Abstract
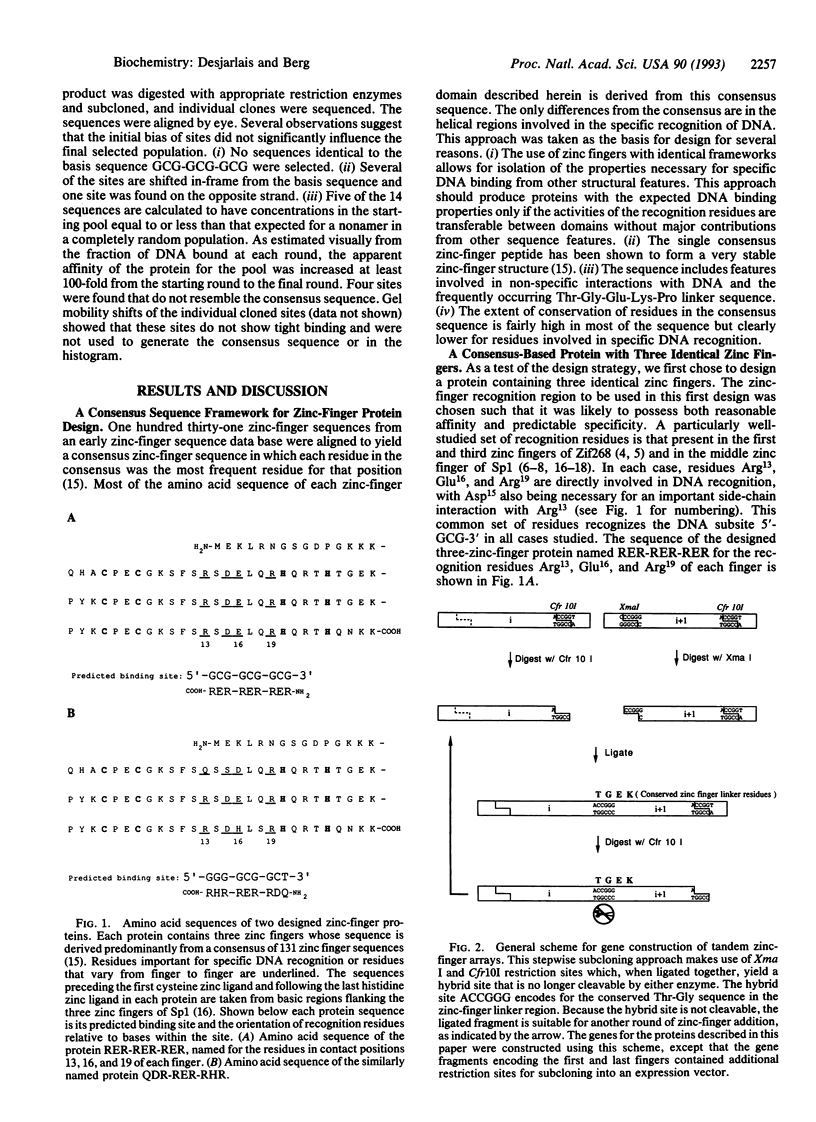
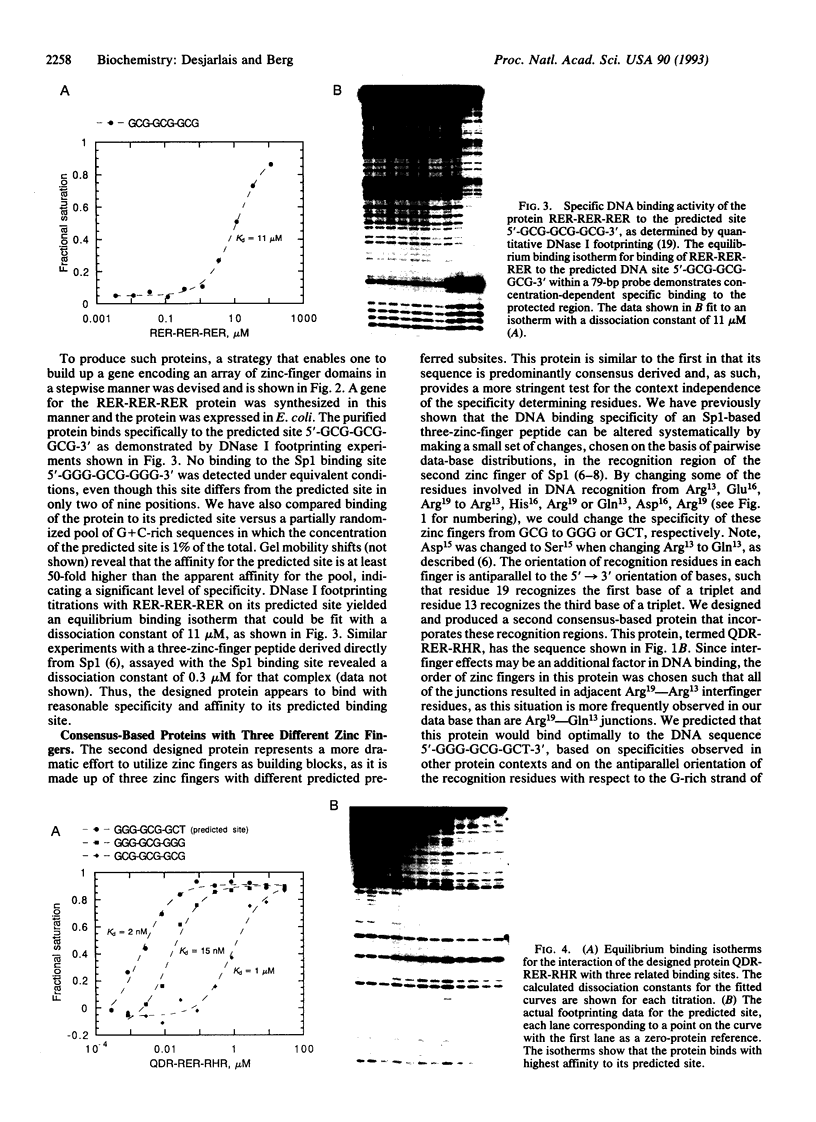
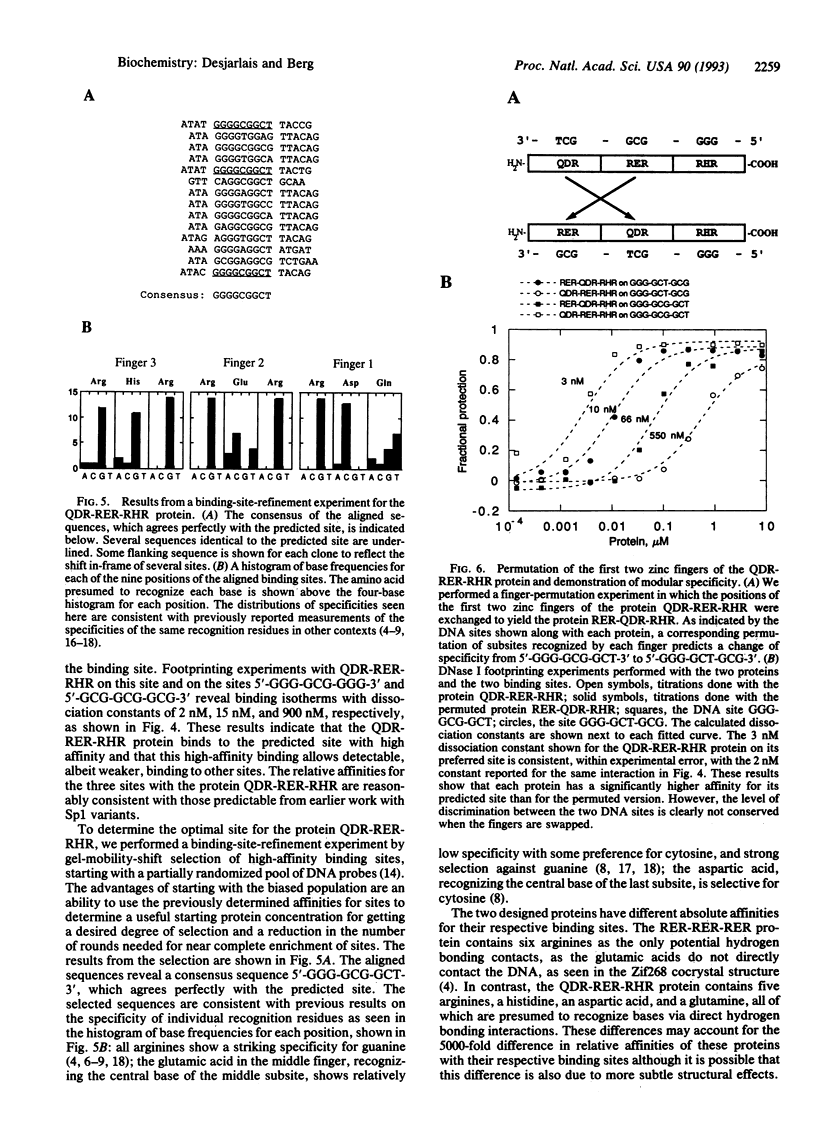
We have designed three zinc-finger proteins with different DNA binding specificities. The design strategy combines a consensus zinc-finger framework sequence with previously characterized recognition regions such that the specificity of each protein is predictable. The first protein consists of three identical zinc fingers, each of which was expected to recognize the subsite GCG. This protein binds specifically to the sequence 5'-GCG-GCG-GCG-3' with a dissociation constant of approximately 11 microM. The second protein has three zinc fingers with different predicted preferred subsites. This protein binds to the predicted recognition site 5'-GGG-GCG-GCT-3' with a dissociation constant of 2 nM. Furthermore, selection experiments indicate that this is the optimal binding site. A permuted version of the second protein was also constructed and shown to preferentially recognize the corresponding permuted site 5'-GGG-GCT-GCG-3' over the non-permuted site. These results indicate that earlier observations on the specificity of zinc fingers can be extended to generalized zinc-finger structures and realize the use of zinc fingers for the design of site-specific DNA binding proteins. This consensus-based design system provides a useful model system with which to study details of zinc-finger-DNA specificity.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander P., Fahnestock S., Lee T., Orban J., Bryan P. Thermodynamic analysis of the folding of the streptococcal protein G IgG-binding domains B1 and B2: why small proteins tend to have high denaturation temperatures. Biochemistry. 1992 Apr 14;31(14):3597–3603. doi: 10.1021/bi00129a007. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Zinc finger domains: hypotheses and current knowledge. Annu Rev Biophys Biophys Chem. 1990;19:405–421. doi: 10.1146/annurev.bb.19.060190.002201. [DOI] [PubMed] [Google Scholar]
- Brenowitz M., Senear D. F., Shea M. A., Ackers G. K. "Footprint" titrations yield valid thermodynamic isotherms. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8462–8466. doi: 10.1073/pnas.83.22.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjarlais, Berg J. M. Redesigning the DNA-binding specificity of a zinc finger protein: a data base-guided approach. Proteins. 1992 Jul;13(3):272–272. doi: 10.1002/prot.340130309. [DOI] [PubMed] [Google Scholar]
- Desjarlais J. R., Berg J. M. Redesigning the DNA-binding specificity of a zinc finger protein: a data base-guided approach. Proteins. 1992 Feb;12(2):101–104. doi: 10.1002/prot.340120202. [DOI] [PubMed] [Google Scholar]
- Desjarlais J. R., Berg J. M. Toward rules relating zinc finger protein sequences and DNA binding site preferences. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7345–7349. doi: 10.1073/pnas.89.16.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A. D., Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990 Aug 30;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Klevit R. E. Recognition of DNA by Cys2,His2 zinc fingers. Science. 1991 Sep 20;253(5026):1367–1393. doi: 10.1126/science.1896847. [DOI] [PubMed] [Google Scholar]
- Letovsky J., Dynan W. S. Measurement of the binding of transcription factor Sp1 to a single GC box recognition sequence. Nucleic Acids Res. 1989 Apr 11;17(7):2639–2653. doi: 10.1093/nar/17.7.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardelli J., Gibson T. J., Vesque C., Charnay P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature. 1991 Jan 10;349(6305):175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Thiesen H. J., Bach C. Target Detection Assay (TDA): a versatile procedure to determine DNA binding sites as demonstrated on SP1 protein. Nucleic Acids Res. 1990 Jun 11;18(11):3203–3209. doi: 10.1093/nar/18.11.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Morrison M. L., Young E. T. Mutations in the zinc fingers of ADR1 that change the specificity of DNA binding and transactivation. Mol Cell Biol. 1992 Jun;12(6):2784–2792. doi: 10.1128/mcb.12.6.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]





