Abstract
Proteomics is the study of structure and function of proteins in a large scale. For any living organism, preteins are considered to be the vital part because of its role in metabolic pathways of cells. These proteins not only play a role in physiological condition of the cell but also in altered manner during pathologic conditions. These altered proteins in diseased conditions are called as biomarkers. Several such biomarkers were identified in oral diseaes. This review is a brief note on proteins involved in odontogenesis and list of altered proteins proteins identified in various dental and oral diseases. The knowledge about the role of proteomics in dentistry and the importance of proteomic studies in early diagnosis and prognostic part of oral diseases helps in appliction of precised and sucessful treatment.
KEY WORDS: Biomarkers, cancer proteomics, proteins
Proteomics is a branch of functional genomics. The term proteomics was first coined in 1994 by Wilkins, to make an analogy with genomics, the study of genes. The perceived ultimate goal of identifying and sequencing the 40,000–60,000 genes of the human genome have come to an end, and functional and structural genomics and proteomics projects have now begun, to investigate their structure and function using the genetic sequences to predict the resultant proteins.[1]
Micro-characterization of proteins for the identification of proteins in large scale and their posttranslational modifications
“Differential display” in proteins and protein levels for comparison with potential application in a wide range of diseases and
Protein-protein interactions.[2]
The activity most often associated with proteomics is fractionating and visualizing large numbers of proteins from cells. These types of experiments have been performed for more than 20 years to build databases of proteins expressed from certain cell or tissue types. This field has expanded due to the development of additional technologies.
Proteomics can be broadly divided into two areas of research: (i) Protein expression mapping, and (ii) protein interaction mapping. Two-dimensional (2D) gel electrophoresis coupled with mass spectrometry is used to protein expression mapping, which involves the quantitative study of global changes in protein expression in cells, tissues or body fluids. The aim of protein expression mapping is to compare the spectrum of proteins expressed in cells or tissues under different environmental conditions or from different disease states and furthermore, an understanding of posttranslational modifications of expressed proteins under this state.
Protein-protein interaction mapping involves determining, on a proteome-wide scale, the interaction partners for each of the encoded proteins of a cell or organism using the yeast two-hybrid system and by mass spectrometry.[3]
Necessity of Proteomics
Complete sequences of genomes are not sufficient to elucidate biological function. There is no strict linear relationship between genes and the protein complement or “proteome” of a cell. It is still difficult to predict genes accurately from genomic data.
Proteomics is complementary to genomics. Proteomics directly contributes to drug development as almost all drugs are directed against proteins. Proteomic methodologies determine isoforms and posttranslational modifications. Proteins across toxicity-induced diseases can be systematically analyzed using proteomics.[2]
The proteome-wide analysis undoubtedly has a major impact on understanding the phenotypes of normal and toxicant exposed cells. Proteomics approach focuses on the identification of potential early protein biomarkers/signatures, which are indicative of the toxicity or carcinogenesis in exposed animals and are being used to substantially reduce the time and costs of toxicity or carcinogenicity testing.[4,5] It has also provided potential throughput in the discovery of new biomarkers/toxicity signatures and mapping the serum/plasma and other biological fluid proteomes. Differential protein expression analysis has also provided a more sensitive indicator of toxicity than the conventional techniques. Proteins control the phenotype of a cell by determining its structure and, above all, by carrying out all function in a cell. Defective proteins are the major causes of diseases and thus serve as a useful indicator for the diagnosis of particular diseases. In addition, proteins are the primary targets of most drugs and thus are the main basis for the development of new drugs. Therefore, the study of proteomics is important for understanding their roles in the cause and control of diseases and in the development of humans, as well as that of other organisms.[6]
History
Genetics has had several major breakthroughs during its development that have made biology a well-established discipline of science. The first major discovery was the rules of inheritance by Mendel. This provided the particulate nature of inheritance and established the presence of genes, which control phenotypes. It also provided genes as the ultimate basis for propelling the process of evolution of organisms and integrated the different branches of the science of biology. In addition, Mendelian genetics transformed biology from a science-based exclusively on observations to an experimental science where certain ideas could be tested by performing experiments.[7]
The second major breakthrough was discovered by Beadle and Tatum with their conceptual one-gene-one-enzyme hypothesis. This proved the biochemical basis for the mechanism of gene action and integrated chemistry into biology and also provided the tool for analyzing metabolic pathways and several complex systems, including the nervous system. It also provided the understanding of the genetic basis of diseases and their possible cures by chemical manipulations and ultimately by gene therapy.[8] The discovery of the structure of DNA by Watson and Crick marked the third major breakthrough in biology.[9,10]
The coming of genomics marked the fourth major breakthrough in biology. It provides molecular insight into the genetic basis for differences in our response to the same drug. The variation in individual DNA sequences is expected to provide the molecular understanding of our several complex traits, including behavior. DNA sequences also provide a better insight into the record of the evolutionary processes in an organism. Genomics is expected to provide a better understanding of a complex organism like humans after the elucidation of the roles of noncoding sequences (introns) of DNA. Understanding the roles of introns is currently a formidable task. It is believed that the interpretation of the roles of introns (a segment of a gene situated between exons that does not function in coding for protein synthesis) will help in reaching a new height to the understanding of biology. The problem with genomics is it is difficult or impossible to identify the gene sequence with the behavior of gene product. Therefore, this entire amount to making the prediction of gene function a very tricky proposition.[11]
Molecular fingerprinting is the next phase in this genomics, in which it correlates the genomic state, the complementary DNA expression pattern, and the protein range with the functional status of the cells or tissue. The promise of this phase is that expressions profiles can uncover clues to functionally important molecules in the development of human disease and generate information to sub-classify human tumors and tailor a treatment to the individual patient.[12]
The fifth breakthrough underway is the development of proteomics. Proteomics conveyed a better understanding of biochemical pathways and the roles of protein interactions. Above all, proteomics provides a clue to answering the big question of how a small number of genes can control several phenotypes in a complex organism like humans. A major abstract pattern emerging from proteomics is that, it is the number of interactions of proteins responsible for the countless phenotypes in an organism and not the number of proteins by itself. Advances in proteomics are expected to integrate the reductionist views into systems biology to show how molecular parts evolved and how they fit together to work as an organism. The latter is expected to provide the ultimate understanding of biology. Development of customized molecular targeted therapies can be implicated for early cancer diagnostics from the drop of a patient's blood/saliva, to the molecular dissection of a patients individual tumor cells to find the atypical expression of protein interaction, with the usage of proteomics.[13,14]
Proteins
The term protein was derived from the Greek word proteios, meaning “first.” Proteins are the center of action in biological processes. Nearly all the molecular transformation that define cellular metabolism are mediated by protein catalysts. Proteins also perform regulatory roles, monitoring extracellular and intracellular conditions and relaying information to other cellular components. Proteins are essential structural components of cells.[15] Proteins are generally regarded as favorable and are a necessary part of the diet of all animals. Most proteins have enzymatic functions, but several of them such as actin and fibrinoactin are structural components of cells. Proteins are also responsible for the mobility of muscle cells. Certain proteins serve as receptors for different molecules or work as immunoglobulins or antigens, or proteins can serve as allergens or participate in the transport of various molecules, such as oxygen or sex hormones. Important metabolic functions in humans and many other living organisms were controlled by hormones, which are originally proteins (insulin, human growth hormone).[15]
A complete list of known protein functions would contain many thousands of entries, including proteins that transport that other molecule and proteins that gene.[15] Protein-based antibiotics and vaccines help to fight disease. The deadly properties of protein toxins and venoms are less prevalent.[16,17]
Proteome
The proteome is the entire set of proteins expressed by a genome, cell, tissue or organism. The term “proteome” was first used by Wilkins in 1994. All the proteins that can be synthesized by the cell or all the proteins encoded by the genome of a species are defined as proteome. The cell's storehouse of proteins with their nature and biological function is regarded as proteome. More specifically, it is the set of expressed proteins in a given type of cells or an organism at a given time under defined conditions. This term can also be used for the set of a subcellular structure or organelle.
Biomarker Identification
Proteomics is playing a major role in the understanding of the disease. Dramatic improvements in the precision of analytical techniques and the availability of the human genome sequence have made it possible to discover, identify, and characterize proteins that are of diagnostic and prognostic value. The sensitivity of proteomic techniques and their capacity to be applied to the analysis of tissue samples and body fluids has resulted in the application of proteomics to an enormous diversity of disease conditions. A large number of putative biomarker proteins have, therefore, been described to date. Although these biomarkers are yet to be adopted in the clinic, their eventual validation and commercialization are likely to lead to a revolution whereby early diagnosis of disease, the choice of treatment.[18]
Identification of Biomarker Proteins
Biomarker proteins can be either qualitative or quantitative. Qualitative biomarkers are those present in one patient group, but not another and are thus easily found. Quantitative biomarkers, by contrast, are present in different patient groups in different degrees. In some cases, a single protein will have sufficiently different expression levels that can be of diagnostic or prognostic importance, but in other cases, the combination of expression levels of a number of proteins – a panel of biomarkers – may be of the highest accuracy and utility. Quantitative biomarkers, whether single proteins or groups of proteins, require the use of statistical tests to assist in their discovery [Figure 1].[4]
Figure 1.
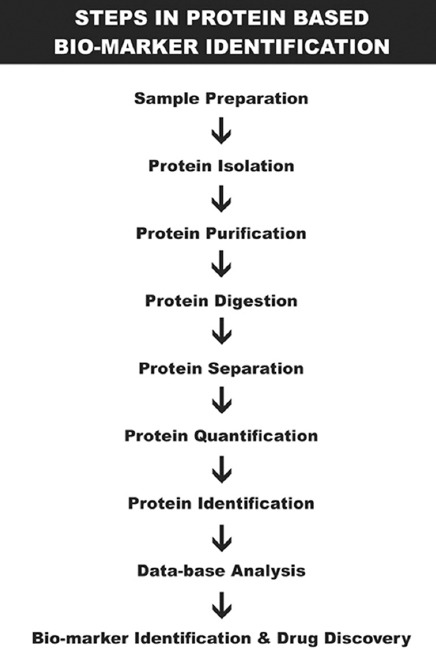
Biomarker identification
Applications of Proteomics in Human Disease, and Medicine
Proteins determine the structure and function of a cell and, thus, that of an organism. Proteins are subject to change depending on the cellular activities during the differentiation and developmental stages in the life of an organism. Proteins also change under diseased conditions as the cause or effect of the disease. Proteins also change in response to medications and other physical activities, and even in response to any other changes in the environment factors (diet, temperature, etc.).[19]
The link among genes, proteins, and diseases, as well as the possible intervention of a disease by medicine, was suggested by the one-gene–one-enzyme theory of Beadle and Tatum in 1941. This theory implied that a specific protein controlling the cellular structure or a metabolic reaction is either missing or defective in a person who has a hereditary disease. The first support to this view, hemoglobin protein alteration was found in persons suffering from sickle cell anemia was found in a demonstration. From then on, many human diseases were revealed to possess a particular defective protein or are missing that protein altogether. Soon, the one-gene–one-enzyme theory became the basis of several kinds of therapies, including the gene therapy. Gene therapy involves adding the accurate type of gene instead of the affected gene to a person with a disease because a defective gene produces defective proteins that cannot control a biochemical reaction. As many diseases were shown to be hereditary and many possess defective proteins, it has been shown that there are two classes of hereditary diseases, one in which one gene controls a particular disease and another in which several genes control a particular disease. These diseases are called monogenic and polygenic, respectively.[20]
An understanding of proteomics is essential for medicine because of the involvements of proteins. Particularly, proteins are the cause of diseases, and they can be used to diagnose prostate cancer; the level of prostate-specific antigen is a good biomarker of the disease. Proteins are used as drugs such as insulin or human growth hormone. Proteins such as protein kinase and human epidermal growth factor receptor 2 are targets for drugs such as Gleevec and Herceptin, which are used to treat cancer. The side-effects of the drug prescribed are recognized by the interactions between proteins and drugs. In view of these facts, proteomics has an indispensible role in medicine. Proteomics is simple in principle but is laborious in practice. It involves many steps, including the sample preparation, protein separation by 2D gel electrophoresis, a series of chromatography, and finally identification of the proteins by matrix-assisted laser desorption ionization-time of flight-mass spectrometry and by a comparison of protein sequence in the protein databank. This process requires concerted efforts by many investigators and a large amount of funding.[5]
Proteomics and Dentistry
Salivary diagnostics like oral fluid biomarkers and proteomics of bone and enamel structures (especially dental enamel) were the two primary areas, which dental proteomics have really made known. Human saliva contains proteins that can be a revealing tool for disease detection and surveillance of oral health. The discovery of salivary protein markers for human disease detection, in particular for oral cancer, Sjogren's syndrome and many other oral diseases were identified by complete analysis and documentation of the proteomic contents in human whole and ductal saliva.[21,22]
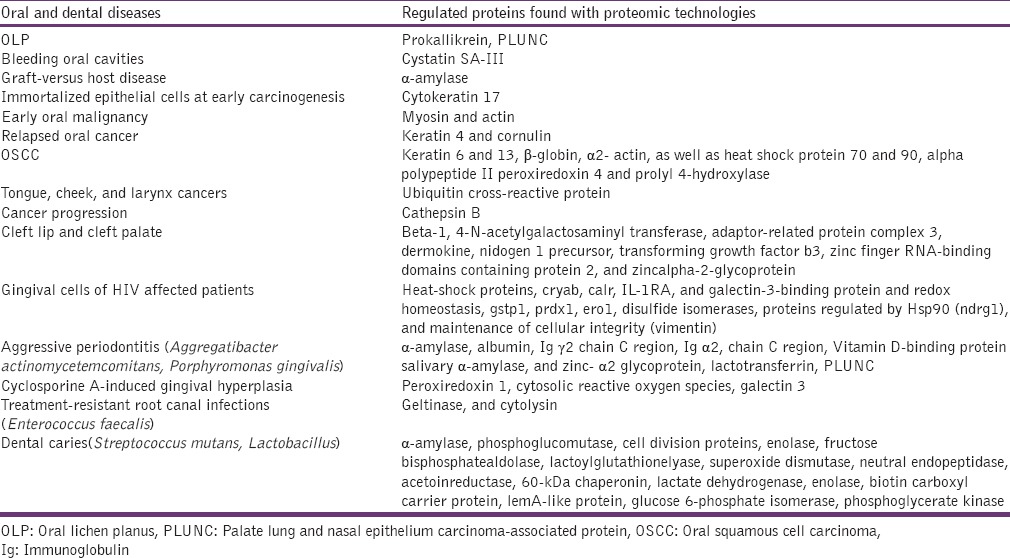
Proteomic studies related to dental structure formation (enamel, dentin, pulp, and periodontal ligament) and malady in different fields of dental pathology have exposed the proteins involved in the physiological function and altered proteins in dental diseases [Tables 1 and 2].[23]
Table 1.
Proteins expression identified with proteomic technique during tooth development[23]
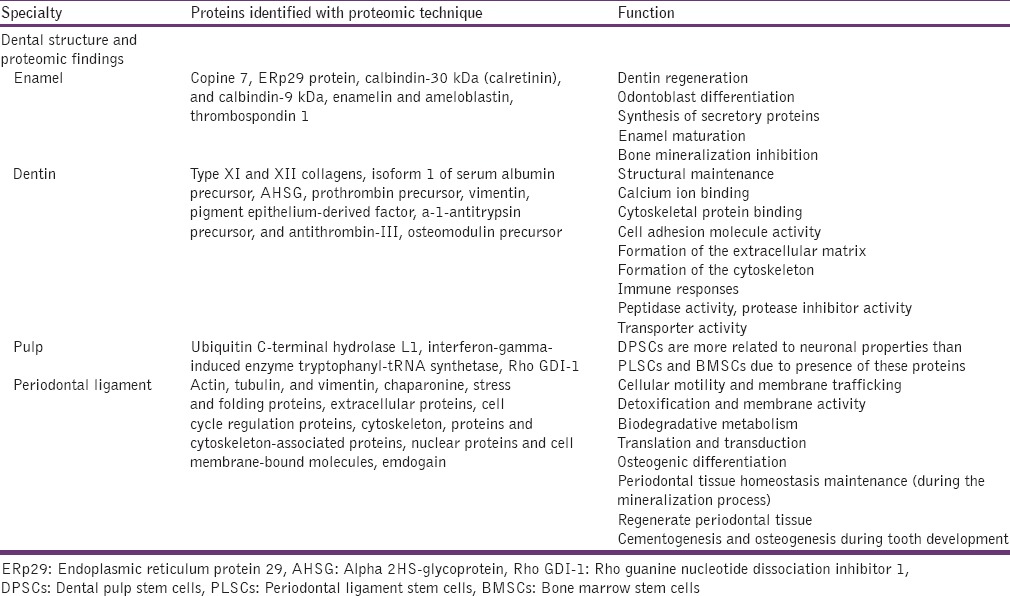
Table 2.
Altered proteins observed with various oral diseases[23]
Cancer Proteomics
Cancer is a manifestation of abnormal alteration of cellular protein molecular networks and cell signaling pathways that are underpinned by genetic changes. Cancer pathways contain various phases such as initiation, promotion, invasion, and metastasis. The major aim of proteomics has been to identify a protein or a group of proteins that can serve as a biomarker for an early detection of a specific cancer before its clinical manifestation, as evidenced by biopsy and/or histological analysis of the tissue involved in a particular kind of cancer. Surface enhanced laser capture micro-dissection (LCM) has been a new protein analysis system, recently applied for the detection, and analysis of multiple proteins in very small amounts of cancer tissue. This system facilitates protein isolation, purification, digestion, quantification, identification, analysis, and processing. LCM also aid in identifying the proteins expressed from squamous cell carcinoma tissue of the oral cavity. It will be analyzed using a high-throughput antibody microarray approach for a better understanding of how patterns of protein expression profile the tissue microenvironment. Correlation between the pattern of proteins expression within epithelial cells and the oral tumors was found with this procedure. Proteomic technologies have high possibility to greatly aid in the development of molecular diagnosis and serve as biomarkers for the early detection of oral cancer and also accelerate the rational drug designing according to the molecular profile of the cancer cell, anticancer drug target discovery to facilitate the development of personalized cancer therapy and execution.[23,24,25,26]
Conclusion
Numerous aspects like drug development, identifying new targets and facilitating assessment of drug action and toxicity both in the preclinical and clinical phases were enhanced with the help of proteomics.
Sequencing the human genome is simple and easy comparing to the framework of the proteome. Such a developing proteome framework will categorically help in obtaining:
The knowledge of all human proteins
Their arrangement, their posttranslational modifications, and their interactions within them
Their distribution in cells, and
Their sequential pattern of expression in healthy and diseased.
If such extensive framework of total proteome framework materialize in the future, more modest goals may well be within reach. In near future the advancement of the above mentioned technologies will likely expand significantly, in particular to meet the need for early detection of diseases, better diagnosis and the path of developing effective and precise therapy for the well-being the mankind.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Banks RE, Dunn MJ, Hochstrasser DF, Sanchez JC, Blackstock W, Pappin DJ, et al. Proteomics: New perspectives, new biomedical opportunities. Lancet. 2000;356:1749–56. doi: 10.1016/S0140-6736(00)03214-1. [DOI] [PubMed] [Google Scholar]
- 2.Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–46. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 3.Palzlill T. Proteomics. Boston: Kluwer Academic Publishers; 2002. pp. 1–4. [Google Scholar]
- 4.Chen R, Pan S, Crispin DA, Brentnall TA. Gene expression and proteomic analysis of pancreatic cancer: A recent update. Cancer Genomics Proteomics. 2006;3:1–10. [PubMed] [Google Scholar]
- 5.Mishra N. Hoboken, NJ, USA: John Wiley and Sons, Inc; 2010. Introduction to Proteomics - Principles and Application. [Google Scholar]
- 6.Boguski MS, McIntosh MW. Biomedical informatics for proteomics. Nature. 2003;422:233–7. doi: 10.1038/nature01515. [DOI] [PubMed] [Google Scholar]
- 7.Druery CT, Bateson W. Experiments in plant hybridization. J R Horticul Soc. 1901;26:1–32. [Google Scholar]
- 8.Beadle GW, Tatum EL. Genetic control of biochemical reactions in neurospora. Proc Natl Acad Sci U S A. 1941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944;79:137–58. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–8. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 11.Dove A. Breaching the barrier. Nat Biotechnol. 2008;26:1213–5. doi: 10.1038/nbt1108-1213. [DOI] [PubMed] [Google Scholar]
- 12.Sharp PA. The discovery of split genes and RNA splicing. Trends Biochem Sci. 2005;30:279–81. doi: 10.1016/j.tibs.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Calvo KR, Petricoin EF, 3rd, Liotta LA. Understanding cancer at the molecular level: An evolving frontier. J Am Chem Soc. 1969;91:501–2. [Google Scholar]
- 14.O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–21. [PMC free article] [PubMed] [Google Scholar]
- 15.Anson ML, Mirsky AE. On some general properties of proteins. J Gen Physiol. 1925;9:169–79. doi: 10.1085/jgp.9.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–30. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 17.Hunt SM, Thomas MR, Sebastian LT, Pedersen SK, Harcourt RL, Sloane AJ, et al. Optimal replication and the importance of experimental design for gel-based quantitative proteomics. J Proteome Res. 2005;4:809–19. doi: 10.1021/pr049758y. [DOI] [PubMed] [Google Scholar]
- 18.Wilkins MR. Renal and urinary proteomics: Current applications and challenges. Proteomics. 2005;5:1033–42. doi: 10.1002/pmic.200401012. [DOI] [PubMed] [Google Scholar]
- 19.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Eyk J, Dunn MJ. Clinical Proteomics. From Diagnosis to Therapy. Weinheim: Wiley-VCH Verlag GmbH and Co. KGaA; 2008. [Google Scholar]
- 21.Sah N, Bhutani H. Proteomics and periodontal diseases. Paripex Indian J Res. 2013;2:242–4. [Google Scholar]
- 22.Ahmed SM, Mubeen, Jigna VR. Molecular biology:An early detector of oral cancers. Ann Diagn Pathol. 2009;13:140–5. doi: 10.1016/j.anndiagpath.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Rezende TM, Lima SM, Petriz BA, Silva ON, Freire MS, Franco OL. Dentistry proteomics: From laboratory development to clinical practice. J Cell Physiol. 2013;228:2271–84. doi: 10.1002/jcp.24410. [DOI] [PubMed] [Google Scholar]
- 24.Chen R, Pan S, Brentnall TA, Aebersold R. Proteomic profiling of pancreatic cancer for biomarker discovery. Mol Cell Proteomics. 2005;4:523–33. doi: 10.1074/mcp.R500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Chen R, Yi EC, Donohoe S, Pan S, Eng J, Cooke K, et al. Pancreatic cancer proteome: The proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology. 2005;129:1187–97. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Chen R, Pan S, Crispin DA, Brentnall TA. Gene expression and proteomic analysis of pancreatic cancer: A recent update. Cancer Genomics Proteomics. 2006;3:1–10. [PubMed] [Google Scholar]


