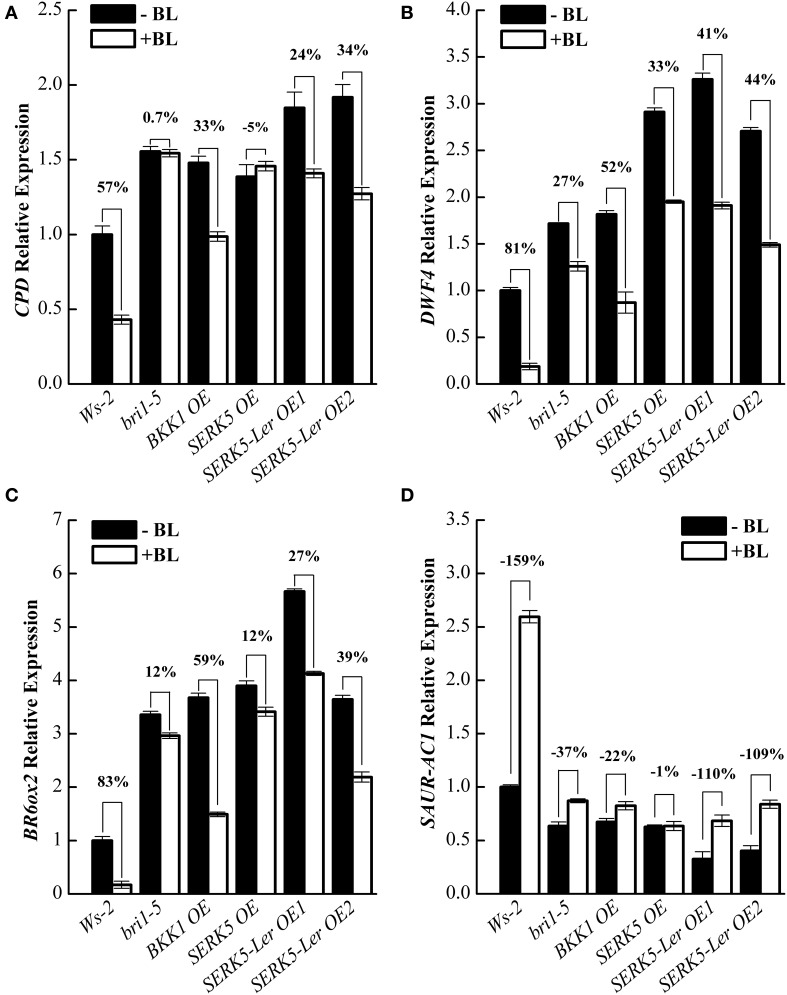
To further validate that SERK5-Ler is truly involved in BR signaling, quantitative RT-PCR (qRT-PCR) experiments were performed to examine the expression levels of BR responsive genes such as CPD, DWF4, BR6ox2, and SAUR-AC1 in wild-type and transgenic plants, when treated with or without BL. In WT plants, upon BL treatments, the expression of CPD, DWF4, and BR6ox2 decreased 57%, 81%, and 83%, respectively, and the expression of SAUR-AC1 increased 159%. Whereas, CPD, DWF4 and BR6ox2 showed only 0.7%, 27%, and 12% expression decrease, and SAURN-AC1 showed 37% expression increase in bri-5 after BL treatment, indicating a hampered BR signaling in bri1-5. The expression levels of CPD, DWF4, and BR6ox2 decreased 24%/34%, 41%/54%, and 27%/39%, respectively, and the expression of SAUR-AC1 increased 110%/109% in two SERK5-Ler OE lines, suggesting the impaired BR signaling in bri1-5 was partly recovered. These BR-regulated genes exhibited similar response to BL treatments in bri1-5 overexpressing BKK1 compared to SERK5-Ler OE lines. The overexpression of SERK5 in bri1-5 mutant led to identical responses to BL treatments with its genetic background, confirming non-function of SERK5 in BR signaling (Figures 4A–D).
Figure 4.
Overexpression of SERK5-Ler partially restores BR signaling in bri1-5. (A–D) 10-day-old seedlings grown on 1/2 MS medium were treated with or without 1 μM 24-epiBL for 3 h. Relative expression levels of CPD, DWF4, BR6ox2, and SAUR-AC1 was examined by qRT-PCR. Overexpression of SERK5-Ler partially recovers the sensitivity of BR-responsive genes in bri1-5 upon BL treatment. ACTIN2 was used as the reference gene. Values are means ± SE (n = 3). Experiments were repeated three times with similar results.
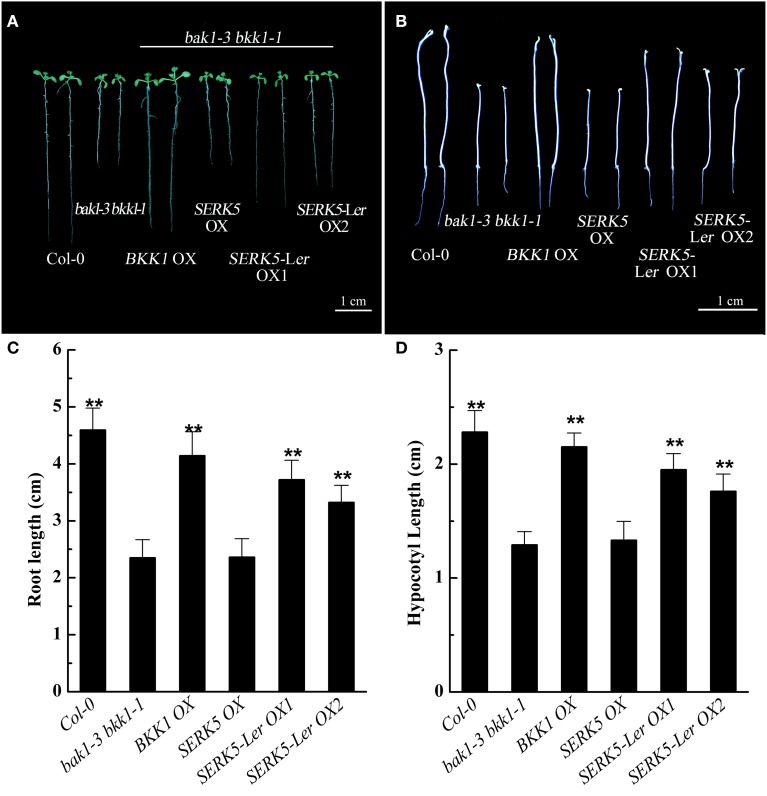
Overexpressing SERK5-Ler can partially rescue the root and hypocotyl phenotypes of bak1-3 bkk1-1 and increase its sensitivity to BR
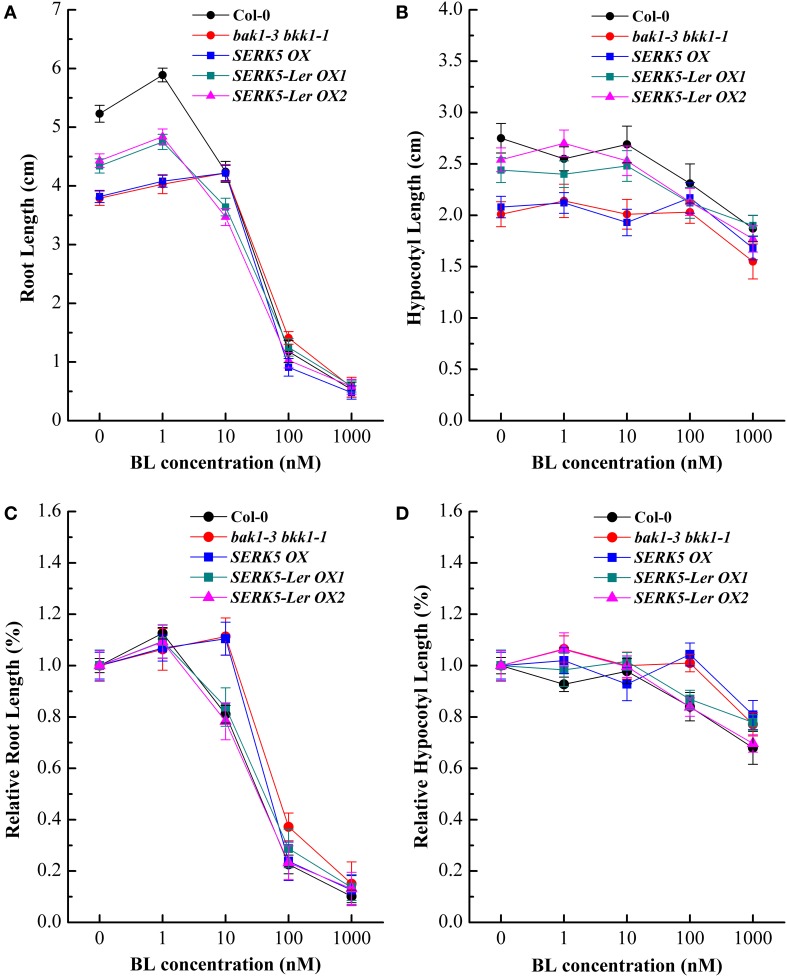
Our previous studies indicated that the spontaneous cell death and BR defective phenotypes could be restored to wild-type-like by overexpressing BAK1 or BKK1 in null double mutant bak1-4 bkk1-1 (He et al., 2007) and a weak double mutant bak1-3 bkk1-1. As the closest paralog of BKK1, SERK5-Ler with an intact RD motif is postulated to function redundantly with BKK1. Transgenic plants of bak1-3 bkk1-1 overexpressing SERK5-Ler, BKK1, and SERK5 fused to FLAG were generated, serving as a complementation assay (Figures 5A,B). Overexpression of the transgenes was confirmed by western-blotting analysis using anti-FLAG antibody (Figure S1B). As shown in Figure 5A, overexpression of SERK5-Ler in bak1-3 bkk1-1 (named as SERK5-Ler OX) exhibited no strikingly different aerial phenotype but showed elongated root lengths compared with bak1-3 bkk1-1 when grown in the light (Figures 5A,C). This complementation phenotype appeared to be weaker than bak1-3 bkk1-1 overexpressing BKK1 (named as BKK1 OX). By contrast, overexpressing SERK5 in bak1-3 bkk1-1 (named as SERK5 OX) caused no phenotypic alterations to bak1-3 bkk1-1 (Figures 5A,B). When grown in the dark, the shorten hypocotyls, a typical BR mutant phenotype referred to “de-etiolation,” of bak1-3 bkk1-1 was partially rescued by overexpressing SERK5-Ler (Figures 5B,D), which, however, is not observed in bri1-5 overexpressing SERK5-Ler (Figures S2B,C). Comparing to bak1-3 bkk1-1, the root and hypocotyl lengths increased about 58%/41% and 51%/36%, respectively, in two SERK5-Ler OX lines. Similarly, BKK1 OX showed 76% and 67% increase in root and hypocotyl lengths, respectively, compared to bak1-3 bkk1-1. The root and hypocotyl growth was identical between SERK5 OX and bak1-3 bkk1-1. Next, we performed root and hypocotyl growth inhibition assays to monitor the sensitivity of SERK5-Ler OX lines in response to exogenously applied BL. The SERK5-Ler OX lines appeared to restore the BR sensitivity in roots, when treated with 10 nM BL, similar to WT and distinct from its background bak1-3 bkk1-1 and SERK5 OX lines (Figures 6A,C). The hypocotyls of SERK5-Ler OX lines also restored its BR sensitivity, when treated with 100 nM BL (Figures 6B,D). These results demonstrated that the expression of SERK5-Ler effectively compensated the function of BAK1 or BKK1 in BR pathway in bak1-3 bkk1-1.
Figure 5.
Phenotypic analyses of bak1-3 bkk1-1 overexpressing SERKs. (A) Overexpression of SERK5-Ler elongates the root lengths of bak1-3 bkk1-1. 8-day-old seedlings grown on 1/2 MS medium in the light were presented. (B) Overexpression of SERK5-Ler elongates the hypocotyl lengths of bak1-3 bkk1-1. 8-day-old seedlings grown on 1/2 MS medium in the dark were presented. (C) Statistic analyses of root lengths of the plants presented in (A). (D) Statistic analyses of hypocotyl lengths of the plants presented in (B). The data are shown as means ± standard deviation (SD) (n = 25). Student's t-test indicated the differences are statistically significant (**P < 0.01). Experiments were repeated three times with similar results.
Figure 6.
Elevated expression of SERK5-Ler in bak1-3 bkk1-1 restore its sensitivity to BL. The root and hypocotyl growth of 8-day-old seedlings grown on 1/2 MS medium containing different concentrations of 24-epiBL in the light and dark were analyzed. (A) Root lengths of the seedlings under 24-epiBL treatments. (B) Hypocotyl lengths of the seedlings under 24-epiBL treatments. (C) Relative root growth of the seedlings presented in (A). Overexpression of SERK5-Ler in bak1-3 bkk1-1 restore its sensitivity when treated with 10 nM 24-epiBL. (D) Relative hypocotyl growth of the seedlings presented in (C). Overexpression of SERK5-Ler in bak1-3 bkk1-1 restores its sensitivity when treated with 100 nM 24-epiBL. The data are shown as means ± standard deviation (SD) (n ≧ 40). Experiments were repeated five times with similar results.
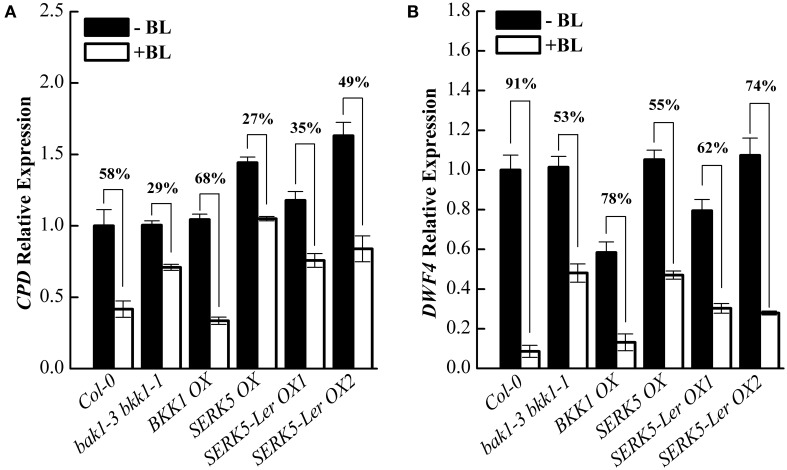
Subsequently, we analyzed the responses of BR-regulated genes CPD and DWF4 in different genetic backgrounds. Upon BL treatment, the expression levels of CPD and DWF4 were decreased about 58% and 91% in Col-0 wild-type but merely 29% and 53% in bak1-3 bkk1-1, respectively (Figures 7A,B). When treated with BL, the expression levels of CPD and DWF4 decreased about 35%/49% and 62%/74% in two SERK5-Ler OX lines, which were greater than those in bak1-3 bkk1-1, implicating an enhanced BR response in bak1-3 bkk1-1 by overexpressing SERK5-Ler.
Figure 7.
Overexpression of SERK5-Ler partially restores BR signaling in bak1-3 bkk1-1. (A,B) 10-day-old seedlings which grown on 1/2 MS medium were treated with or without 1 μM 24-epiBL for 3 h. Relative expression level of CPD and DWF4 were measured by qRT-PCR. bak1-3 bkk1-1 partially restores the sensitivity of BR-responsive genes to BL treatment by overexpressing SERK5-Ler. ACTIN2 was used as the reference gene. Values are means ± SE (n = 3). Experiments were repeated three times with similar results.
Collectively, our genetic, physiological and molecular evidence verified that SERK5-Ler but not SERK5 plays a role in BR signaling.
SERK5-Ler interacts with BRI1 in vivo
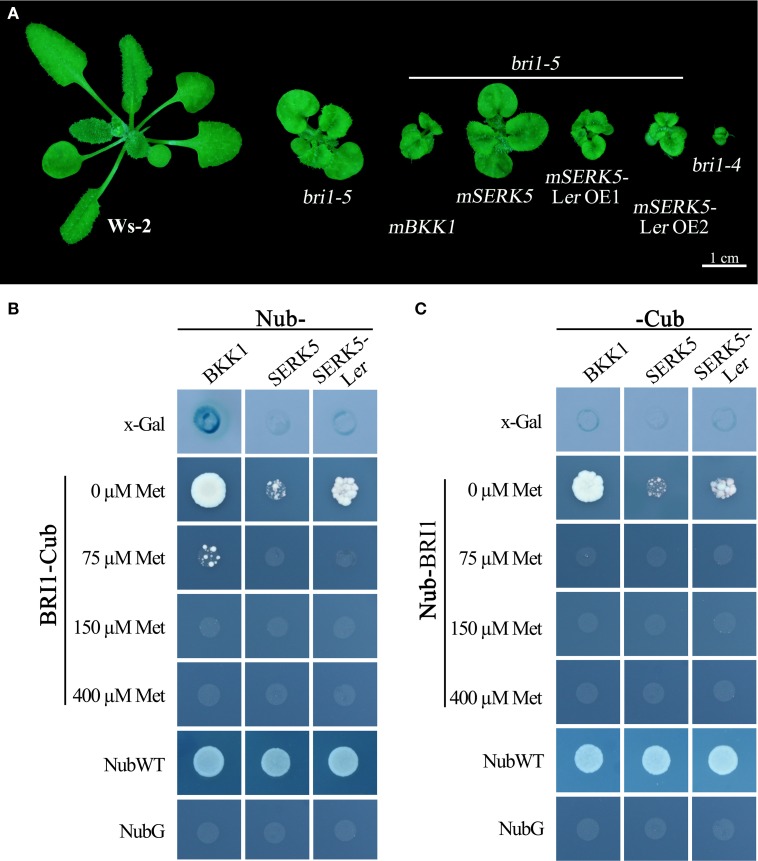
BR molecules are perceived by ligand-binding receptor BRI1 and co-receptor SERKs. BR-induced formation of BRI1-SERKs heterodimer initiates downstream signaling cascades. If SERK5-Ler truly participates in BR pathway, it is postulated to associate with BRI1. Our previous studies indicated that overexpression of a kinase-inactive version of SERK1 to SERK4 in bri1-5 caused a typical dominant-negative phenotype which resembles a null BRI1 mutant, bri1-4 (He et al., 2007; Gou et al., 2012). Similarly, as shown in Figure 8A, overexpression of a kinase-dead form of SERK5-Ler (K301E), not SERK5 (K303E), in bri1-5 resulted in typical dominant-negative phenotype, characterized by extreme dwarfism and severely compacted rosette leaves (Figure 8A). These results served as a piece of genetic evidence that SERK5-Ler associates with BRI1 to mediate BR signaling in planta and that the kinase activity of SERK5-Ler is crucial for its role in this process.
Figure 8.
SERK5-Ler interacts with BRI1. (A) Overexpression of a kinase-inactive variant of SERK5-Ler (K301E) led to a dominant-negative phenotype resembling bri1-4. Mating-based split ubiquitin system (mbSUS) to show BRI1-SERK5-Ler interaction in vivo. Interaction was determined by the growth ability of mated yeast on selective medium (-AHLT) supplemented with 0, 75, 150, and 400 μM methionine. NubWT and NubG were used as positive and negative control, respectively. (B) BRI1 was cloned as a -Cub fusion, and SERK5-Ler, BKK1, and SERK5 were cloned as a Nub- fusion. (C) BRI1 was cloned as a Nub- fusion, and SERK5-Ler, BKK1, and SERK5 were cloned as a -Cub fusion.
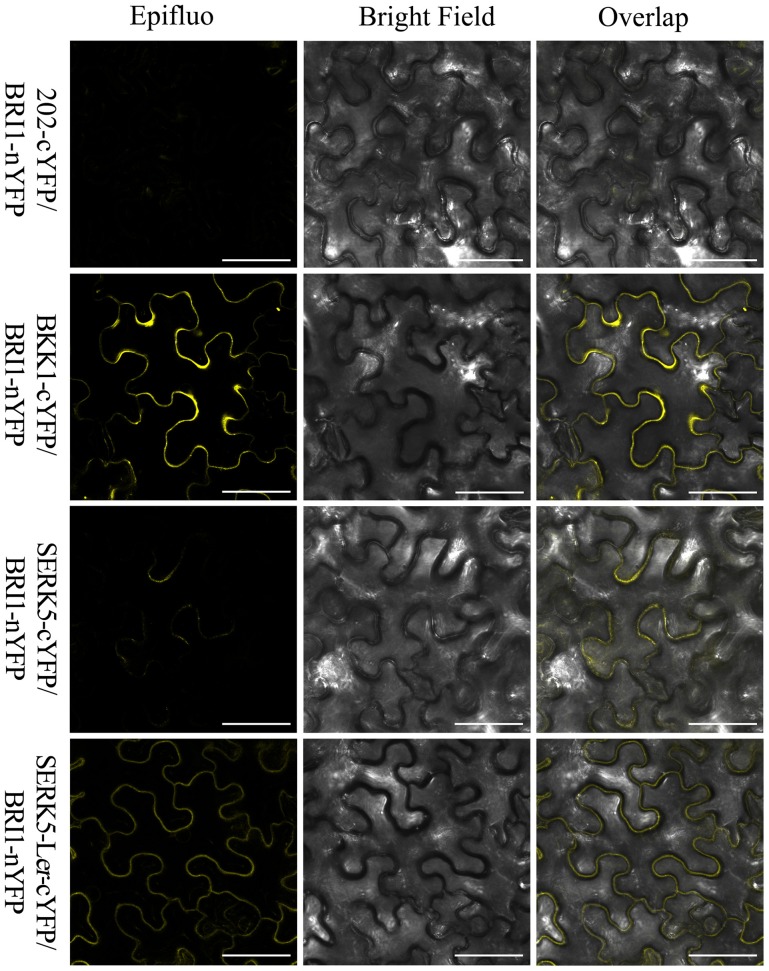
Furthermore, a mating-based yeast split ubiquitin system (mbSUS) was used to examine the interaction between BRI1 and SERK5-Ler/SERK5/BKK1. Consistent to the genetic data, the yeast two-hybrid result indicated that BRI1 strongly interacted with BKK1 (Figures 8B,C). Despite a slightly weaker interaction with BRI1 compared to BKK1-BRI1, SERK5-Ler showed a stronger association with BRI1 in yeast system whereas SERK5 showed a much weaker interaction with BRI1. Moreover, the complex formed by BRI1 and SERK5-Ler was verified by bimolecular fluorescence complementation (BiFC) assay. BRI1 was fused to the N-terminal half of YFP (BRI1-nYFP), while SERK5-Ler, SERK5, and BKK1 were fused to the C-terminal half of YFP (SERK5-Ler-cYFP, SERK5-cYFP, and BKK1-cYFP). Strong YFP fluorescence signals were detected at plasma membrane when BRI1-nYFP and SERK5-Ler-cYFP/BKK1-cYFP were co-transformed in Nicotiana benthamiana epidermal cells, confirming the interaction between BRI1 and SERK5-Ler/BKK1 in planta. Although detectable, the YFP fluorescence emitted by co-expression of BRI1-nYFP and SERK5-cYFP was very weak (Figure 9).
Figure 9.
BiFC assay to show the association of BRI1 and SERK5-Ler in planta. BRI1-nYFP was co-expressed with SERK5-Ler-cYFP, BKK1-cYFP, and SERK5-cYFP in epidermal cells of Nicotiana benthamiana. Representative confocal images showing the YFP fluorescence signal indicate the physical interaction between BRI1 and SERK5-Ler, BKK1, SERK5 at plasma membrane. BKK1 and SERK5-Ler strongly interact with BRI1, while SERK5 only shows very weak binding with BRI1. Co-expression of 202-cYFP and BRI1-nYFP was used as a negative control. Similar results were obtained in more than three replicates. Scale bars = 50 μm.
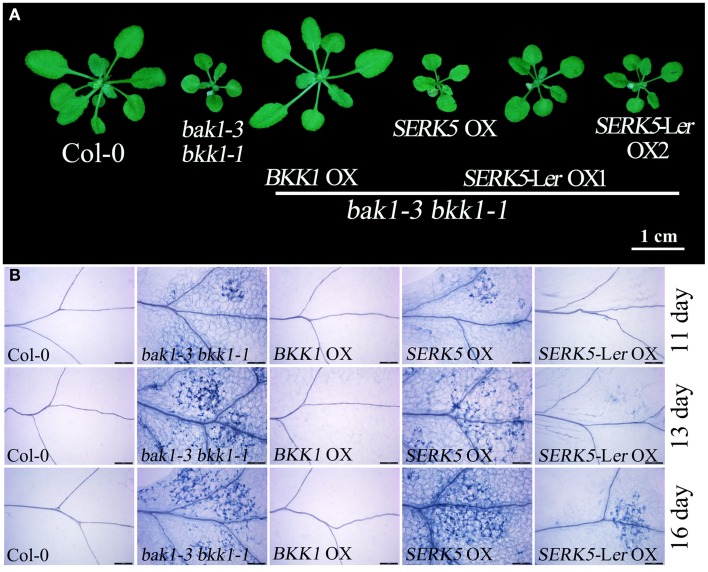
Overexpression of SERK5-Ler can delay the cell death phenotype of bak1-3 bkk1-1
Our previous work has indicated that BKK1 plays a central role in a BR-independent cell death control in addition to regulating BR signaling (He et al., 2007). Given that SERK5-Ler, the closest paralog of BKK1, is indeed involved in BR pathway, it is logically hypothesized that SERK5-Ler would also function in cell death control. As shown in Figure 10, overexpressing BKK1 in bak1-3 bkk1-1 completely suppressed its cell death symptom. However, overexpression of SERK5-Ler failed to fully rescue the cell death phenotype of bak1-3 bkk1-1 (Figure 10A). Subsequently, the cell death in the cotyledons of 11, 13, and 16-day-old seedlings grown on 1/2 MS medium were visualized by trypan-blue staining. The result showed that overexpression of SERK5-Ler is capable of delaying the cell death symptom but is unable to completely suppress the cell death in bak1-3 bkk1-1 (Figure 10B).
Figure 10.
Overexpression of SERK5-Ler can delay the cell death in bak1-3 bkk1-1. (A) The aerial phenotype of 25-day-old seedlings. (B) The cell death was indicated by trypan-blue staining in the cotyledons of 11, 13, and 16-day-old seedlings grown at 22°C under long-day conditions (16 h light/8 h dark). Scale bars = 200 μm.
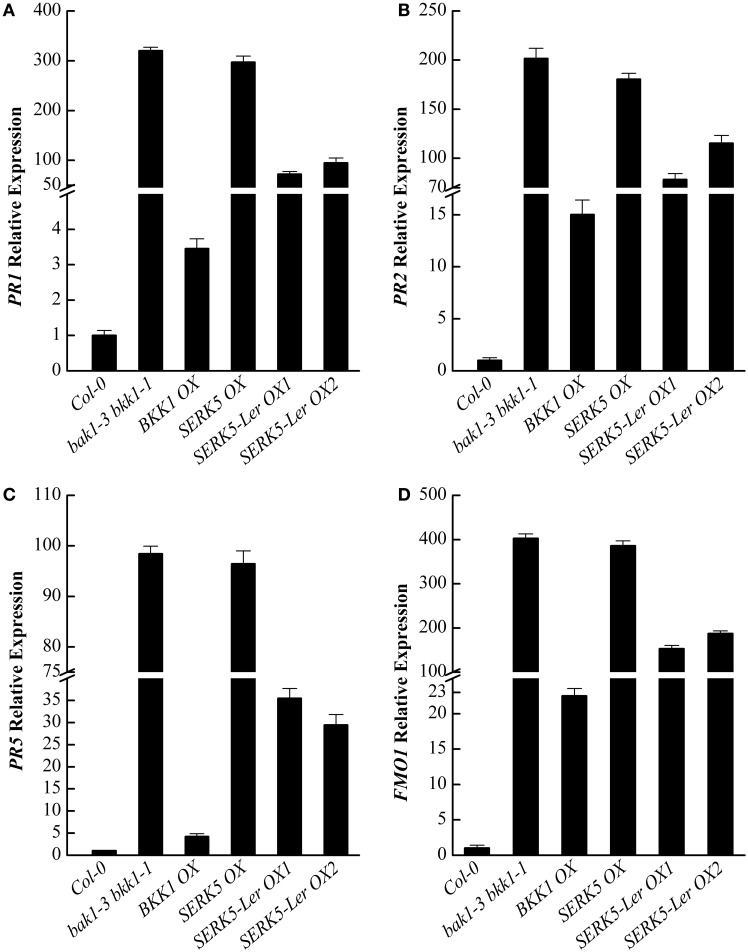
It has been reported that some defense-related genes are highly up-regulated in bak1 bkk1 (He et al., 2007; Yang et al., 2010). Therefore, the expression levels of pathogenesis-related genes PR1, PR2, PR5, and cell death marker gene FMO1 in 13-day-old seedlings were analyzed. qPCR results indicated that the overexpression of SERK5-Ler can slightly reduce the expression levels of defense and senesces-related genes in bak1-3 bkk1-1 (Figures 11A–D), which is consistent with the phenotypic and trypan-blue staining results. Thus, our data suggested that SERK5-Ler may play a role in cell death control, albeit less significantly than in BR signaling.
Figure 11.
qRT-PCR analysis of the PR1, PR2, PR5, and FMO1 gene expression levels in WT, bak1-3 bkk1-1 and bak1-3 bkk1-1 lines overexpressing SERKs. (A–D) Defense-related genes PR1, PR2, PR5, and senescence-related gene FMO1 were down-regulated in bak1-3 bkk1-1 when SERK5-Ler is overexpressed.
Discussion
Perception of external stimuli and initiation of appropriate cellular responses are essential for plant growth and development, where RLKs act as crucial components. As a group of highly conserved LRR-RLK genes, SERKs are found to exist in all known plant genomes forming four major clusters including non-vascular SERKs, monocot SERKs, dicot SERK1/2, and dicot SERK3/4 (Aan Den Toorn et al., 2015). It is indicated that the whole SERK family in Arabidopsis has originated from gene duplications followed by functional differentiation (Aan Den Toorn et al., 2015). The members of the SERK family in Arabidopsis have been identified to play multiple roles in a variety of developmental events via interacting with different ligand-binding RLKs. SERK1, SERK3/BAK1, and SERK4/BKK1 act as co-receptors for BR perception and signaling initiation (He et al., 2013). SERK1 was reported to be involved in organ separation in flowers (Lewis et al., 2010). SERK1 and SERK2 also participate in male sporogenesis (Albrecht et al., 2005; Colcombet et al., 2005). BAK1 was found to form complexes with FLS2, EFR1, and BIR1, respectively, involved in plant immunity, and play overlapping role with BKK1 to regulate cell death (He et al., 2007; Kemmerling et al., 2007).
Despite numerous detailed functional analyses of SERKs, the biological roles of SERK5 in Arabidopsis still remain blurred. In Arabidopsis ecotype Col-0, SERK5 bears an amino acid substitution of Arg (R) by Leu (L) within the critical arginine-aspartate (RD) motif in comparison with other four members in the SERK family (He et al., 2007). Therefore, it is interesting to explore whether the genomes of Arabidopsis ecotypes other than Col-0 contain SERK5 genes encoding a LRR-RLK with intact RD motif that makes SERK5 a biologically functional regulator. To answer the question, we cloned SERK5 cDNAs from Ler, Col-0, Ws-2, Lan-0, and C24 accessions. Sequencing analysis indicated that SERK5 in Ler encodes a normal RD LRR-RLK (Figures 1A,C). The sequences of SERK5 in Ws-2, Lan-0, and C24 contain a deletion of two nucleotides (TA), presumably resulting in a disruption of SERK5 protein translation (Figure 1B). Gene duplication serves as a key mechanism of expanding the gene number and diversifying the gene functions in plants and animals during evolution. Lehti-Shiu et al. reported that 30% of RLK family members are found in tandem repeats in Arabidopsis, suggesting higher frequency of RLK to be copied and clustered compared with other gene families (Lehti-Shiu et al., 2009). BKK1 (At2G13790) and SERK5 (At2G13800) are found as tandem repeats on chromosome 2 and share very high identity to each other, suggesting a recent duplication and high functional redundancy. As a result, one of these genes, SERK5 may undergo low selection pressures during evolution, leading to a mutation in an extremely important region, RD motif, and frequent occurrences of nucleotide deletion. It is indicated that all plant SERKs, except SERK5 (in Col-0), contain RD motif (Aan Den Toorn et al., 2015). This result suggested that RD motif may be under purifying selection, the selection attempting to minimize amino acid change throughout evolution. Nevertheless, the possibility has not been ruled out that RD motif mutation, together with other SNPs, in SERK5-Col may confer novel functions yet to be characterized. Whether SERK5 in Col-0 is a pseudogene needs further validation.
The natural genetic variations often shape the phenotypes of specific Arabidopsis strains. Numerous studies have described that different Arabidopsis populations vary in a number of traits (Weigel, 2012). A genetic method, quantitative trait locus (QTL) mapping, is widely utilized to identify loci responsible for specific traits in diverse Arabidopsis accessions. For instance, through QTL mapping in genetic combination of accessions displaying distinct trichome phenotypes, transcription factors ETC2 was identified to regulate trichome density (Weigel, 2012). Accession Gr-1 displays a low-trichome-number phenotype, which is modulated by ETC2, a major determining factor in trichome patterning. ETC2 in strain Can-0 contains an E19K substitution, leading to high trichome density (Hilscher et al., 2009). In addition to SERK5, other LRR-RLKs showing variations among Arabidopsis accessions are well documented, including non-RD kinase FLS2 and RD kinase ERECTA (ER). FLS2 perceives bacterial flagellin to initiate PAMP-triggered immunity (PTI) in plant. A polymorphism of FLS2 in Ws-0 accession leads to a stop-codon-mutation in the kinase domain, which makes Ws-0 insensitive to flg22, a 22 amino acid elicitor conserved in bacterial flagellin (Gómez-Gómez and Boller, 2002). ER functions as a key regulator in multiple physiological processes such as cell patterning, pathogenesis, and light and hormone signaling. It has been reported that several Arabidopsis accessions contain natural loss-of-function mutation in ER including Ler, Hiroshima-1 (Hir-1) and Vancouver-0 (Van-0) (Van Zanten et al., 2009).
To assess whether SERK5-Ler, like other SERKs, is involved in BR signaling, we overexpressed SERK5-Ler in the bri1-5 background. It has been validated to be an efficient approach to identify BR-related genes. When overexpressed, the genes involved in BR signaling pathway can either suppress or enhance the mutant phenotype of bri1-5 (Ws-2 background) and the candidate genes can be isolated from the ecotypes other than Ws-2 (Li et al., 2002; He et al., 2007; Gou et al., 2012). Our results indicated that elevated expression of SERK5-Ler but not SERK5 can suppress the BR mutant phenotypes of bri1-5 (Figure 2A). Furthermore, we introduced SERK5-Ler into bak1-3 bkk1-1, which caused partially rescued root and hypocotyl lengths of bak1-3 bkk1-1 and increased its sensitivity to exogenous BL (Figures 5, 6). The interaction of BRI1 and SERK5-Ler was verified by different approaches (Figures 8, 9). Overexpression of a kinase death variant of SERK5-Ler in bri1-5 resulted in a null bri1-like phenotype, indicating SERK5-Ler and BRI1 form complex that can be disrupted by occupation of BRI1 with highly accumulated kinase-inactive mSERK5-Ler. In addition, yeast two-hybrid and BiFC analyses provided direct evidence that SERK5-Ler is associated with BRI1 in vivo (Figures 8, 9). Our previous work showed that overexpression of mSERK5 is unable to cause a dominant-negative phenotype in bri1-5, which is explicated by current results that SERK5 can barely associate with BRI1. Similar results were reported in an earlier study of allelic variation of MYC1. MYC1 is a bHLH transcription factor participating in trichome determination by associating with a scaffold protein, TTG1. MYC contains a P or A at amino acid position 189 in Col-0 and Ler, respectively. In a yeast two-hybrid assay, MYC-Col interacts with TGG1 while the interaction is abolished when P189 is replaced by A. MYC-Ler is not associated with TGG1, but an A189P substitution confers its interaction with TGG1 (Symonds et al., 2011). These results suggested that natural variation in MYC1 changes its biochemical nature and eventually causes distinct trichome density phenotype in Col-0 and Ler. These results, together with our data, indicated that allelic variations may affect the function of a protein by altering the affinities with its partners when specific amino acids are changed.
It is worth noting that overexpressing SERK5-Ler in bri1-5 can partially rescue the BR defective phenotype of bri1-5 by enlarging the size of rosette leaves. However, overexpression of SERK5-Ler in bri1-5 did not increase the lengths of petioles, roots, and hypocotyls, or recover the sensitivity of roots and hypocotyls of bri1-5 to BL treatment (Figure 3, Figures S2, S3). This result is reminiscent of an earlier reports that overexpression of SERK1, SERK2, BAK1, and BKK1 in bri1-301, a weak bri1 mutant allele, led to partial restored rosette leaves, yet none of the transgenic lines is able to alter the hypocotyl and root phenotype (Albrecht et al., 2008). Interestingly, overexpression of SERK5-Ler in bak1-3 bkk1-1 appeared to rescue the BR defective phenotype by increasing the lengths of roots and hypocotyls of bak1-3 bkk1-1. These observations may suggest that BR signaling is more severely impaired in bri1-5 mutant than that in bak1-3 bkk1-1 double mutant (Gou et al., 2012). As a result, overexpression of SERK5-Ler is insufficient to significantly rescue the root and hypocotyl phenotypes in bri1-5, yet it can effectively restore the root and hypocotyl phenotypes of bak1-3 bkk1-1.
Previous reports have demonstrated that BKK1 is not only involved in the BR signaling, it also participated in the cell death control (He et al., 2007; Albrecht et al., 2008; Jeong et al., 2010). Our results revealed that SERK5-Ler can only slightly rescue the cell death phenotype of bak1-3 bkk1-1 when overexpressed. It suggested that SERK5-Ler is preferentially engaged in BR signaling pathway than in cell death control pathway.
BKK1 and SERK5-Ler are not fully functionally redundant in a variety of signaling pathways. This divergence may be caused by the specific allelic variations in these two SERKs. The future studies will focus on the functional analyses of different amino acid residues in addition to RD motif in BKK1 and SERK5-Ler. This may provide novel insights into the biological significance of specific residues of BKK1 and SERK5-Ler in regulating diverse signaling pathways. The further investigations on BKK1 and SERK5-Ler can also serve as a paradigm for better understanding of the detailed molecular mechanisms of gene duplications and mutations, non-RD/RD kinase conversion, biological consequences of polymorphisms, and differentiations in gene functionality during evolution.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
These studies were supported by the grant from the National Natural Science Foundation of China to JL (91317311 and 31470380), the grant from the National Natural Science Foundation of China to KH (31471305), and the Fundamental Research Funds for the Central Universities to KH (lzujbky-2014-84, lzujbky-2015-k17, and lzujbky-2015-ct).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00852
Western blotting to confirm the expressed protein by transgenes in different genetic background by using FLAG antibody. (A) Overexpression of SERK5-Ler in bri1-5. (B) Overexpression of SERK5-Ler in bak1-3 bkk1-1. (C) Overexpression of kinase-death forms of SERK5-Ler in bri1-5.
Overexpression of SERK5-Ler in bri1-5 is unable to rescue the root and hypocotyl phenotypes of bri1-5. (A) Statistic analyses of the average petiole lengths of 30-day-old WT, bri1-5 and bri1-5 lines overexpressing SERKs. (B,C) Statistic analyses of root and hypocotyl lengths of 8-day-old seedlings grown on 1/2 MS medium in light and dark, respectively. Overexpression of BKK1 can partly recue the petiole phenotype, but not the root and hypocotyl phenotypes of bri1-5. Overexpression of SERK5-Ler recues none of the petiole, root and hypocotyl phenotypes of bri1-5. The data are shown as means ± standard deviation (SD) (n ≧ 40). Student's t-test indicated the differences are statistically significant (***P < 0.001, **P < 0.01). Experiments were repeated three times with similar results.
Overexpression of SERK5-Ler in bri1-5 is unable to enhance the sensitive of bri1-5 to BL. (A) Root growth analysis of 8-day-old seedlings grown on 1/2 MS medium containing different concentrations of 24-epiBL in the light. (B) Hypocotyl growth analysis of 8-day-old seedlings grown on 1/2 MS medium containing different concentrations of 24-epiBL in the dark. The data are shown as means ± standard deviation (SD) (n ≧ 40). Experiments were repeated three times with similar results.
Primers used for gene cloning, mutagenesis, real-time RT-PCR and 24 RT-PCR. 25.
References
- Aan Den Toorn M., Albrecht C., de Vries S. (2015). On the origin of serks: bioinformatics analysis of the somatic embryogenesis receptor kinases. Mol. Plant 8, 762–782. 10.1016/j.molp.2015.03.015 [DOI] [PubMed] [Google Scholar]
- Albrecht C., Russinova E., Hecht V., Baaijens E., de Vries S. (2005). The Arabidopsis thaliana somatic embryogenesis receptor-like kinases1 and 2 control male sporogenesis. Plant Cell 17, 3337–3349. 10.1105/tpc.105.036814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C., Russinova E., Kemmerling B., Kwaaitaal M., de Vries S. C. (2008). Arabidopsis somatic embryogenesis receptor kinase proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol. 148, 611–619. 10.1104/pp.108.123216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y., Jaillais Y. (2015). The molecular circuitry of brassinosteroid signaling. New Phytol. 206, 522–540. 10.1111/nph.13269 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Clouse S. D., Sasse J. M. (1998). BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451. 10.1146/annurev.arplant.49.1.427 [DOI] [PubMed] [Google Scholar]
- Colcombet J., Boisson-Dernier A., Ros-Palau R., Vera C. E., Schroeder J. I. (2005). Arabidopsis somatic embryogenesis receptor kinases1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17, 3350–3361. 10.1105/tpc.105.036731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi U. K., Krishna P. (2009). Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. New Biotechnol. 26, 131–136. 10.1016/j.nbt.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Du J., Yin H., Zhang S, Wei Z. Y., Zhao B. L., Zhang J. H., et al. (2012). Somatic embryogenesis receptor kinases control root development mainly via brassinosteroid-independent actions in Arabidopsis thaliana. J. Integr. Plant. Biol. 54, 388–399. 10.1111/j.1744-7909.2012.01124.x [DOI] [PubMed] [Google Scholar]
- Gao M., Wang X., Wang D., Xu F., Ding X., Zhang Z., et al. (2009). Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6, 34–44. 10.1016/j.chom.2009.05.019 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2002). Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 7, 251–256. 10.1016/S1360-1385(02)02261-6 [DOI] [PubMed] [Google Scholar]
- Gou X., Yin H., He K., Du J., Yi J., Xu S., et al. (2012). Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8:e1002452. 10.1371/journal.pgen.1002452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K., Gou X., Yuan T., Lin H., Asami T., Yoshida S., et al. (2007). BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17, 1109–1115. 10.1016/j.cub.2007.05.036 [DOI] [PubMed] [Google Scholar]
- He K., Xu S., Li J. (2013). BAK1 directly regulates brassinosteroid perception and BRI1 activation. J. Integr. Plant. Biol. 55, 1264–1270. 10.1111/jipb.12122 [DOI] [PubMed] [Google Scholar]
- He Y., Tang W., Swain J. D., Green A. L., Jack T. P., Gan S. (2001). Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol. 126, 707–716. 10.1104/Pp.126.2.707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Wang Z. Y., Li J., Zhu Q., Lamb C., Ronald P., et al. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288, 2360–2363. 10.1126/science.288.5475.2360 [DOI] [PubMed] [Google Scholar]
- Hecht V., Vielle-Calzada J. P., Hartog M. V., Schmidt E. D. L., Boutilier K., Grossniklaus U., et al. (2001). The Arabidopsis somatic embryogenesis receptor kinase 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–816. 10.1104/Pp.010324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A., Hann D. R., Gimenez-Ibanez S., Jones A. M. E., He K., Li J., et al. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 12217–12222. 10.1073/pnas.0705306104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilscher J., Schlötterer C., Hauser M. T. (2009). A single amino acid replacement in ETC2 shapes trichome patterning in natural Arabidopsis populations. Curr. Biol. 19, 1747–1751. 10.1016/j.cub.2009.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M., Belkhadir Y., Dreux M., Dabi T., Noel J. P., Wilson I. A., et al. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474, 467–471. 10.1038/nature10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y. J., Shang Y., Kim B. H., Kim S. Y., Song J. H., Lee J. S., et al. (2010). BAK7 displays unequal genetic redundancy with BAK1 in brassinosteroid signaling and early senescence in Arabidopsis. Mol. Cell 29, 259–266. 10.1007/s10059-010-0024-0 [DOI] [PubMed] [Google Scholar]
- Johnson L. N., Noble M. E., Owen D. J. (1996). Active and inactive protein kinases: structural basis for regulation. Cell 85, 149–158. 10.1016/S0092-8674(00)81092-2 [DOI] [PubMed] [Google Scholar]
- Karlova R., Boeren S., Russinova E., Aker J., Vervoort J., de Vries S. (2006). The Arabidopsis somatic embryogenesis receptor-like kinase1 protein complex includes brassinosteroid-insensitive1. Plant Cell 18, 626–638. 10.1105/tpc.105.039412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerling B., Schwedt A., Rodriguez P., Mazzotta S., Frank M., Qamar S. A., et al. (2007). The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 17, 1116–1122. 10.1016/j.cub.2007.05.046 [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Caño-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., et al. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433, 167–171. 10.1038/nature03227 [DOI] [PubMed] [Google Scholar]
- Kobe B., Heierhorst J., Feil S. C., Parker M. W., Benian G. M., Weiss K. R., et al. (1996). Giant protein kinases: domain interactions and structural basis of autoregulation. EMBO J. 15, 6810–6821 [PMC free article] [PubMed] [Google Scholar]
- Krupa A., Preethi G., Srinivasan N. (2004). Structural modes of stabilization of permissive phosphorylation sites in protein kinases: distinct strategies in Ser/Thr and Tyr kinases. J. Mol. Biol. 339, 1025–1039. 10.1016/j.jmb.2004.04.043 [DOI] [PubMed] [Google Scholar]
- Lehti-Shiu M. D., Zou C., Hanada K., Shiu S. H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150, 12–26. 10.1104/pp.108.134353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. W., Leslie M. E., Fulcher E. H., Darnielle L., Healy P. N., Youn J. Y., et al. (2010). The SERK1 receptor-like kinase regulates organ separation in Arabidopsis flowers. Plant J. 62, 817–828. 10.1111/j.1365-313X.2010.04194.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. (2010). Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr. Opin. Plant Biol. 13, 509–514. 10.1016/j.pbi.2010.09.004 [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. 10.1016/S0092-8674(00)80357-8 [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K. A., Doke J. T., Tax F. E., Walker J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. 10.1016/S0092-8674(02)00812-7 [DOI] [PubMed] [Google Scholar]
- Mayans O., van der Ven P. F. M., Wilm M., Mues A., Young P., Fürst D. O., et al. (1998). Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature 395, 863–869. 10.1038/27603 [DOI] [PubMed] [Google Scholar]
- Nakaya M., Tsukaya H., Murakami N., Kato M. (2002). Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol. 43, 239–244. 10.1093/pcp/pcf024 [DOI] [PubMed] [Google Scholar]
- Nam K. H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. 10.1016/S0092-8674(02)00814-0 [DOI] [PubMed] [Google Scholar]
- Nolen B., Taylor S., Ghosh G. (2004). Regulation of protein kinases: controlling activity through activation segment conformation. Mol. Cell 15, 661–675. 10.1016/j.molcel.2004.08.024 [DOI] [PubMed] [Google Scholar]
- Nolen B., Yun C. Y., Wong C. F., McCammon J. A., Fu X. D., Ghosh G. (2001). The structure of Sky1p reveals a novel mechanism for constitutive activity. Nat. Struct. Biol. 8, 176–183. 10.1038/84178 [DOI] [PubMed] [Google Scholar]
- Obrdlik P., El-Bakkoury M., Hamacher T., Cappellaro C., Vilarino C., Fleischer C., et al. (2004). K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. U.S.A. 101, 12242–12247. 10.1073/pnas.0404467101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., et al. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440–2455. 10.1105/tpc.111.084301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Henzler C., Hothorn M. (2013). Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341, 889–892. 10.1126/science.1242468 [DOI] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A. K., Mueller K., Beeler S., Boller T., et al. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285, 9444–9451. 10.1074/jbc.M109.096842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., Jones A., et al. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. Plos Genet 7, e1002046. 10.1371/journal.pgen.1002046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J., Han Z., Kim T. W., Wang J. J., Cheng W., Chang J. B., et al. (2011). Structural insight into brassinosteroid perception by BRI1. Nature 474, 472–U496. 10.1038/nature10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S. H., Bleecker A. B. (2001a). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 2001:re22. 10.1126/stke.2001.113.re22 [DOI] [PubMed] [Google Scholar]
- Shiu S. H., Bleecker A. B. (2001b). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768. 10.1073/pnas.181141598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Han Z., Tang J., Hu Z., Chai C., Zhou B., et al. (2013b). Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 23, 1326–1329. 10.1038/cr.2013.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li L., Macho A. P., Han Z., Hu Z., Zipfel C., et al. (2013a). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628. 10.1126/science.1243825 [DOI] [PubMed] [Google Scholar]
- Symonds V. V., Hatlestad G., Lloyd A. M. (2011). Natural allelic variation defines a role for ATMYC1: trichome cell fate determination. PLoS Genet 7:e1002069. 10.1371/journal.pgen.1002069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G. M., Davies C., Shavrukov Y., Dry I. B., Reid J. B., Thomas M. R. (2006). Grapes on steroids. Brassinosteroids are involved in grape berry ripening. Plant Physiol. 140, 150–158. 10.1104/pp.105.070706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zanten M., Snoek L. B., Proveniers M. C., Peeters A. J. (2009). The many functions of ERECTA. Trends Plant Sci. 14, 214–218. 10.1016/j.tplants.2009.01.010 [DOI] [PubMed] [Google Scholar]
- Wang X., Goshe M. B., Soderblom E. J., Phinney B. S., Kuchar J. A., Li J., et al. (2005). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis brassinosteroid-insensitive1 receptor kinase. Plant Cell 17, 1685–1703. 10.1105/tpc.105.031393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y., Seto H., Fujioka S., Yoshida S., Chory J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. 10.1038/35075629 [DOI] [PubMed] [Google Scholar]
- Weigel D. (2012). Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol. 158, 2–22. 10.1104/pp.111.189845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Gou X. P., He K., Xi D. H., Du J. B., Lin H. H., et al. (2010). BAK1 and BKK1 in Arabidopsis thaliana confer reduced susceptibility to turnip crinkle virus. Eur. J. Plant Pathol. 127, 149–156. 10.1007/s10658-010-9581-5 [DOI] [Google Scholar]
- Ye Q., Zhu W., Li L., Zhang S., Yin Y., Ma H., et al. (2010). Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. U.S.A. 107, 6100–6105. 10.1073/pnas.0912333107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western blotting to confirm the expressed protein by transgenes in different genetic background by using FLAG antibody. (A) Overexpression of SERK5-Ler in bri1-5. (B) Overexpression of SERK5-Ler in bak1-3 bkk1-1. (C) Overexpression of kinase-death forms of SERK5-Ler in bri1-5.
Overexpression of SERK5-Ler in bri1-5 is unable to rescue the root and hypocotyl phenotypes of bri1-5. (A) Statistic analyses of the average petiole lengths of 30-day-old WT, bri1-5 and bri1-5 lines overexpressing SERKs. (B,C) Statistic analyses of root and hypocotyl lengths of 8-day-old seedlings grown on 1/2 MS medium in light and dark, respectively. Overexpression of BKK1 can partly recue the petiole phenotype, but not the root and hypocotyl phenotypes of bri1-5. Overexpression of SERK5-Ler recues none of the petiole, root and hypocotyl phenotypes of bri1-5. The data are shown as means ± standard deviation (SD) (n ≧ 40). Student's t-test indicated the differences are statistically significant (***P < 0.001, **P < 0.01). Experiments were repeated three times with similar results.
Overexpression of SERK5-Ler in bri1-5 is unable to enhance the sensitive of bri1-5 to BL. (A) Root growth analysis of 8-day-old seedlings grown on 1/2 MS medium containing different concentrations of 24-epiBL in the light. (B) Hypocotyl growth analysis of 8-day-old seedlings grown on 1/2 MS medium containing different concentrations of 24-epiBL in the dark. The data are shown as means ± standard deviation (SD) (n ≧ 40). Experiments were repeated three times with similar results.
Primers used for gene cloning, mutagenesis, real-time RT-PCR and 24 RT-PCR. 25.