Abstract
D-glucaric acid can be used as a building block for biopolymers as well as in the formulation of detergents and corrosion inhibitors. A biosynthetic route for production in Escherichia coli has been developed (Moon et al., 2009), but previous work with the glucaric acid pathway has indicated that competition with endogenous metabolism may limit carbon flux into the pathway. Our group has recently developed an E. coli strain where phosphofructokinase (Pfk) activity can be dynamically controlled and demonstrated its use for improving yields and titers of the glucaric acid precursor myo-inositol on glucose minimal medium. In this work, we have explored the further applicability of this strain for glucaric acid production in a supplemented medium more relevant for scale-up studies, both under batch conditions and with glucose feeding via in situ enzymatic starch hydrolysis. It was found that glucaric acid titers could be improved by up to 42% with appropriately timed knockdown of Pfk activity during glucose feeding. The glucose feeding protocol could also be used for reduction of acetate production in the wild type and modified E. coli strains.
Abbreviations: aTc, anhydrotetracycline; G6P, glucose-6-phosphate; INO1, myo-inositol-1-phosphate synthase; IPTG, β-D-1-thiogalactopyranoside; MIOX, myo-inositol oxygenase; Pfk, phosphofructokinase
Keywords: Dynamic metabolic engineering, Glucaric acid, Protein degradation, Synthetic biology
Highlights
-
•
Dynamic growth/production switching for glucaric acid production in E. coli.
-
•
Optimal time for switching via Pfk knockdown was screened in microtiter format.
-
•
Production from glucose was improved under batch conditions and with starch feeding.
-
•
Glucaric acid yield and titer improvements of up to 42% were achieved.
1. Introduction
D-glucaric acid was identified by the United State Department of Energy as a top value-added chemical for production from biomass (Werpy and Petersen, 2004). It has a number of potential applications including use in biopolymers (Kiely and Chen, 1994) and as a detergent builder and corrosion inhibitor (Smith et al., 2012). Glucaric acid can be produced through nitric acid oxidation of glucose (Mehltretter and Rist, 1953) but a biological route to glucaric acid production could potentially provide several advantages, including mild processing conditions and high selectivity for the product of interest.
Production of D-glucaric acid in Escherichia coli was previously demonstrated by our group via expression of heterologous enzymes from three different organisms (Moon et al., 2009). Titers of 1.13 g/L glucaric acid were achieved in strain BL21(DE3) in LB medium supplemented with 10 g/L glucose. Following demonstration of the initial pathway, some increases in glucaric acid titers were achieved through improved strategies for expression of the myo-inositol oxygenase (MIOX) enzyme, one of the limiting factors in glucaric acid production in LB supplemented with glucose or myo-inositol (Moon et al., 2010, Shiue and Prather, 2014). However, competition for glucose-6-phosphate (G6P) between native E. coli enzymes (phosphoglucosisomerase and glucose-6-phosphate dehydrogenase) and the first enzyme in the glucaric acid pathway, myo-inositol-1-phosphate synthase (INO1), is also a concern. High level expression of INO1 is required for detectable myo-inositol and glucaric acid production, indicating it competes poorly with endogenous metabolism for substrate (Moon et al., 2009). Additionally, the second pathway enzyme, MIOX, appears to be stabilized by its substrate, myo-inositol, so more rapid accumulation of myo-inositol may help reduce limitations in MIOX activity as well (Moon et al., 2010).
With this in mind, our group has explored strategies for development of strains capable of accumulating G6P and directing greater fluxes of this metabolite into production of glucaric acid and myo-inositol. By eliminating the pathways for glucose catabolism in the production strain, and feeding alternative carbon sources, higher yields of glucaric acid from glucose could be achieved (Shiue et al., 2015). However, the rate of glucose uptake in this K-12 host strain was quite slow, especially in minimal medium, and its use was limited to mixed sugar substrates.
While gene knockouts provide a static solution for redirecting fluxes in the cell (Kogure et al., 2007, Shiue et al., 2015), under many conditions, it may be advantageous to develop cells where dynamic changes in enzyme levels can be used to switch between substrate consumption for biomass formation and substrate conversion into product. Dynamic control of key enzymes can be used to facilitate more rapid initial accumulation of biomass, overcoming potential reductions in growth rate, and can eliminate the need for supplementation of the medium or addition of secondary carbon sources required with some gene knockouts (Anesiadis et al., 2008, Gadkar et al., 2005). At the desired time, activity of the target enzyme(s) can be reduced through decreasing transcription (Scalcinati et al., 2012, Solomon et al., 2012, Soma et al., 2014) or translation (Williams et al., 2015) of the enzyme, or initiating rapid degradation (Brockman and Prather, 2015, Torella et al., 2013). Coupling such controls with sensors capable of reporting on intracellular metabolite levels allows for the development of more complex systems capable of continuously adjusting enzyme levels to balance metabolite pools or maintain cellular state (Dahl et al., 2013, Farmer and Liao, 2000, Xu et al., 2014, Zhang et al., 2012).
It was recently shown that by inducing degradation of phosphofructokinase I (Pfk-I) activity in the cell, the pools of G6P could be increased during growth on glucose minimal medium, along with the yields and titers of the glucaric acid precursor myo-inositol (Brockman and Prather, 2015). In this work, we explore the expanded utility of this system for production of glucaric acid from glucose in a semi-defined medium under batch conditions and a fed-batch condition simulated by glucose release from in situ enzymatic starch hydrolysis. To explore the interplay of production conditions with metabolic intervention through Pfk-I degradation, initial screening runs were carried out in 48-well plates in a BioLector benchtop bioreactor. Follow-up experiments were then carried out at altered conditions or altered scale (shake flask) to understand the robustness of the results. Improvements in glucaric acid titer of up to 42% were achieved through appropriately timed induction of Pfk activity knockdown during the fermentation.
2. Materials and methods
2.1. Strains and plasmids
E. coli strains and plasmids used in this study are listed in Table 1. Strains IB1863 and IB1379 were constructed by our group previously (Brockman and Prather, 2015). To eliminate catabolism of glucaric acid and the pathway intermediate glucuronic acid in strain IB1863, knockouts of gudD and uxaC were carried out via sequential P1 transduction from Keio collection donor strains (Baba et al., 2006). The kanamycin resistance cassette was removed after each transduction via expression of FLP recombinase from pCP20 (Datsenko and Wanner, 2000). The λDE3 lysogen was integrated into this strain using a λDE3 Lysogenization Kit (Novagen, Darmstadt, Germany), generating strain IB1486. To generate the ΔpfkA control strain IB2255, serial P1 transductions were also carried out in IB1379 to knock out pfkA, gudD, and uxaC, and the λDE3 lysogen was integrated as described above. An additional control strain without any degradation tag on pfkA, IB2472, was generated using lambda red recombination combined with Cas9-based counterselection (Reisch and Prather, in press). Using this method, the tag sequence was removed from the pfkA locus without any addition of antibiotic resistance cassettes or FRT scars. Construction of plasmids for production of glucaric acid, pRSFD-IN-MI and pTrc-udh, was described previously (Moon et al., 2009, Yoon et al., 2009).
Table 1.
Strains and plasmids used in this study.
| Strain/plasmid | Genotype | Reference/source |
|---|---|---|
| Strains | ||
| LG1458 | MG1655(DE3) ΔgudD ΔuxaC | Prather Lab |
| IB1863 | MG1655 ΔendA Δzwf ΔpfkB ΔsspB pfkA::114-pfkA(DAS+4) HK022::tetR-Ptet-sspB | (Brockman and Prather, 2015) |
| IB1379 | MG1655 ΔendA Δzwf ΔpfkB | (Brockman and Prather, 2015) |
| IB1486 | MG1655(DE3) ΔendA Δzwf ΔpfkB ΔsspB pfkA::114-pfkA(DAS+4) HK022::tetR-Ptet-sspB ΔgudD ΔuxaC | This study |
| IB2255 | MG1655(DE3) ΔendA Δzwf ΔpfkB ΔpfkA ΔgudD ΔuxaC | This study |
| IB2472 | MG1655(DE3) ΔendA Δzwf ΔpfkB ΔsspB pfkA::114-pfkA HK022::tetR-Ptet-sspB ΔgudD ΔuxaC | This study |
| JW2758-5 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔgudD785::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | (Baba et al., 2006) |
| JW3887-1 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔpfkA775::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | (Baba et al., 2006) |
| JW3603-2 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔuxaC782::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | (Baba et al., 2006) |
| IB1486-GA | IB1486 / pRSFD-IN-MI / pTrc-udh | This study |
| LG1458-GA | LG1458 / pRSFD-IN-MI / pTrc-udh | This study |
| IB2255-GA | IB2255 / pRSFD-IN-MI / pTrc-udh | This study |
| IB2472-GA | IB2472 / pRSFD-IN-MI / pTrc-udh | This study |
| Plasmids | ||
| pCP20 | Repa, AmpR, CmR, FLP recombinase expressed by λ pr under control of λ cI857 | CGSC #7629 |
| pRSFD-IN-MI | pRSR1030 ori, lacI, KanR, INO1 (S. cerevisiae) and MIOX (M. musculus) expressed under control of T7 promoter | (Moon et al., 2009) |
| pTrc-udh | pBR322 ori, lacI, AmpR, Udh (P. syringae) expressed under control of Trc promoter | (Yoon et al., 2009) |
2.2. Culture medium and conditions
For plasmid preparation and genetic manipulations, strains were cultured in Luria–Bertani (LB) medium at either 30° or 37 °C. Temperature sensitive plasmids were cured at 42 °C.
Glucaric acid production experiments were carried out in T12 medium containing 7.5 g/L yeast extract, 7.5 g/L soy peptone, 7 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 3 g/L (NH4)2SO4, 4 mM MgSO4, 100 μg/ml carbenicillin, 50 μg/ ml kanamycin, and the indicated amount of glucose and/or soluble starch (Sigma-Aldrich S9765) plus amyloglucosidase (Sigma-Aldrich A7095). For experiments in the BioLector (m2p-labs, Baesweiler, Germany), starter cultures were incubated in culture tubes at 30 °C and 250 rpm overnight in T12 supplemented with 10 g/L glucose and diluted 1:100 into working cultures. Working cultures were incubated at 30 °C, 1200 rpm (3 mm orbit), and 80% relative humidity in the BioLector. A working volume of 1 ml was used in the BioLector 48-well flower plate, sealed with gas-permeable sealing film with evaporation reduction layer (m2p-labs). To induce expression of enzymes required for glucaric acid production, 100 μM β-D-1-thiogalactopyranoside (IPTG) was added to production cultures at inoculation. For induction of SspB in strain IB1486 to knock down Pfk activity, 100 ng/ml anhydrotetracycline (aTc) was added at the times indicated in the Results section. At the indicated time points, the contents of the sample well were removed for measurement of glucaric acid production and residual glucose levels. All conditions were tested in triplicate wells. For example, to generate the data set illustrated in Fig. 1, 18 wells of IB1486 were inoculated (3 with aTc added at inoculation, 3 with aTc added at 5 h, etc.). At the times where aTc addition was indicated, the BioLector was briefly stopped, the plate opened, and aTc was added to the 3 appropriate wells. After 48 h, all 18 wells were collected for analysis of glucaric acid production.
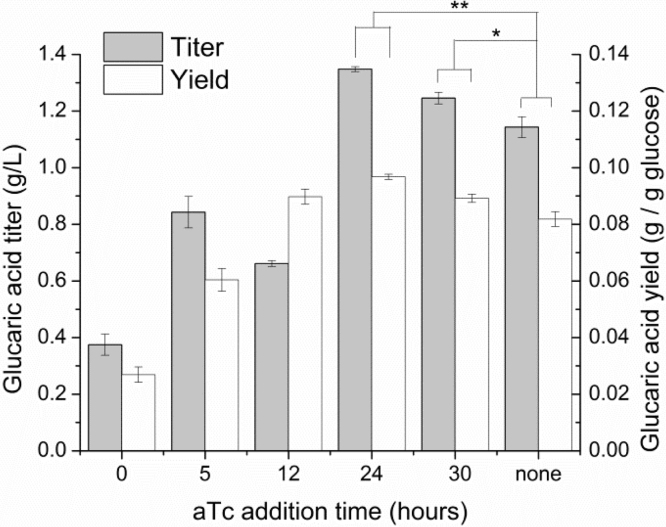
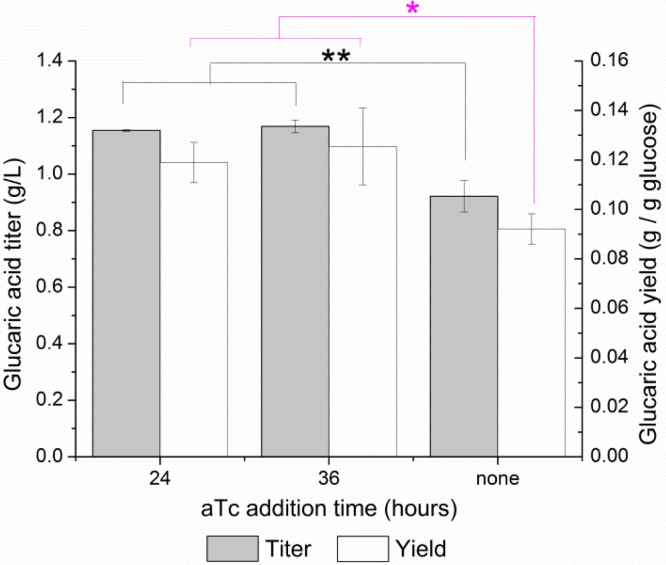
Fig. 1.
Yields (white bars) and titers (gray bars) of glucaric acid produced by IB1486-GA as a function of aTc addition time. Glucaric acid production was measured at 48 h in T12 medium supplemented with 15 g/L glucose. Error bars represent triplicate mean±SD. *, p<0.05; **, p<0.005; between yield or titer values as indicated.
For experiments in shake flasks, starter cultures were grown to mid-exponential phase (OD600~5) in 250 ml baffled flasks containing 30 ml T12+10 g/L glucose and used to inoculate 30 ml working cultures to a starting OD600=0.05. Production cultures were incubated in 250 ml baffled shake flasks at 30 °C, 80% humidity, and 250 rpm. IPTG (100 μM) was added at inoculation and aTc (100 ng/ml) was added at the indicated time points. Flasks were sampled periodically for measurement of optical density, as well as for HPLC and biomass samples. Samples from all experiments were stored at −20 °C until analysis.
2.3. Measurement of extracellular metabolites and starch
Glucose, glucaric acid, acetate, and myo-inositol levels were quantified by high performance liquid chromatography (HPLC) on an Agilent 1100 or 1200 series instrument (Santa Clara, CA) with an Aminex HPX-87H column (300 mm by 7.8 mm; Bio-Rad Laboratories, Hercules, CA). Sulfuric acid (5 mM) at a flow rate of 0.6 mL/min was used as the mobile phase. Compounds were quantified from 10 μL sample injections using refractive index (glucose, myo-inositol, glucaric acid, acetate) and diode array detectors (glucaric acid, 210 nm). Column and refractive index detector temperatures were held at 65 °C and 35 °C respectively.
To quantify the amount of starch hydrolysis in fed-batch samples, samples were split at collection. Half of the sample was centrifuged at 15000xg for 15 min and used for HPLC analysis as described above. The remaining portion of the sample was treated with 15 U/ml amyloglucosidase for 15 min at room temperature for full hydrolysis of remaining starch. After treatment, the sample was centrifuged for 5 min at 15000xg and the glucose concentration in the supernatant was measured using a YSI 2900 Biochemistry Analyzer (YSI Life Sciences, Yellow Springs, OH). The difference between the glucose content measured in the fully hydrolyzed sample and the glucose content measured via HPLC in the sample without an additional hydrolysis step was used to calculate the content of un-hydrolyzed starch.
The maximum amount of glucose that could be liberated from starch in the medium was determined by full hydrolysis of the starting medium with amyloglucosidase. To calculate glucose utilized by the cell, the amount of free glucose and the amount glucose generated from full hydrolysis of residual starch in a sample were subtracted from the maximum amount available in the medium. This value for consumed glucose was then used in the calculation of glucaric acid yield from glucose.
Titers and yields are reported in the text as the average of triplicate measurements ±1 STD and error bars in figures also represent average values±1 STD. To test for statistically significant differences between conditions, an unpaired two-tailed Student's t test was applied assuming equal variance. The level of significance is indicated in figures by the following: *p<0.05, ** p<0.005. At other points were significance is discussed, p values are indicated in the text.
2.4. Phosphofructokinase activity measurements
Phosphofructokinase activity assays were carried out as described previously (Brockman and Prather, 2015).
3. Results
Glucaric acid production was screened in strain IB1486-GA. This strain was derived from a previously developed strain, IB1863, where Pfk activity can be dynamically controlled through addition of aTc (Brockman and Prather, 2015). A modified SsrA tag was added to the coding sequence of pfkA in this strain, which results in slow degradation of the phosphofructokinase-I (Pfk-I) protein in the absence of the adapter protein SspB, but more rapid degradation of Pfk-I in the presence of SspB (McGinness et al., 2006). In IB1863, aTc addition induces SspB expression, resulting in rapid depletion of Pfk-I and buildup of intracellular G6P in glucose minimal medium. IB1486 contains the same modifications, along with additional knockouts of gudD and uxaC to prevent glucaric acid catabolism (Yoon et al., 2009) and the DE3 lysogen for expression of T7 RNA polymerase. In previous work with IB1863, it was shown that dynamic knockdown of Pfk activity could result in increased production of the glucaric acid precursor myo-inositol in glucose minimal medium, but that correct timing of aTc addition was required to achieve maximum yields and titers. Very early switching to “production mode” by aTc addition may result in insufficient time for protein expression and formation of biomass. However, very late switching results in more utilization of glucose for growth, and less remaining glucose to be redirected to product formation.
In moving to glucaric acid production, screening for optimal aTc addition time was again required, due to changes in cellular growth rate from the burden of expression of the complete glucaric pathway and the change in medium composition compared to the glucose minimal medium previously tested. The modified MOPS glucose minimal medium previously used for myo-inositol production (Brockman and Prather, 2015) was initially explored for glucaric acid production, but lag times of approximately 48 hours were observed, likely due to the burden associated with expression of all three pathway proteins. In addition to glucose, the T12 medium used in this work for testing of glucaric acid production contains yeast extract and soy peptone, which provide supplemental carbon sources. While glucose would primarily be used as a feedstock for glucaric acid production, some additional carbon supplementation was desired in the medium to reduce batch time and simulate a potential semi-defined, scale-up medium.
To facilitate rapid screening of a variety of aTc addition times in triplicate, cultures were grown in 48-well flower plates in a BioLector microbioreactor system. Glucaric acid production was screened in T12 medium in both batch conditions (15 g/L glucose at inoculation) and simulated fed batch conditions, where 3–5 g/L glucose was added at inoculation, and additional glucose was released slowly from 10–12 g/L starch by addition of amyloglucosidase.
3.1. Screening batch conditions for timing of Pfk knockdown
For screening of IB1486-GA under batch conditions, working cultures consisted of T12 medium with 15 g/L glucose and with 100 μM IPTG added at inoculation for induction of the glucaric acid pathway enzymes. Additions of aTc were made at times varying from 0–30 h after inoculation. Fig. 1 illustrates the yields and titers of glucaric acid observed after 48 hours.
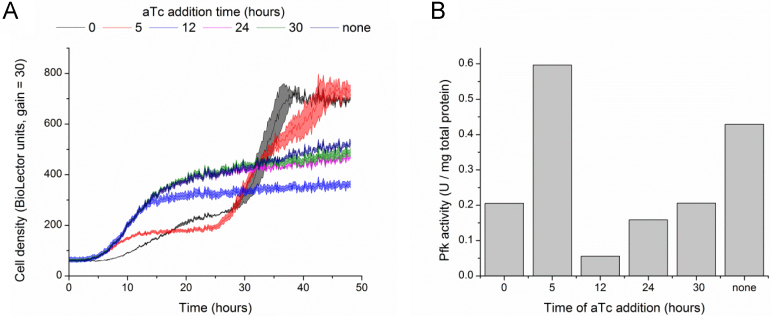
Glucose was fully consumed in all cultures at 48 h, except the culture with aTc addition at 12 h, which still contained 6.6±0.1 g/L glucose. The highest titers of glucaric acid, 1.35 g/L (9.7% yield from glucose), were achieved with aTc addition at 24 h, representing an 18% improvement in both yields and titers over the case of no Pfk switching. As expected, switching after 24 h resulted in somewhat lower titers, as more glucose had already been consumed for biomass and could not be redirected to glucaric acid production. Earlier switching resulted in lower titers either due to incomplete consumption of glucose, as in the case of 12 h aTc addition, or due to an “escape” phenotype. The escape phenotype correlates with rapid growth to higher cell densities (Fig. 2A) and an increase in Pfk activity (Fig. 2B). Studies of the system based on IB1863 have indicated that the increase in Pfk activity is likely linked to disruption of SspB expression, which is required for rapid Pfk-I degradation, through mutation or mobile element insertion (Supplementary methods and Supplementary Fig. 1). Very early addition of aTc results in high stress on the cell from a combination of limited glucose uptake and high protein expression for the glucaric acid pathway enzymes, which may result in more rapid selection for the escape phenotype.
Fig. 2.
Growth profiles and Pfk activity at 48 h for IB1486-GA in T12+15 g/L glucose. (A) Growth of IB1486-GA with aTc addition at the times indicated. Error bars represent triplicate mean±SD. (B) Pfk activity measured in selected wells from screening plate at 48 h.
3.2. Screening fed-batch conditions for timing of Pfk knockdown
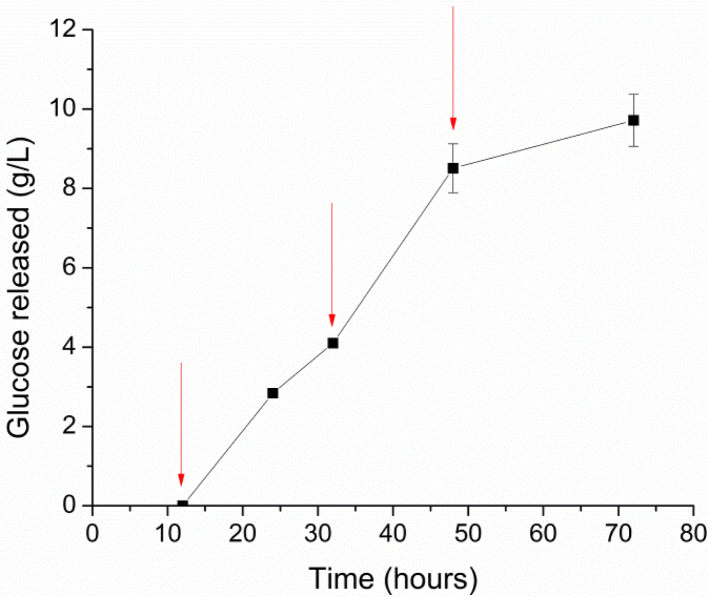
Fed batch conditions were initially screened in T12 medium with 3 g/L free glucose and 12 g/L starch, with 100 μM IPTG added at inoculation for induction of the glucaric acid pathway enzymes. Feeding was started at 12 hours by addition of 0.006 U/ml amyloglucosidase. As the starch hydrolysis rate declines with time, secondary additions of 0.006 U/ml and 0.012 U/ml amyloglucosidase were carried out at 36 and 48 hours, giving the glucose release profile shown in Fig. 3. At the conclusion of the experiment, in addition to the initial 3 g/L glucose, another 9.7±0.7 g/L free glucose had been released in the cultures on average. (For calculation of yield, unhydrolyzed starch was measured in individual wells via the method outlined in Section 2.3).
Fig. 3.
Glucose release via starch hydrolysis in T12 medium, with initial starch addition of 12 g/L. Amyloglucosidase additions are indicated by red arrows: 0.006 U/ml at 12 and 36 hours, 0.012 U/ml at 48 h. Error bars represent triplicate mean±SD. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
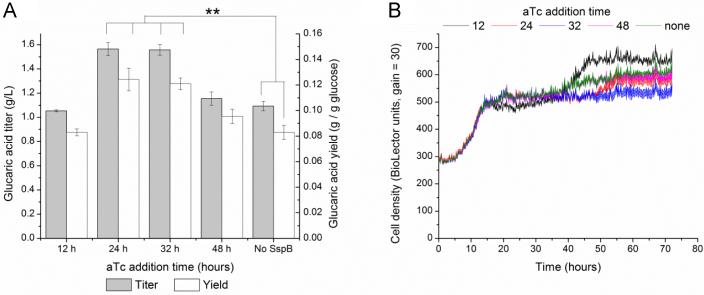
In this system, maximum titers of 1.56 g/L could be achieved with aTc addition at either 24 or 32 h (Fig. 4A), a 42% improvement over no aTc addition. The yields of glucaric acid were also improved by up to 50% with aTc addition at 24 h, with a maximum yield of 12.4% on glucose (based on available glucose added and hydrolyzed from starch). This improvement was larger than the batch condition, likely due to differences in the amount of glucose still available for consumption after addition of aTc. Early aTc addition at 12 h did not result in improved titers, with the shape of the growth curve indicative of escape (Fig. 4B). Measurement of Pfk activity also showed activity recovery to levels at or above the case of no aTc addition for this condition.
Fig. 4.
Growth and glucaric acid production in IB1486-GA in T12+3 g/L glucose+12 g/L starch. (A) Titers and yields of glucaric acid at 72 hours for aTc addition times from 12–48 h. (B) Growth of IB1486-GA in T12+3 g/L glucose+12 g/L starch. Starch addition resulted in higher opacity of medium at start of fermentation, and changes in OD600 after amylase addition represent both cell growth and changes in opacity as starch was broken down. Error bars represent triplicate mean±SD. *, p<0.05; **, p<0.005; between yield or titer values as indicated.
3.3. Shake flask studies under batch conditions
Consistent improvements in glucaric acid titers could be observed by timed knockdown of Pfk activity under the conditions tested in the BioLector. To determine whether these improvements would be robust to moderate changes in culture conditions, a set of batch experiments was carried out in shake flasks, along with feeding experiments testing alternative starch hydrolysis conditions in both the BioLector and in shake flasks.
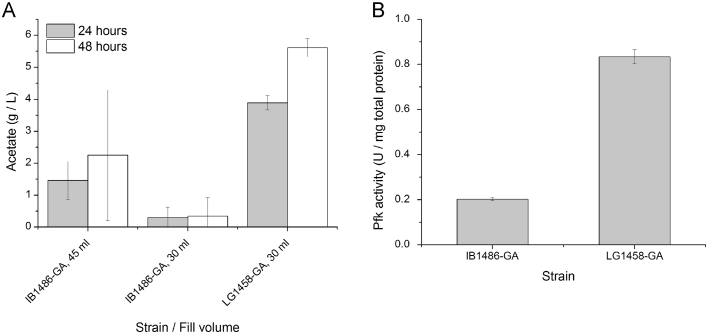
Initially, glucaric acid production was tested in shake flasks under batch conditions in both IB1486-GA and in LG1458-GA, a wild-type MG1655 background with only gudD and uxaC knockouts. In shake flasks, batch testing resulted in high baseline variability in titers that made it difficult to validate improvements in the 20–40% range that were previously achieved through Pfk knockdown in IB1486-GA. However, the shake flask testing did provide some interesting insights into the metabolism of IB1486-GA versus the wild-type strain LG1458-GA, in the absence of any aTc addition. Significantly, under batch conditions, acetate production varied greatly between the two strains, with LG1458-GA producing much higher levels of acetate. The fill volume of flasks in batch testing appeared to have an effect on acetate production from IB1486-GA, potentially due to changes in aeration. In IB1486-GA, several cultures showed residual acetate at 48 hours in shake flasks with a 45 ml fill volume, while in previous testing in the BioLector, no acetate was observed at 48 h. Testing at a lower fill volume (30 ml) resulted in reduced acetate accumulation at both 24 and 48 h. A summary of observed acetate production in IB1486-GA and LG1458-GA in shake flasks is presented in Fig. 5A.
Fig. 5.
Acetate production and Pfk activity in IB1486-GA and LG1458-GA in T12+15 g/L glucose. (A) Acetate production at 24 and 48 h in IB1486-GA and LG1458-GA in T12+15 g/L glucose. Cultures were carried out in 250 ml baffled shake flasks with the fill volume noted and 250 rpm shaking at 30 °C and 80% relative humidity. (B) Pfk activity at 24 hours after inoculation in T12+15 g/L glucose with 30 ml fill volume in 250 ml flasks. Error bars represent triplicate mean±SD.
While improved aeration could alleviate moderate acetate accumulation in IB1486-GA, acetate accumulation in LG1458-GA remained high. Pfk activity measurements at 24 h in IB1486-GA and LG1458-GA (Fig. 5B) showed that Pfk activity in LG1458-GA was significantly higher than in IB1486-GA, likely leading to metabolic overflow and greater acetate production. The low baseline activity of IB1486-GA in T12 medium was unexpected, given that the parent strain, IB1863, always showed Pfk activity higher than the wild type in MOPS minimal medium with glucose (Brockman and Prather, 2015).
In addition to exhibiting higher acetate accumulation, glucaric acid production was very poor for LG1458-GA under batch conditions, with titers below the limit of detection in T12+15 g/L glucose (30 ml fill volume). LG1458-GA showed incomplete glucose consumption as well, with 3.0±0.1 g/L remaining at 48 hours. Although titers in IB1486-GA showed high variability, glucaric acid production was clear in all samples in T12+15 g/L glucose, with measured titers of 0.9±0.3 g/L glucaric acid in shake flasks with 30 ml fill volume. Glucose was also completely consumed in the cultures.
Although a static condition of low Pfk activity can clearly be tolerated in T12 medium and can provide a benefit in glucaric acid production under batch conditions, the previous screening work in Section 3.1 indicated that there is a limit to what could be gained in this manner, as complete knockdown of Pfk activity by aTc addition at inoculation resulted in poor growth and eventual “escape” of the culture. A ΔpfkA ΔpfkB double knockout strain, IB2255, was tested to assess the productivity that could be expected in T12 medium under batch conditions with no Pfk activity. After 48 hours in T12 with 15 g/L glucose, 0.18±0.01 g/L glucaric acid was produced, significantly lower than the 0.9±0.3 g/L produced in IB1486-GA in shake flasks in batch (p<0.05, Student’s t test). To maximize productivity in a 48 h batch, at least some period of culture growth needs to be provided with higher Pfk activity.
3.4. Validation of fed-batch results under altered feeding conditions
The high variability in batch shake flask experiments made it difficult to validate whether Pfk knockdown could be used to increase titers at that scale, as well as in the BioLector. Larger titer improvements were seen with fed-batch conditions, so a set of validation experiments was run under those conditions, testing both an alternate starch hydrolysis strategy in the BioLector and application at shake flask scale.
As the highest fed-batch titers were achieved previously with Pfk knockdown at 24–32 h, it would be expected that growth up to that point could be carried out under either batch or fed-batch conditions without significant changes to the outcomes. An alternative feeding strategy was tested with the initial free glucose increased to 5 g/L and feeding started at 24 h, from a reservoir of 10 g/L starch. This was initially tested in 48 well plates in the BioLector microbioreactor, with amyloglucosidase additions of 0.006 U/ml at 24 h and 48 h. The highest titers of 1.17 g/L were achieved with aTc addition at 36 hours, a 27% improvement over no aTc addition (Fig. 6), validating that Pfk knockdown could be used to improve titers under alternative feeding conditions. A maximum glucaric acid yield of 12.5% from glucose was also achieved with aTc addition at 36 h (36% improvement over no aTc addition). Although titers were lower than in the original fed-batch screening, maximum yields were similar, as un-hydrolyzed starch contents were higher under this feeding strategy, leaving only 9.7 g/L free glucose (initial dose and feeding) available for conversion to glucaric acid, versus 12.7 g/L in the previous test.
Fig. 6.
Titers and yields of glucaric acid at 72 h for IB1486-GA in T12+5 g/L glucose+10 g/L starch. Amyloglucosidase additions were carried out at 24 and 48 hours. Error bars represent triplicate mean±SD. *, p<0.05; **, p<0.005; between yield (purple) or titer (black) values as indicated. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
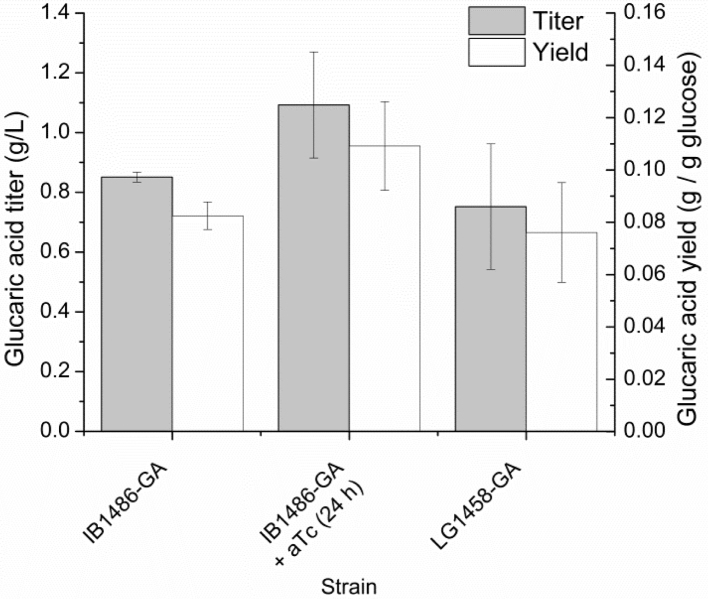
The starch fed-batch strategy was then tested in shake flasks, both for IB1486-GA and for LG1458-GA. The cultures contained 5 g/L free glucose and 10 g/L starch. Free glucose was measured at 18 h, and since glucose was already found to be exhausted, amyloglucosidase addition was started at that time. Secondary additions were carried out at 40 and 48 h. Despite the extra amyloglucosidase addition, starch hydrolysis was again poorer in this condition, and resulted in 10.1±0.5 g/L total free glucose available in the cultures on average. Baseline yields for IB1486-GA were still comparable with the previous tests. Addition of aTc for Pfk knockdown at 24 hours resulted in a 28% improvement in titers and a 32% improvement in yield over no aTc addition (Fig. 7). Variability was higher in shake flasks than in previous testing in the BioLector, and statistical significance at 95% confidence was not achieved for this result, although it would be significant at a 90% confidence level (0.05<p<0.1, Student's t test).
Fig. 7.
Yields and titers of glucaric acid in IB1486-GA and LB1458-GA in shake flasks with T12+5 g/L glucose+10 g/L starch. Amyloglucosidase additions were carried out at 18, 40, and 48 h. Error bars represent triplicate mean±SD.
The results of fed-batch testing in shake flasks also illustrated some potential benefits of slower glucose feeding for the wild type strain, LG1458-GA. In contrast to the batch condition, under the starch hydrolysis (glucose feeding) condition shown in Fig. 7, glucaric acid titers of 0.75±0.2 g/L were achieved in LG1458-GA, more comparable to the 0.85±0.02 g/L produced in IB1468-GA without aTc addition, and any acetate produced was fully consumed by the conclusion of the experiment for both strains. Yields were also more comparable between the two strains under glucose feeding, with 7.6% for LG1458-GA versus 8.3% for IB1458-GA. One of the clear advantages of the fed-batch condition is the elimination of acetate buildup and carbon loss to acetate formation. Additional changes in metabolism, such as upregulation of genes associated with sugar transport (Franchini and Egli, 2006, Raman et al., 2005), could also be favorably changing relative metabolite pools. While a fed-batch production strategy provides a benefit for LG1458-GA, IB1468-GA does not benefit as strongly from slow glucose feeding, likely due to the fact that acetate production is already much lower in this strain. Pfk activity may also be low enough in this strain that other changes in metabolism related to glucose limitation are not significant.
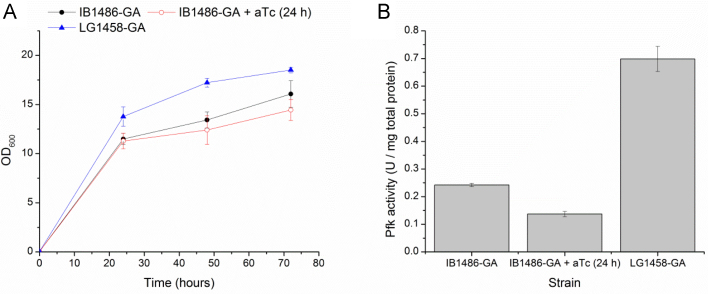
Growth (Fig. 8A) and Pfk activity measurements (Fig. 8B) during fed-batch testing in shake flasks showed trends similar to previous tests. As under batch conditions, IB1486-GA had significantly lower baseline Pfk activity than LG1458-GA before aTc addition, with aTc addition resulting in an additional decrease in activity of about 50%. While the mismatch in baseline activity did not cause a significant difference in titers between IB1486-GA and LG1458-GA under fed-batch conditions (p>0.4, Student's t test), this did affect batch performance strongly, as discussed previously in Section 3.3.
Fig. 8.
Growth and Pfk activity in IB1486-GA and LG1458-GA in T12+5 g/L glucose+10 g/L starch with amyloglucosidase addition at 18, 40, and 48 h. (A) Growth of LG1458-GA and IB1486-GA with and without aTc addition at 24 h after inoculation. (B) Pfk activity in these strains at 48 h after inoculation. Error bars represent triplicate mean±SD.
An additional control experiment was carried out under fed-batch conditions to analyze potential effects of SspB expression on glucaric acid production in the absence of a modified ssrA-tagged degradation target. The IB1486 strain background includes the deletion of the native copy of sspB, and it is possible that restoring its expression through aTc addition could normalize native protein degradation and affect glucaric acid production independent of Pfk knockdown. To control for this possible broader effect of SspB expression on production, strain IB2472 was constructed, which is identical to IB1486, except that the modified ssrA tag was removed from the sequence of pfkA. In this strain, Pfk is not degraded upon SspB expression, so the effect of SspB expression alone can be observed. This strain was tested under the same culture conditions used to generate Fig. 7. There was no significant difference in the titers or yields of glucaric acid in IB2472-GA between flasks with SspB expressed and those without (p>0.9, Supplementary Table 1). This is consistent with reports in the literature that deletion of sspB alone does not result in a large buildup of native ssrA tagged proteins in the cell, in contrast to deletions of the unfoldase ClpX or protease ClpP, which result in clear buildup of tagged proteins (Lies and Maurizi, 2008). The glucaric acid titers in IB2472-GA were lower than those observed in IB1486, likely due to higher baseline Pfk activity in this strain. The removal of the DAS+4 tag in the control strain also eliminates the low level background degradation of the protein that takes place even in the absence of SspB, resulting in higher Pfk activity.
4. Discussion
Results with IB1486-GA indicate that dynamic control of Pfk activity can be utilized to improve titers of glucaric acid, a product requiring several enzymatic conversions starting from G6P. The system was applicable for use with a semi-defined medium under both batch and simulated fed-batch conditions. While gains in titer were consistent across multiple conditions, the maximum gains were smaller than those observed previously for myo-inositol production in glucose minimal medium (Brockman and Prather, 2015). Previous work with myo-inositol production in glucose minimal medium showed that switching at low cell density was optimal for the largest gains in titer. In T12 medium, these earlier switching times resulted in more rapid escape and little time for conversion of glucose to glucaric acid, perhaps due to the greater expression burden of the complete glucaric acid pathway and the higher availability of nutrients in T12 that “escapers” could use to rapidly grow and overtake the population. The later switching times result in higher usage of glucose for biomass formation, so the amount of glucose processed after switching to production mode is relatively low. While genetic stability can be achieved out to at least 72 h for aTc addition at 24 h, future work may be needed to address genetic stability in longer fermentations and expand the usefulness of switching between growth and production modes.
Activity of the downstream enzymes in the glucaric acid pathway is another potential limitation, but in this particular medium, it does not appear that the activity of MIOX, a bottleneck under some other conditions, was limiting overall pathway yield, as minimal buildup of myo-inositol was observed in the cultures. However, balancing of expression between the three pathway enzymes could be an issue, since high level INO1 expression is required for any myo-inositol to be produced for further conversion (Moon et al., 2009). Reductions in INO1 expression upon expression of other enzymes in the glucaric acid pathway would be expected to limit maximum fluxes into the pathway, also limiting the glucose that could be effectively redirected in a given time period.
Importantly, IB1486-GA showed titers that were comparable with a wild-type control strain under fed-batch conditions and superior under batch conditions, indicating the genetic modifications required for control of Pfk activity were not detrimental to baseline glucaric acid production and could potentially be transferred into high-performing strains as well. Although the baseline Pfk activity was low in T12 medium, it was still sufficient for rapid growth without excessive overflow metabolism. More consistent success with chromosomal modifications in the K strains led to the initial construction of the Pfk-I control system in that background, but additional improvements in glucaric acid titer, both with and without Pfk knockdown, can likely be achieved by transferring the genetic modifications of IB1486 to an E. coli B strain. In previous work, wild-type BL21 has outperformed MG1655 containing the same pathway genes (Moon et al., 2009, Raman et al., 2014, Shiue et al., 2015).
5. Conclusions
Glucaric acid titers and yields could be improved under multiple culture conditions through timed knockdown of Pfk activity, with maximum improvements of up to 42% observed. In the absence of aTc, the switchable strain IB1486 shows titers comparable to or above those observed with wild-type MG1655, indicating the genetic modifications in IB1486 do not result in degradation of baseline performance and could potentially be applied to high-performing strains for increases in titer. Optimization of strain background and pathway enzyme expression levels may lead to both higher baseline titers and to greater gains from dynamic control of Pfk activity.
Conflict of Interest Statement
N.C.C. and K.L.J.P. are founders of Kalion, Inc.
Acknowledgments
This work was supported by the US National Science Foundation through the CAREER program (I.M.B.R. and K.L.J.P., Grant No. CBET-0954986), the Graduate Research Fellowship program (A.G.), and the Synthetic Biology Engineering Research Center (SynBERC; A.G. and K.L.J.P., Grant No. EEC-0540879); and through the Biotechnology Training Program of the National Institutes of Health (I.M.B.R., Grant No. T32GM008334). This work was also supported by a Dean Paolo Cucchi Research Grant from Drew University awarded to A.R.S. We thank m2p-labs for the loan of a BioLector unit.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2015.09.002.
Appendix A. Supplementary material
Supplementary material
References
- Anesiadis N., Cluett W.R., Mahadevan R. Dynamic metabolic engineering for increasing bioprocess productivity. Metab. Eng. 2008;10:255–266. doi: 10.1016/j.ymben.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman I.M., Prather K.L. Dynamic knockdown of E. coli central metabolism for redirecting fluxes of primary metabolites. Metab. Eng. 2015;28:104–113. doi: 10.1016/j.ymben.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R.H., Zhang F., Alonso-Gutierrez J., Baidoo E., Batth T.S., Redding-Johanson A.M., Petzold C.J., Mukhopadhyay A., Lee T.S., Adams P.D., Keasling J.D. Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotech. 2013;31:1039–1046. doi: 10.1038/nbt.2689. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer W.R., Liao J.C. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat. Biotechnol. 2000;18:533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- Franchini A.G., Egli T. Global gene expression in Escherichia coli K-12 during short-term and long-term adaptation to glucose-limited continuous culture conditions. Microbiology. 2006;152:2111–2127. doi: 10.1099/mic.0.28939-0. [DOI] [PubMed] [Google Scholar]
- Gadkar K.G., Doyle F.J., III, Edwards J.S., Mahadevan R. Estimating optimal profiles of genetic alterations using constraint-based models. Biotechnol. Bioeng. 2005;89:243–251. doi: 10.1002/bit.20349. [DOI] [PubMed] [Google Scholar]
- Kiely D.E., Chen L. Glucaric acid monoamides and their use to prepare poly(glucaramides) Google Pat. 1994 [Google Scholar]
- Kogure T., Wakisaka N., Takaku H., Takagi M. Efficient production of 2-deoxy-scyllo-inosose from D-glucose by metabolically engineered recombinant Escherichia coli. J. Biotechnol. 2007;129:502–509. doi: 10.1016/j.jbiotec.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Lies M., Maurizi M.R. Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J. Biol. Chem. 2008;283:22918–22929. doi: 10.1074/jbc.M801692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness K.E., Baker T.A., Sauer R.T. Engineering controllable protein degradation. Mol. Cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Mehltretter C.L., Rist C.E. Sugar oxidation, saccharic and oxalic acids by the nitric acid oxidation of dextrose. J. Agric. Food Chem. 1953;1:779–783. [Google Scholar]
- Moon T.S., Dueber J.E., Shiue E., Prather K.L.J. Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab. Eng. 2010;12:298–305. doi: 10.1016/j.ymben.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Moon T.S., Yoon S.H., Lanza A.M., Roy-Mayhew J.D., Prather K.L. Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl. Environ. Microbiol. 2009;75:589–595. doi: 10.1128/AEM.00973-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman B., Nandakumar M.P., Muthuvijayan V., Marten M.R. Proteome analysis to assess physiological changes in Escherichia coli grown under glucose-limited fed-batch conditions. Biotechnol. Bioeng. 2005;92:384–392. doi: 10.1002/bit.20570. [DOI] [PubMed] [Google Scholar]
- Raman S., Rogers J.K., Taylor N.D., Church G.M. Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. 2014;111:17803–17808. doi: 10.1073/pnas.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisch, C.R., Prather, K.L.J., in press. The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli. Sci. Rep. [DOI] [PMC free article] [PubMed]
- Scalcinati G., Knuf C., Partow S., Chen Y., Maury J., Schalk M., Daviet L., Nielsen J., Siewers V. Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquitepene alpha-santalene in a fed-batch mode. Metab. Eng. 2012;14:91–103. doi: 10.1016/j.ymben.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Shiue E., Brockman I.M., Prather K.L. Improving product yields on D-glucose in Escherichia coli via knockout of pgi and zwf and feeding of supplemental carbon sources. Biotechnol. Bioeng. 2015;112:579–587. doi: 10.1002/bit.25470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiue E., Prather K.L. Improving D-glucaric acid production from myo-inositol in E. coli by increasing MIOX stability and myo-inositol transport. Metab. Eng. 2014;22:22–31. doi: 10.1016/j.ymben.2013.12.002. [DOI] [PubMed] [Google Scholar]
- Smith T.N., Kiely D.E., Kramer-Presta K. Corrosion inhibiting composition. Google Pat. 2012 [Google Scholar]
- Solomon K.V., Sanders T.M., Prather K.L. A dynamic metabolite valve for the control of central carbon metabolism. Metab. Eng. 2012;14:661–671. doi: 10.1016/j.ymben.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Soma Y., Tsuruno K., Wada M., Yokota A., Hanai T. Metabolic flux redirection from a central metabolic pathway toward a synthetic pathway using a metabolic toggle switch. Metab. Eng. 2014;23:175–184. doi: 10.1016/j.ymben.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Torella J.P., Ford T.J., Kim S.N., Chen A.M., Way J.C., Silver P.A. Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proc. Natl. Acad. Sci. 2013;110:11290–11295. doi: 10.1073/pnas.1307129110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werpy T., Petersen G. U.S. Department of Energy; Washington, D.C.: 2004. Top value added chemical from biomass, volume 1-results of screening for potential candidates from sugars and synthetic gas. [Google Scholar]
- Williams T.C., Averesch N.J.H., Winter G., Plan M.R., Vickers C.E., Nielsen L.K., Krömer J.O. Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae. Metab. Eng. 2015;29:124–134. doi: 10.1016/j.ymben.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Xu P., Li L., Zhang F., Stephanopoulos G., Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc. Natl. Acad. Sci. 2014;111:11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.-H., Moon T.S., Iranpour P., Lanza A.M., Prather K.J. Cloning and characterization of uronate dehydrogenases from two pseudomonads and Agrobacterium tumefaciens strain C58. J. Bacteriol. 2009;191:1565–1573. doi: 10.1128/JB.00586-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Carothers J.M., Keasling J.D. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotech. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material