Abstract
The Ras GTPase activating proteins (RasGAPs) are regulators of the conserved Ras/MAPK pathway. Various roles of some of the RasGAPs in learning and memory have been reported in different model systems, yet, there is no comprehensive study to characterize all gap genes in any organism. Here, using reverse genetics and neurobehavioural tests, we studied the role of all known genes of the rasgap family in C. elegans in associative learning and memory. We demonstrated that their proteins are implicated in different parts of the learning and memory processes. We show that gap-1 contribute redundantly with gap-3 to the chemosensation of volatile compounds, gap-1 plays a major role in associative learning, while gap-2 and gap-3 are predominantly required for short- and long-term associative memory. Our results also suggest that the C. elegans Ras orthologue let-60 is involved in multiple processes during learning and memory. Thus, we show that the different classes of RasGAP proteins are all involved in cognitive function and their complex interplay ensures the proper formation and storage of novel information in C. elegans.
The Ras family of small GTPase proteins plays a central role in many biological processes including cell proliferation, differentiation, cell migration, as well as in synaptic transmission and memory formation1,2,3,4. The activity of Ras is regulated by its guanine nucleotide-binding state. The active GTP-Ras slowly becomes inactive due to its intrinsic GTPase activity that hydrolyzes GTP to GDP. In addition, the activity of Ras is also regulated by two groups of proteins. Ras guanine exchange factors (GEFs) promote the dissociation of GDP from Ras and help the exchange of GDP to GTP, thus RasGEFs increase Ras activity. Contrarily, Ras GTPase activating proteins (RasGAPs) are negative regulators by enhancing the intrinsic GTPase activity of Ras. In contrast to vertebrates, the C. elegans genome encodes only three RasGAPs (gap-1, gap-2 and gap-3) each orthologous to respective major vertebrate subgroups5. gap-1, the representative of rasal subfamily, was originally described as a negative regulator of the LET-60/Ras pathway during vulval development5,6. gap-2, the homolog of the syngap subfamily, was shown to suppress the larval lethality of the let-23/EGF receptor and let-60 hypomorphic mutants7. Finally, gap-3 corresponding to the vertebrate rasa1 (also known as p120RasGAP), is a predominant negative regulator of let-60 signaling during germ cell development5. Interestingly, deletion of any single gap gene has no visible phenotype in C. elegans. In addition to the conservation of rasgap genes in nematodes, C. elegans also exhibits different forms of associative learning, and short-term and long-term memory8,9,10. Thus, C. elegans represents an ideal system to understand the role and possible interplay of the different rasgap genes during learning and memory.
In the current study, we investigated the roles of the gap gene family members in locomotion, olfaction, associative learning and memory formation. Our results indicate that gap-1 contributes to olfaction and associative learning; gap-2 is involved in short-term and long-term associative memory with a minor, redundant role in associative learning, while gap-3 plays a major role in olfaction, short- and long-term memory, and act redundantly with gap-2 during associative learning.
Results
C. elegans RasGAPs are dispensable for chemotaxis to olfactory cues
Aversive olfactory associative learning and memory rely on chemotaxis because environmental cues are acquired by olfaction. To test the role of the gap genes in these processes, we first analysed the chemotaxis towards three chemoattractants (diacetyl, benzaldehyde, and isoamyl-alcohol) of single gap mutant worms or of double mutant animals carrying mutations in gap genes in different combination. Chemotaxis assays were conducted with chemicals at two different concentrations in order to be able to identify even mild sensory deficits of the mutant strains. The volatile compounds used in the assay represent bacterial metabolic molecules that can be nutriments, therefore, induce attraction11.
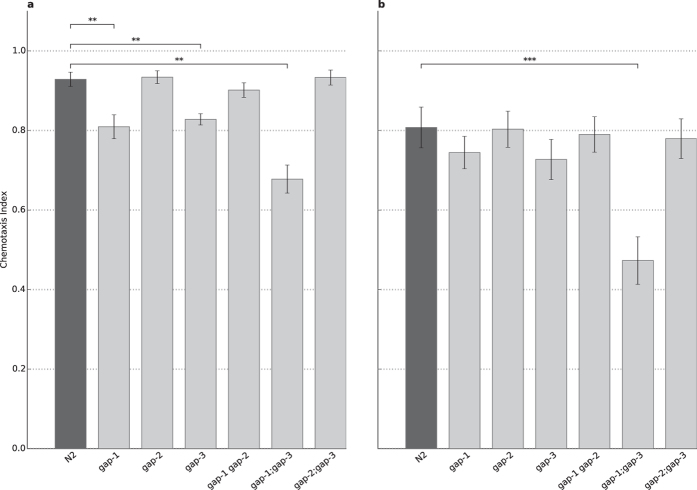
We found that all the single gap mutants as well as most of the double mutant worms demonstrated an attraction to olfactory cues similar to the wild type across the conditions used. The gap-2(tm748), gap-3(ga139), gap-1(ga133) gap-2(tm748) and gap-2(tm748);gap-3(ga139) mutants showed normal chemotaxis towards diacetyl, benzaldehyde and isoamyl-alcohol at both high and low concentrations (Fig. 1 and Suppl. Fig. 1). Interestingly, gap-1(ga133) mutants demonstrated decreased attraction towards high concentration of diacetyl (p = 7.88 × 10−3) and both concentrations of isoamylalcohol (p = 2.14 × 10−2 for high and p = 1.57 × 10−2 for low concentration) while chemotaxis to benzaldehyde remained unaffected (Suppl. Fig. 1). The gap-3(ga139) mutant had a decreased attraction towards high concentration of diacetyl (p = 2.45 × 10−3), which was not observable in any other condition. The gap-1(ga133);gap-3(ga139) mutant had a strong chemosensory defect towards all compounds tested (Fig. 1 and Suppl. Fig. 1).
Figure 1. Involvement of various RasGAP isoforms in chemotaxis towards diacetyl.
(A) Chemotaxis to 1:100 diluted diacetyl, attraction of N2 wild type (n = 33) and animals carrying the mutation(s) gap-1(ga133) (n = 9, p = 7.88 × 10−3), gap-2(tm748) (n = 18), gap-3(ga139) (n = 6, p = 2.45 × 10−3), gap-1(ga133) gap-2(tm748) (n = 9), gap-1(ga133);gap-3(ga139) (n = 7, p = 1,80 × 10-3) and gap-2(tm748);gap-3(ga139) (n = 15). (B) Chemotaxis to 1:1000 diluted diacetyl, attraction of N2 wild type (n = 31) and animals carrying the mutation(s) gap-1(ga133) (n = 31), gap-2(tm748) (n = 52), gap-3(ga139) (n = 31), gap-1(ga133) gap-2(tm748) (n = 24), gap-1(ga133);gap-3(ga139) (n = 29, p = 1.17 × 10-14) and gap-2(tm748);gap-3(ga139) (n = 36). Error bars indicate SD and asterisks indicate Bonferroni-corrected significant differences (**P < 0.01, ***P < 0.001).
In summary, strains carrying rasgap mutations all respond to chemotactile stimuli. GAP-1 is involved in the chemosensation of isoamyl-alcohol and high concentration of diacetyl, while GAP-3 plays a role in the olfaction of high concentration of diacetyl only. For both single mutants we observed a statistically significant but only marginal decrease in the odor response. Futhermore, none of the mutations caused significant changes in chemosensation to low concentrations of diacetyl, and therefore these do not interfere with negative olfactory associative learning and memory tests. The only exception was the gap-1(ga133);gap-3(ga139) mutant, which had a chemosensory defect to both concentrations of all tested attractants.
RasGAPs have no impact on locomotor behaviour and response to food
Similar to olfactory defects, impaired locomotion and lack of response to starvation could interfere with the assays addressing learning and memory functions. Therefore motility tests were conducted following three different feeding conditions to exclude locomotor or starvation response defects of the different rasgap single or double mutant strains.
We first tested the motility of well-fed worms on bacterial lawn to determine the baseline activity. Next we tested the food seeking behaviour by placing well-fed worms on empty plates. Finally, we investigated the starvation response of the different mutants by placing 1-hour-long starved worms on food source. No difference was observed in the mean body bends for fed (Fig. 2A) and starved (Fig. 2C) animals having gap mutations compared to the N2 reference strain. Food seeking animals showed similar behaviour except the gap-1(ga133);gap-3(ga139) mutant, which showed decreased motility (p = 2.54 × 10−12, Fig. 2B). A possible explanation of this difference is the deteriorated food searching motivation rooting in the chemosensory defect described above, because no locomotor defect was found in the ‘Fed’ and ‘Empty’ conditions.
Figure 2. Locomotion is not affected by gap mutations.
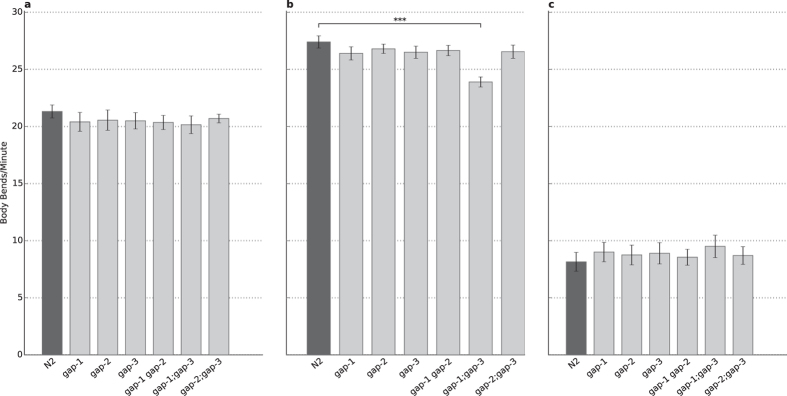
(A) Baseline feeding activity of gap mutant worms compared to the wild type N2. (B) Food searching activity of the N2 wild type and the gap mutants. The gap-1;gap-3 double mutant shows decreased locomotion (p = 3.62 × 10−13). (C) gap mutations do not affect the feeding activity of the worms after one hour of starvation. n = 20 for each strain in each condition, error bars indicate SD and asterisks indicate Bonferroni-corrected significant differences (***P < 0.001).
From these experiments we concluded that locomotion is intact for the gap-1(ga133), gap-2(tm748), gap-3(ga139), gap-1(ga133) gap-2(tm748), gap-2(tm748);gap-3(ga139) strains in any experimental condition. The gap-1(ga133);gap-3(ga139) strain was excluded from further testing due to its chemosensory and possible locomotory defect.
Complex regulation of learning and short-term associative memory by RasGAP proteins
In the previous experiments we tested all gap single and double mutants for defects that would interfere with negative olfactory associative learning. Since all gap single mutants and all gap double mutant worms (except the gap-1;gap-3 mutants) showed no or minor defect in chemotaxis or motility, next the role of different RasGAPs during negative olfactory learning and short-term memory (STAM) was investigated12. Naïve worms are attracted to diacetyl, which can be turned into aversion during a 1-hour conditioning period that associates starvation with the previously attractive diacetyl. This association deteriorates over time, therefore the effectiveness of conditioning itself informs about learning, while the amount of deterioration characterizes the memory function.
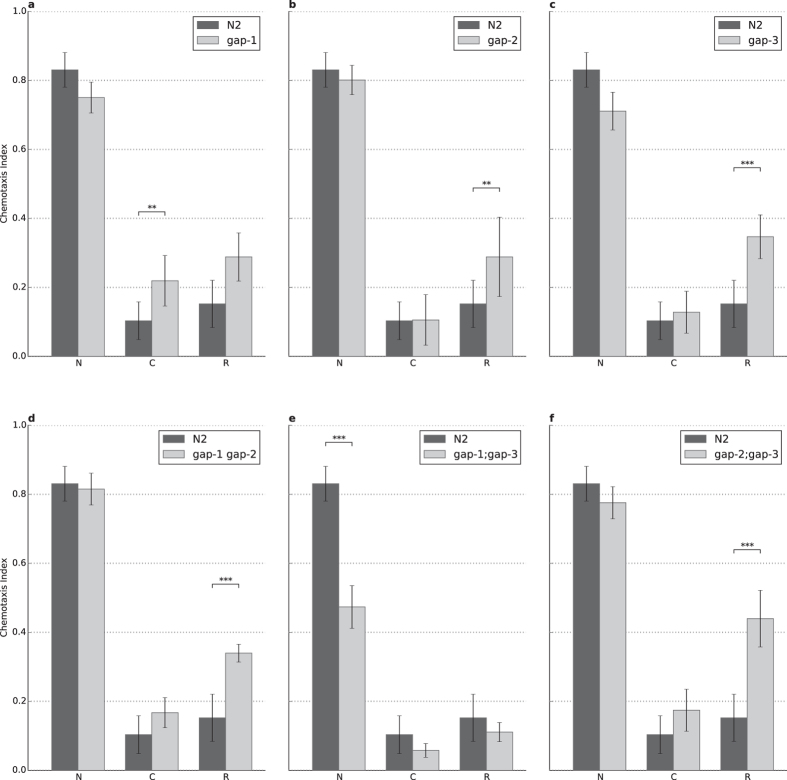
First, we tested all three single mutants and compared chemotaxis to diacetyl in naïve or conditioned worms. The combination of a 1-h starvation period in the presence of diacetyl (conditioning stimulus) dramatically reduced the attraction towards the chemoattractant in wild-type animals and in gap-2 and gap-3 single mutants to similar extent, but we found acquisition defects in the gap-1(ga133) (p = 0.00139) worms (Fig. 3 and Table 1). gap-1 mutant worms also showed a significant memory defect when compared to wild types (p = 9.7 × 10−6), however, this may be due to the learning defect. To address that, we analyzed the rate of memory loss by comparing the difference between conditioned and 30 min delay attraction (also called recovery phase) to diacetyl in wild type and gap-1 mutant worms and we found a statistically not significant difference between the genotypes. Thus, the memory difference was likely a consequence of the acquisition defect and this suggests that gap-1 is playing a role in the learning process. In the case of the other two gap mutants, we observed a defect in both gap-2 and gap-3 only in memory (Fig. 3) and this suggests that gap-2 and gap-3 are both regulating memory consolidation.
Figure 3. A complex interplay of RasGAPs is involved in associative learning and short-term memory.
(A) gap-1 mutation leads to a defect in learning (n = 23, p = 6.18 × 10−3) without short-term memory being affected. (B–D,F) Strong defect in short-term memory was observable for the (B) gap-2 (n = 33, p = 1.34 × 10−2), (C) gap-3 (n = 23, p = 2.75 × 10−7), (D) gap-1 gap-2 (n = 15, p = 3.14 × 10−12) and (F) gap-2;gap-3 (n = 21, p = 3.45 × 10−7) mutant animals without significant defect in learning. (E) The gap-1;gap-3 double mutant could not be assessed due to its chemosensory defect (n = 24, p = 8.02 × 10−13). N: naïve, C: conditioned, R: recovered animals (see Materials and Methods for details). Error bars indicate SD and asterisks indicate significant differences (**P < 0.01, ***P < 0.001).
Table 1. Phenotypic features of gap-1(ga133), gap-2(tm748), gap-3(ga139), gap-1(ga133) gap-2(tm748), gap-1(ga133);gap-3(ga139), gap-2(tm748);gap-3(ga139) strains, respectively, regarding chemosensation of diacetyl, motility, learning, short-term associative memory (STAM) and long-term associative memory (LTAM).
| Chemosensation | Motility | Learning | STAM | LTAM | |
|---|---|---|---|---|---|
| gap-1(ga133) | Intact | Intact | Defect | Intact | Intact |
| gap-2(tm748) | Intact | Intact | Intact | Defect | Defect |
| gap-3(ga139) | Intact | Intact | Intact | Defect | Defect |
| gap-1(ga133) gap-2(tm748) | Intact | Intact | Intact | Defect | Defect |
| gap-1(ga133);gap-3(ga139) | Defect | Intact* | N/A | N/A | N/A |
| gap-2(tm748);gap-3(ga-139) | Intact | Intact | Defect | Defect | Defect |
Defect = the strain behaves significantly different compared to the N2 wild-type reference. Intact = no significant difference. N/A: not assessed.
*The baseline fed and feeding locomotory rate of the gap-1(ga133); gap-3(ga139) line are intact. The difference of the food searching activity is probably due to a chemosensory defect.
Besides the single mutants we also tested the effect of simultaneous removal of multiple rasgaps. We found that the gap-1(ga133) gap-2(tm748) (p = 0.0016) and the gap-2(tm748);gap-3(ga139) double mutants (p = 1.5 × 10−7) displayed specific impairment of short-term memory (Fig. 3). Finally, the gap-1(ga133);gap-3(ga139) double mutant displayed a strong chemotaxis defect as described earlier and the strain could not be assessed further due to this sensory deficit (Fig. 3).
In order to exclude possible background mutations that could contribute to the observed phenotypes, we performed RNAi silencing of the different rasgaps in RNAi hypersensitive eri-1(mg366);lin-15B(n744) strain. As control, eri-1(mg366);lin-15B(n744) was fed with bacteria expressing GFP dsRNA (Fig. 4). Double stranded RNA targeting gap-2 and gap-3 mRNA caused a phenotype matching the gap-2 and gap-3 loss-of-function mutants, respectively (Fig. 4). In case of gap-1 RNAi we did not observe this effect, probably due to insufficient RNAi silencing. Thus, we performed a rescue experiment by reintroducing the gap-1 genomic fragment in gap-1(ga133) mutant worms and negative STAM was assessed in three parallel lines (see Materials and Methods for details). Genomic fragment encompassing the gap-1 locus restored learning and memory to levels comparable to wild type in all three lines (Fig. 5). Thus, the observed phenotypes are attributable to the loss-of-function of various rasgap genes.
Figure 4. RNAi silencing of GAP-2 and GAP-3 phenocopy the mutant phenotypes.
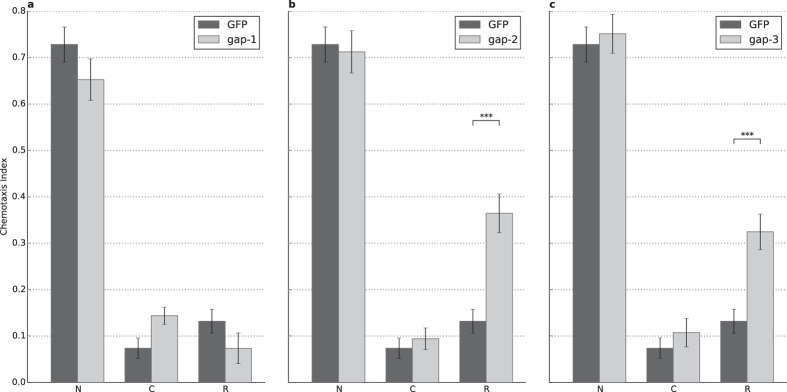
Negative conditioning assays were combined with RNA interference experiments using eri-1(mg366);lin-15B(n744) RNA sensitized worms, fed against (A) gap-1 (n = 3), (B) gap-2 (n = 13, p = 2.72 × 10−7), and (C) gap-3 (n = 9, p = 3.75 × 10−5) dsRNA carrying bacteria. Dark grey represents the same strain fed with bacteria carrying an empty GFP marker dsRNA as reference. N: naïve, C: conditioned, R: recovered animals (see Materials and Methods for details). Error bars indicate SD and asterisks indicate significant differences (***P < 0.001).
Figure 5. Re-introduction of gap-1 gene rescues the loss-of-function gap-1 phenotype.
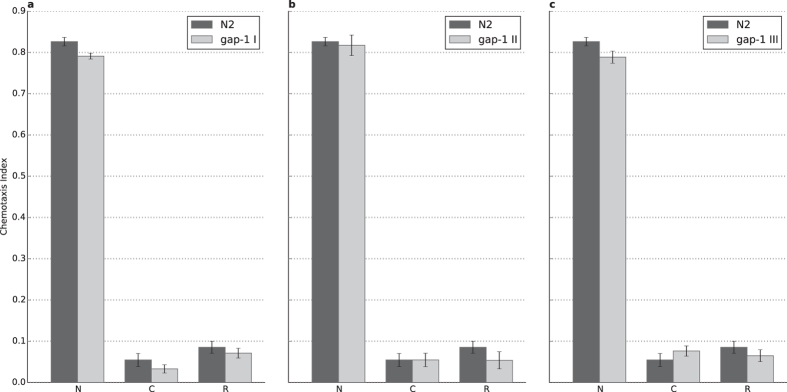
(A–C) Negative conditioning assays were performed with three independent gap-1 rescue lines. Light grey bars represent the rescue lines and dark grey bars represent the reference N2 strain. n = 6 for all lines in each condition. N: naïve, C: conditioned, R: recovered animals (see Materials and Methods for details). Error bars indicate SD.
Interplay of RasGAP proteins during long-term associative memory (LTAM)
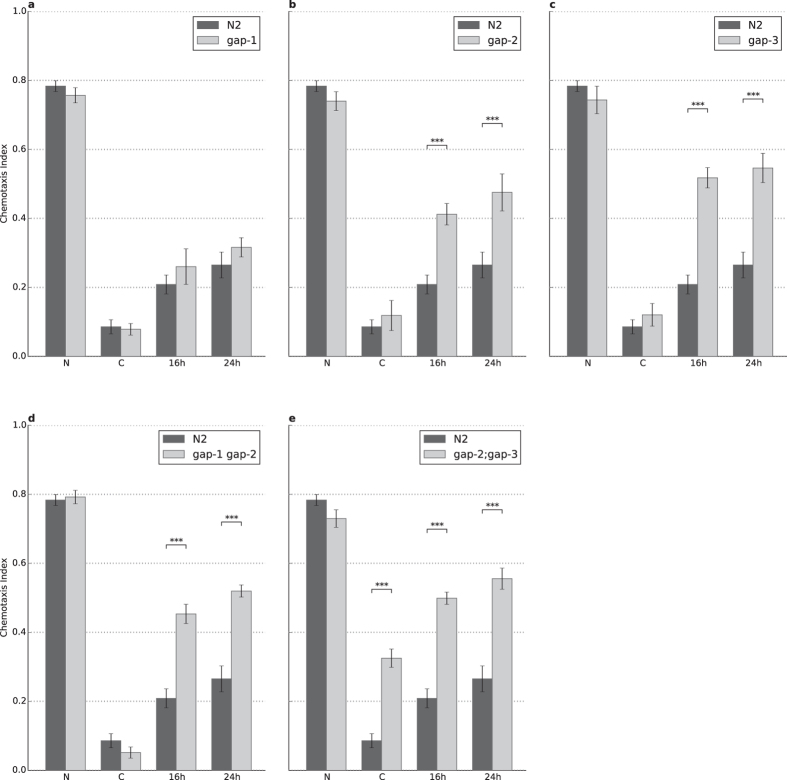
Beside the role of RasGAPs in short-term memory, we also tested the impact of the different C. elegans rasgaps in long-term associative memory. LTAM was assessed by testing worms 16 and 24 hours after negative olfactory conditioning, as described previously10. In this experimental setup we used a reinforcement training consisting of three rounds of conditioning cycles. Briefly, starvation and diacetyl were associated for 1 hour, which turned the attraction towards diacetyl into aversion. Each conditioning step was followed by a feeding phase without diacetyl for 30 minutes to let the worms regenerate. Such rounds of conditioning and feeding were repeated 3 times. Using this training we could not detect a significant difference in acquisition in most gap mutant genotypes. A possible explanation of this phenomenon is that the naïve animals were conditioned 3 times in these assays compared to the single conditioning in assays testing STAM. Interestingly, the learning defect was still present in the gap-2(tm748);gap-3(ga139) double mutant strain (p = 1.24 × 10−7).
Strikingly, gap-1(ga133) did not show any impairment in learning or in long-term memory while all other two single mutants had a specific memory defect similar to their short-term associative memory phenotype (Fig. 6, Table 1). As shown in Fig. 6, remaining two double mutants, gap-1(ga133) gap-2(tm748) (p = 6.75 × 10−7) and gap-2(tm748);gap-3(ga139) (p = 2.00 × 10−6) had similarly strong long-term associative memory defects. In the gap-1(ga133);gap-3(ga139) double mutant strain, the chemotaxis defect prevented any assessment of learning and memory formation. Altogether, the rasgap genes are redundantly regulating long-term memory in nematodes.
Figure 6. RasGAPs are involved in long-term associative memory.
(A) gap-1(ga133) mutants show no significant defect in learning or in long-term associative memory (n = 5). (B) gap-2(tm748) (n = 12, p16h = 5.61 × 10−7, p24h = 6.89 × 10−5), (C) gap-3(ga139) (n = 15, p16h = 9.33 × 10−15, p24h = 1.67 × 10−10), (D) gap-1(ga133) gap-2(tm748) (n = 6, p16h = 2.53 × 10−4, p24h = 5.18 × 10−8) mutants all have long-term associative memory defect together with (E) gap-2(tm748);gap-3(ga139) (n = 9, p16h = 3.57 × 10−13, p24h = 7.93 × 10−7) mutants, which also display learning defect (p = 8.70 × 10−7). The N2 wild type represents the reference on all charts. Naïve (N) animals were conditioned (C), then tested after 0.5 hour recovery (R), and 16 hours (16h) and 24 hours (24h) after conditioning (see Materials and Methods for details). Error bars indicate SD and asterisks indicate significant differences (***P < 0.001).
let-60 is involved in the gap–related learning and memory phenotypes
RasGAPs are known to increase the intrinsic GTPase activity of LET-60 and by that they regulate different biological processes in C. elegans13. Based on these earlier observations we hypothesized that gap mutations combined with a reduction of function let-60 mutation would result in a learning and memory phenotype similar to the wild type. The let-60 mutation itself causes a strong chemosensory defect14, which was in part restored in all gap;let-60 double mutant. Furthermore, in agreement with our assumptions, we found that the let-60(n2021) hypomorph mutation restored the learning and memory defects observed in the rasgap mutants (Fig. 7). This suggests that all three rasgaps act at least in part through let-60 to regulate learning and memory in C. elegans.
Figure 7. let-60 is required for the gap(lf) learning and memory phenotypes.
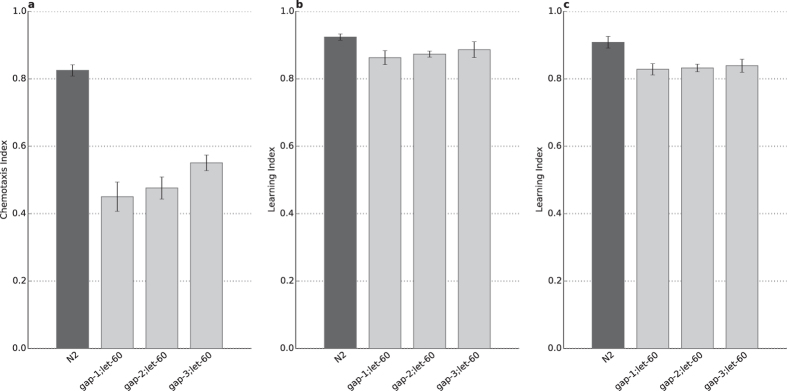
. Naïve gap(lf);let-60(n2021hf) double mutants (A) were conditioned to assess learning (B) and short term associative memory (C). Naïve mutants are characterized by lowered chemotaxis index, e.g. chemosensory defect due to the let-60(n2021hf) mutation. Conditioned worms have no significant learning defect (B) and recovery phase has not revealed significant memory defect either (C). Both conditioned and recovery phases were assessed by calculating learning indices (LI = [CIconditioned – CInaïve]/CInaïve) to ensure comparability with the N2 wild type.
Discussion
Here, we investigated the role of each member of the RasGAP family in the learning and memory process in C. elegans. We found that all RasGAP forms are regulating cognitive functions, and strikingly, they exhibit specific roles during learning and memory. While the exact molecular machinery connecting RasGAPs to learning and memory formation in C. elegans is unclear yet, our results show that RasGAPs are likely involved in multiple distinct processes during acquisition and storage of new informations.
In C. elegans, synaptic cytoskeletal reorganization is widely accepted as a form of synaptic plasticity, and was suggested recently as a molecular process of forgetting15. Fast activation and inactivation of Ras is important for the olfactory behaviour16, although the negative feedback loop for inactivation has not been elucidated yet.
In humans, analysis of molecular signaling networks revealed potential cross-talks between the Ras/MAPK pathway and the possible cytoskeletal rearrangments17, which is further supported by the earlier finding that RasGAPs control Rho-mediated cytoskeletal reorganization18. In rat hippocampal neurons, synaptic Ras GTPase activating protein (SynGAP), an orthologue of C. elegans GAP-2, can be phosphorylated by the calcium/calmodulin-dependent protein kinase II (CaMKII) upon long-term potentiation induction. This leads to the dispersion of SynGAP from the dendritic spines19. Ras family proteins, the major effectors downstream of GAPs, have also been associated with Ca2 + -dependent synaptic crosstalk after NMDA receptor activation in rat hippocampal slices20. Neurofibromin 1 (NF1), a GAP orthologue not found in C. elegans yet, can also inactivate Ras in rat hippocampal dendritic spines21. In mouse model systems, homozygous knock-out of syngap leads to postnatal lethality, while heterozygous mice exhibit specific defects in hippocampal long-term potentiation and glutamate receptor trafficking, although the molecular relation between these two processes is difficult to assess at phenotypic level22. In a mouse schizophrenia model, reduced expression of SynGAP leads to non-habituating mice showing persistent hyperactivity, lack of social memory, impaired fear conditioning and working memory, probably due to defected interaction between SynGAP and NMDA receptor23. Neurofibromin 1 also regulates GABA release, long-term potentiation and learning in mice24. Furthermore, in zebrafish, loss of Neurofibromin 1 results in learning and memory defects25. Altogether, these findings suggest a) a conserved role for different RasGAPs in learning and memory, b) a significant role for SynGAP in signal transmission during learning and memory formation. In addition to the NF1 and SynGAPs we show here that RASAL and p120RasGAP subfamily members are also regulating learning and memory and that different RasGAP subclasses are redundantly modulating different parts of the acquisition and storage processes.
Furthermore, our results are in good agreement with clinical findings. Rasopathies are human pathological conditions associated with germline mutations of the Ras/MAPK pathway26,27. The overlapping phenotypic features are developmental and cutaneous abnormalities, predisposition to malignancies and varying degree of neurocognitive impairment including learning disability. Rasopathies involving RasGAPs are neurofibromatosis type 1 (NF1, also known as von Recklinghausen disease)28,29 and capillary malformation – arteriovenous malformation syndrome (CM–AVM)30, caused by germline mutations in the nf1 and rasa1 genes, respectively. The exact molecular mechanisms underneath the symptoms are poorly understood yet.
In conclusion, we characterized all three known genes of the gap gene family in C. elegans to show that gap genes are involved in olfaction, associative learning, short- and long-term memory formation. Our results are in good agreement with the characteristics known partially from other model systems, and are in accordance with the clinical features of Rasopathies, further supporting that the molecular mechanisms of learning, memory and the role of the Ras/MAPK pathway in these are well conserved. This work also opens perspectives for neurocognitive and neurobehavioural studies of RasGAPs in C. elegans.
Materials and Methods
The reagents were obtained from Sigma (Sigma-Aldrich, St Louis, MO) unless otherwise indicated. Standard methods were used for maintaining and manipulating C. elegans strains31. Worms were grown at 20 °C on E. coli OP50 bacteria. Behavioural tests were performed with well fed, 1-day-old adult worms. The C. elegans Bristol strain, variety N2 was used as wild type reference otherwise indicated. C. elegans strains used were: gap-1(ga133), gap-2(tm748), gap-3(ga139), gap-1(ga133) gap-2(tm748), gap-1(ga133);gap-3(ga139), gap-2(tm748);gap-3(ga139), let-60(n2021), and eri-1(mg366);lin-15B(n744).
For the gap-1 rescue, the fosmid clone WRM0629aG09 was digested with AvrII/SbfI restriction endonucleases and the 9.5 kb fragment encompassing the gap-1 gene was microinjected at a concentration of 100 ng/μl into both arms of the syncytial gonads of gap-1(ga133) worms as described earlier32. Sur-5::dsRed at 10 ng/μl concentration was coinjected as transformation marker.
RNA interference (RNAi)
RNAi was performed with eri-1(mg366);lin-15B(n744) worms as previously described33. Animals in L3 stage were placed for 1 day on NGM plates containing 5 mM Isopropyl-D-thiogalactopyranoside (IPTG) and HT115(DE3) E. coli strain producing dsRNA against gap-1, gap-2 and gap-3, respectively, as described34,35. Bacteria containing vector with GFP were used as control in all RNA interference experiments.
Motility assay
Motility was characterized by the number of body bends per minute as described earlier36,37. Briefly, well fed single young adult worms were transferred onto seeded NGM plates (baseline activity), onto empty plates (food searching activity) or animals were starved for 1 hour and transferred to seeded NGM plates (feeding activity). Body bends were counted for 1 minute after a 2-minute resting phase. At least 20 animals were recorded per condition and per genotype.
C. elegans behavior assays
Chemotaxis to different compounds was assessed as described previously38. Briefly, worms were washed three times in CTX buffer (5 mM KH2PO4/K2HPO4 pH 6.0, 1 mM CaCl2, 1 mM MgSO4) and were allowed to settle down by gravity. 50 to 200 worms were placed to the middle of 10 cm CTX test plates (5 mM KH2PO4/K2HPO4 pH 6.0, 1 mM CaCl2, 1 mM MgSO4, 2% agar). Worms were given a choice between a spot of attractant (diacetyl, benzaldehyde and isoamylalcohol) in ethanol at the indicated dilutions with 20 mM sodium-azide and a counter spot with ethanol and sodium-azide. The distribution of the worms over the plate was determined after 1 hour and the chemotaxis index was calculated as described earlier38.
Negative olfactory associative conditioning was performed with modifications of the original protocol12. For the conditioning, 1 hour long starvation was coupled with 2 μl of undiluted diacetyl dropped on the lid of 10 cm CTX plates. A subpopulation of worms was washed for half an hour in CTX buffer after conditioning to allow recovery. Chemotaxis of naïve, conditioned and recovery worms were tested with diluted diacetyl (1:1000) as described above.
Long-term associative memory assays were performed as described earlier10. Briefly, worms were conditioned three times by repeating the cycles of conditioning described above and by allowing the worms to regenerate for 30 mins in presence of food after each conditioning. Memory function was assessed after 16 and 24 hours as described for the negative olfactory associative conditioning.
Learning index was calculated as the difference of chemotaxis indices of conditioned and naïve worms normalized by the chemotaxis index of naïve animals:
 |
Computational tools and statistics
The scripting language Python 3.4, the numerical library numpy 1.8 and the plotting library matplotlib 1.4.1 were used to analyze and visualize the experimental results. Welch’s test39, as implemented in the statistical module of SciPy 0.13.3, was used to calculate statistical significances. P-values always refer to results of two-tailed tests, in multiple comparisons p-values are always Bonferroni corrected; *p ≤ 0.05, **p < 0.01, ***p < 0.001. Two-way ANOVA with Bonferroni corrected post-hoc t-test was used for interaction analysis of learning and memory assays, these tests were performed with the R project (http://www.r-project.org/). Error bars represent standard deviation. All computational tools are open source and were designed and implemented by experts to follow the best practices and to ensure scientific reproducibility.
Additional Information
How to cite this article: Gyurkó, M. D. et al. Distinct roles of the RasGAP family proteins in C. elegans associative learning and memory. Sci. Rep. 5, 15084; doi: 10.1038/srep15084 (2015).
Supplementary Material
Acknowledgments
The current work was supported by two research grants from the Hungarian National Science Foundation (OTKA-K83314 and OTKA-K115378) to P.C., (OTKA K116525) to C.S., by the Forschungsfonds of the University of Basel (DPE2112), and the Swiss National Foundation (SNF) (NPE1581) to A.S., and by a fellowship from the Gedeon Richter Centennial Foundation to M.D.G. C.S. acknowledge support from the Merit Prize of the Semmelweis University.
Footnotes
Author Contributions Experiments were designed by all authors. The assays were conducted by M.D.G., molecular biological work was done by M.D.G. and A.S. Equipment, strains and materials were provided by P.C., S.C. and A.S. The main manuscript text and the figures were prepared by M.D.G. and A.S. All authors reviewed the manuscript.
References
- Brambilla R., Gnesutta N. & Minichiello L. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature 390, 281–286 (1997). [DOI] [PubMed] [Google Scholar]
- Sternberg P. W. P. & Han M. Genetics of RAS signaling in C. elegans. Trends Genet. 14, 466–472 (1998). [DOI] [PubMed] [Google Scholar]
- Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta - Rev. Cancer 1603, 47–82 (2003). [DOI] [PubMed] [Google Scholar]
- Ye X. & Carew T. J. Small G Protein Signaling in Neuronal Plasticity and Memory Formation: The Specific Role of Ras Family Proteins. Neuron 68, 340–361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetak A., Gutierrez P. & Hajnal A. Tissue-specific functions of the Caenorhabditis elegans p120 Ras GTPase activating protein GAP-3. Dev. Biol. 323, 166–176 (2008). [DOI] [PubMed] [Google Scholar]
- Hajnal A., Whitfield C. & Kim S. Inhibition of Caenorhabditis elegans vulval induction bygap-1 and by let-23 receptor tyrosine kinase. Genes Dev. 2, 2715–2728 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashizaki S., Iino Y. & Yamamoto M. Characterization of the C. elegans gap-2 gene encoding a novel Ras-GTPase activating protein and its possible role in larval development. Genes Cells 3, 189–202 (1998). [DOI] [PubMed] [Google Scholar]
- Ardiel E. L. & Rankin C. H. An elegant mind: learning and memory in Caenorhabditis elegans. Learn. Mem. 17, 191–201 (2010). [DOI] [PubMed] [Google Scholar]
- Kauffman A., Parsons L., Stein G., Wills A., Kaletsky R. & Murphy C. C. Elegans positive butanone learning, short-term, and long-term associative memory assays. J. Vis. Exp. 49, 2490 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukojevic V., Gschwind L., Vogler C., Demougin P., de Quervain D. J.-F., Papassotiropoulos A. & Stetak A. A role for α-adducin (ADD-1) in nematode and human memory. EMBO J. 31, 1453–66 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I. Chemosensation in C. elegans. WormBook 1–19 (2006). 10.1895/wormbook.1.123.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttley W. M., Atkinson-Leadbeater K. P. & Van Der Kooy D. Serotonin mediates food-odor associative learning in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 99, 12449–12454 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M. V. RTK/Ras/MAPK signaling. WormBook 1–19 (2006). 10.1895/wormbook.1.80.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu T., Saeki S., Yamamoto M. & Iino Y. The Ras-MAPK pathway is important for olfaction in Caenorhabditis elegans. Nature 404, 289–293 (2000). [DOI] [PubMed] [Google Scholar]
- Hadziselimovic N., Vukojevic V., Peter F., Milnik A., Fastenrach M., Fenyves B., Hieber P., Demougin P., Vogler C., de Quervain D. J.-F., Papassotiropoulos A. & Attila S. Forgetting in C. elegans is regulated via Musashi (msi-1) mediated translational control of the Arp2/3 complex. Cell 156, 1153–1166 (2014). [DOI] [PubMed] [Google Scholar]
- Uozumi T., Hirotsu T., Yoshida K., Yamada R., Suzuki A., Taniguchi G., Iino Y. & Ishihara T. Temporally-regulated quick activation and inactivation of Ras is important for olfactory behaviour. Sci. Rep. 2, 500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurkó M. D., Steták A., Sőti C. & Csermely P. Multitarget Network Strategies to Influence Memory and Forgetting: The Ras/Mapk Pathway as a Novel Option. Mini-Reviews Med. Chem. 15, 696–704 (2015). [DOI] [PubMed] [Google Scholar]
- LeBlanc V., Tocque B. & Delumeau I. Ras-GAP controls Rho-mediated cytoskeletal reorganization through its SH3 domain. Mol. Cell. Biol. 18, 5567–78 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y., Zeng M., Zhang M. & Huganir R. L. Rapid Dispersion of SynGAP from Synaptic Spines Triggers AMPA Receptor Insertion and Spine Enlargement during LTP. Neuron 85, 173–189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey C. D., Yasuda R., Zhong H. & Svoboda K. The spread of Ras activity triggered by activation of a single dendritic spine. Science 321, 136–140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A. F. & Yasuda R. Neurofibromin is the major ras inactivator in dendritic spines. J. Neurosci. 34, 776–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Lee H.-K., Takamiya K. & Huganir R. L. The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity. J. Neurosci. 23, 1119–1124 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H. F., Tong J., Hannan F., Luo L. & Zhong Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature 403, 895–898 (2000). [DOI] [PubMed] [Google Scholar]
- Cui Y., Costa R. M., Murphy G. G., Elgersma Y., Zhu Y., Guttmann D. H. et al. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell 135, 549–60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolman M., de Groh E., McBride S., Jongens T., Granato M. & Epstein J. Modulation of cAMP and ras signaling pathways improves distinct behavioral deficits in a zebrafish model of neurofibromatosis type 1. Cell Rep. 8, 1265–70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidyman W. E. & Rauen K. A. The RASopathies: Developmental syndromes of Ras/MAPK pathway dysregulation. Nat. Genet. 19, 230–236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauen K. The RASopathies. Annu. Rev. Genomics Hum. Genet. 14, 355–369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace M. R., Marchuk D., Andersen L., Letcher R., Odeh H. M., Saulino M. et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science 249, 181–186 (1990). [DOI] [PubMed] [Google Scholar]
- Williams V., Lucas J. & Babcock M. Neurofibromatosis type 1 revisited. Pediatrics 123, 124–133 (2009). [DOI] [PubMed] [Google Scholar]
- Eerola I., Boon L. M., Mulliken J. B., Burrows P. E., Dompmartin A., Watanabe S. et al. Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am. J. Hum. Genet. 73, 1240–1249 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D. & Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Costas S. A., Driver S. E. & Mello C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser G. & Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Kennedy S., Conte D. & Kim J. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436, 593–597 (2005). [DOI] [PubMed] [Google Scholar]
- Sawin E. R., Ranganathan R. & Horvitz H. R. C. Elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26, 619–31 (2000). [DOI] [PubMed] [Google Scholar]
- Mohri A., Kodama E., Kimura K., Koike M., Mizuno T. & Mori I. Genetic control of temperature preference in the nematode Caenorhabditis elegans. Genetics 169, 1437–50 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I., Hartwieg E. & Horvitz H. R. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74, 515–527 (1993). [DOI] [PubMed] [Google Scholar]
- Welch B. L. The Generalization of ‘Student’s’ Problem When Several Different Population Variances Are Involved. Biometrika 34, 28–35 (1947). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.