Abstract
Shiga toxins (Stx) are commonly produced by Shigella dysenteriae serotype 1 and Stx-producing Escherichia coli. However, the toxin genes have been detected in additional Shigella species. We recently reported the emergence of Stx-producing Shigella in travelers in the United States and France who had recently visited Hispaniola (Haiti and the Dominican Republic). In this study, we confirm this epidemiological link by identifying Stx-producing Shigella from Haitian patients attending clinics near Port-au-Prince. We also demonstrate that the bacteriophage encoding Stx is capable of dissemination to stx-negative Shigella species found in Haiti, suggesting that Stx-producing Shigella may become more widespread within that region.
Keywords: bacteriophage, Haiti, Shiga toxin, Shigella
Shigella species are the causative agent of an inflammatory diarrhea called bacillary dysentery or shigellosis. Shigella dysenteriae serotype 1 is the only one of the 4 Shigella species that has been recognized as producing Shiga toxins (Stx), which are potent cytotoxins that produce hemorrhagic colitis that can lead to the serious hemolytic uremic syndrome (HUS) [1]. In S dysenteriae 1, the genes encoding stx are found on the S dysenteriae 1 chromosome in an operon consisting of stxA and stxB and are flanked by DNA sequence homologous to lambdoid bacteriophages; however, the toxin genes are not associated with a complete prophage genome [2, 3]. In contrast, in Stx-producing Escherichia coli (STEC), stx is commonly carried by intact lambdoid prophages, which allow for horizontal spread of the toxin genes [4].
In recent studies, stx-encoding prophages have been identified in non-S dysenteriae 1 Shigella species [5–7]. In 2 previous reports, we characterized a new stx-encoding phage, φPOC-J13, in Shigella isolated from travelers returning to the United States and France from Haiti or the Dominican Republic [8, 9]. Those findings suggested that emergence of stx-positive Shigella is linked to that region. In this study, we confirmed that Stx-producing Shigella are circulating in Haiti: stools from Haitian patients were screened for Shigella and the presence of stx and the stx-encoding lambdoid phage. In addition, we demonstrate that φPOC-J13 is capable of lysogenizing stx-negative Shigella species isolated from Haitian patients, indicating that this stx-encoding bacteriophage has the ability to infect other Shigella strains circulating within Haiti.
METHODS
Isolation of Clinical Samples and Growth Conditions
Shigella species were isolated from stools as described by Livio et al [10]. After isolation, Shigella strains were grown in Tryptic Soy Broth (TSB) (BD Difco, Franklin Lakes, NJ) at 37°C with aeration or on TSB plates containing 1.5% agar (TSB agar) with or without 0.025% Congo red (Sigma-Aldrich, St. Louis, MO). Escherichia coli K-12 strain MG1655 was grown in Luria-Bertani (LB) broth and on LB agar plates.
Polymerase Chain Reaction Analysis of Shigella Clinical Isolates
Cell lysates were used for polymerase chain reaction (PCR) to detect stx with primers stx1-det-F1 and stx1-seq-R1 [9]. The stx-encoding isolates were analyzed by PCR to show that stx was phage encoded using primers Stx1R2/Phage_stxR2 and Phage_stx1F2/Stx1F2 [9]. The insertion site of the phage into locus S1742 or a homologous gene was determined by PCR using primers to the region upstream of S1742 and an early phage gene (primers S1742_up/Stx_phage_up) and by amplifying a late phage gene and the region downstream of S1742 with primers S1742_dn/Stx_phage_dn [9].
Toxin and Phage Production
Production and release of Stx was measured using bacterial lysates and supernatants in a Vero cell cytotoxicity assay as previously described [9]. Production of viable phage progeny was determined in a plaque-forming assay by absorbing overnight supernatants from the stx-encoding isolates onto E coli MG1655 as previously described [9].
Construction of φPOC-J13Δstx
The stx operon in φPOC-J13 was replaced with a kanamycin resistance cassette using lambda red recombination in a laboratory strain of Shigella flexneri, 2457T, lysogenized with φPOC-J13 and transformed with pKM208 [11]. Primers Stxko-site1 (5-aaaataattatttttagagtgctaacttttttctttgttatcttttcagttgtgtaggctggagctgcttc-3) and Stxko-site2 (5-acgaaaaataacttcgctgaatccccctccattatgacaggcattagttttcatatgaatatcctcctta-3) were used to amplify kan from pKD4 with 5′ and 3′ overhangs homologous to internal regions of the stx operon. Kanamycin-resistant recombinants were double purified and screened by PCR with primers Stxup (5-ccgtaaaacgccgtccttca-3) and Stxdn (5-catttatgaatctccgcctgc-3). A P1L4 lysate was grown on this mutant and used to transduce the stx::kan mutation into BS937, an S flexneri clinical isolate harboring φPOC-J13 [9], to create strain BS1076. Kanamycin-resistant transductants were purified and confirmed by PCR as described above.
Lysogeny of stx-Negative Shigella Isolates
Supernatants from overnight cultures of BS1076 were absorbed with log phase cultures of the stx-negative isolates in a 1:2 ratio for 20 minutes at 37°C as described previously [9, 12]. After incubation, 10 mM sodium acetate was added to stop phage absorption. Isolates were allowed to grow in 2 mL TSB for 90 minutes at 37°C before being plated on TSB agar-Congo red plates containing 50 µg/mL kanamycin. Kanamycin-resistant colonies were twice colony purified and confirmed to be φPOC-J13Δstx lysogens using primers Stxup and Stxdn. BS1058 (Table 1) was lysogenized with wild-type φPOC-J13 using the soft agar spotting method previously described [9]. Two independently isolated lysogens of each strain were used for further analysis.
Table 1.
Clinical Isolates of Shigella From Haitian Patients Characterized in This Study
| Strain | Species | stx-Positive | Clinic Site | Isolation Date | Age | Gender | GenBank Accession No. |
|---|---|---|---|---|---|---|---|
| BS1039 | Shigella flexneri 2a | Yes | Christianville School, Gressier | 21 November 2012 | 16 | F | LAJQ00000000 |
| BS1057 | S flexneri 2a | Yes | Notre-Dame Hospital, Petit Goâve | 17 December 2013 | 10 | M | LAJS00000000 |
| BS1059 | S flexneri 2a | Yes | Notre-Dame Hospital, Petit Goâve | 26 March 2014 | 6 | F | LAJU00000000 |
| BS1060 | S flexneri Y | Yes | Notre-Dame Hospital, Petit Goâve | 1 April 2014 | 20 | M | LAJV00000000 |
| BS1038 | S flexneri 3a | No | Saint Croix Hospital, Léogâne | 2 July 2013 | 6 | M | LAJP00000000 |
| BS1061 | S flexneri 3a | No | Notre-Dame Hospital, Petit Goâve | 7 April 2014 | 70 | M | LAJW00000000 |
| BS1040 | S flexneri 6 | No | Christianville School, Gressier | 21 November 2012 | 9 | M | LAJR00000000 |
| BS1058 | Shigella sonnei | No | George Guavan Hospital, Grand Goâve | 23 January 2014 | 47 | M | LAJT00000000 |
Determination of Antibiotic Resistance
Isolates were grown overnight on Tryptic Soy Agar at 37°C. Isolated colonies were processed for antibiotic resistance profiles using the Sensititre Antimicrobial Sensitivity Testing assay according to manufacturer's instructions (Sensititre, Thermo Scientific, Waltham, MA).
Whole-Genome Sequencing and Analysis
Genomic DNA was extracted from overnight cultures using the DNeasy Blood and Tissue Kit (QIAGEN, Germantown, MD). Sequencing libraries were prepared with either the TruSeq DNA Sample Prep Kit (Illumina, San Diego, CA) or the Nextera DNA Sample Prep Kit (Illumina) and sequenced on the Illumina MiSeq Platform, generating paired-end 250 base-pair reads in sufficient quantity to provide >49× coverage for each genome. Raw reads were trimmed, and draft genome sequences were assembled de novo with CLC Genomics Workbench version 7.0.3 or 7.0.4 (CLC bio, Boston, MA). In strains carrying the stx gene, the entire stx-encoding phage was contained on 1 contig. The stx-encoding prophage sequences were extracted from the genomic assemblies of the strains investigated and aligned to the phage φPOC-J13 reference sequence (GenBank accession no. KJ603229) using the SeqMan Pro module of the Lasergene software package (DNASTAR Inc., Madison, WI).
Nucleotide Sequence Accession Numbers
Whole draft genome sequences have been deposited at DDBJ/EMBL/GenBank under the accession numbers listed in Table 1.
RESULTS
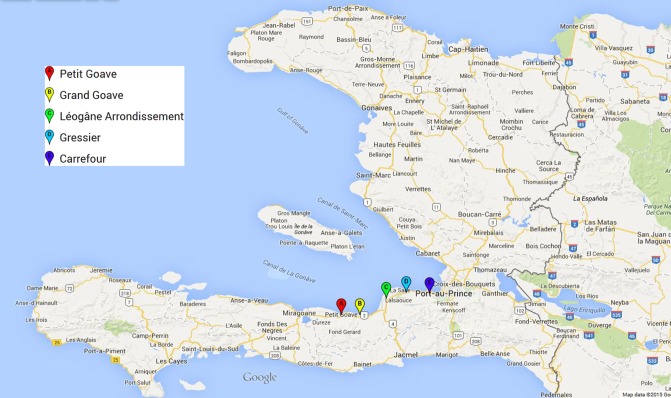
Fourteen Shigella isolates were obtained between 2012 and 2014 from patients at 5 different clinics located west of Port-au-Prince in the Ouest Department (Figure 1). Serological analysis determined that the isolates consisted of 7 S flexneri and 7 Shigella sonnei strains. This classification was confirmed by whole-genome sequencing. Four of the S flexneri isolates were positive for stx by PCR, whereas all of the S sonnei isolates were negative. Although the majority of the patients presented with diarrhea characterized as unremarkable, 2 of the patients had bloody and mucoid stools. One of those patients had been infected with a stx-positive S flexneri 2a (BS1059; Table 1), whereas the other patient was infected with a stx-negative S flexneri 6 (BS1040). In addition, none of the patients were reported to have developed HUS as a complication of infection.
Figure 1.
Location of clinics in Haiti where Shigella was isolated from patients. Map created in Google custom maps; Map Data@2015 Google.
All 7 S flexneri isolates and 1 of the S sonnei strains were further characterized, and epidemiological data for these strains are listed in Table 1. The 4 stx-positive S flexneri isolates (BS1039, BS1057, BS1059, and BS1060) produced and released Stx as shown in a Vero cell cytotoxicity assay. None of the stx-negative bacterial lysates (BS1038, BS1040, BS1058, and BS1061) produced any proteins that were cytotoxic to Vero cells. We also determined that the 4 Stx-producing isolates produced viable prophage by a plaque-forming unit assay on the E coli indicator strain MG1655.
To test whether the phage observed was a stx-encoding phage, we performed PCR using primers based on the φPOC-J13 sequence to demonstrate that the stx operon was flanked by sequence homologous to φPOC-J13. In our previous studies, we found that φPOC-J13 is inserted into the S flexneri chromosome at locus S1742. Polymerase chain reaction analysis using the primers from those studies demonstrated that the phage in the Stx-producing Haiti isolates was also inserted into S1742 or a homologous gene. Whole-genome sequencing of the isolates confirmed that the stx genes were encoded by a bacteriophage. The stx-encoding prophage sequences from the 4 strains were aligned and compared with phage φPOC-J13. The 5 prophages were nearly identical in sequence with only 3 single nucleotide polymorphisms among them.
Bacteriophages are powerful agents of bacterial evolution due to their ability to mediate horizontal transfer of genes. When phages such as φPOC-J13 infect a susceptible bacterium, they can enter a lysogenic state whereby the phage DNA inserts into the bacterial chromosome and replicates along with it. In this lysogenic state, the bacterium can express genes from the integrated phage and display new properties [4]. Therefore, we wanted to determine whether the stx-negative Shigella isolates could be lysogenized with φPOC-J13. We used φPOC-J13Δstx, a mutant of φPOC-J13 where the stx operon has been replaced with a kanamycin-resistant cassette, and screened for kanamycin resistance as described under Methods. All 4 of the stx-negative Shigella isolates yielded kanamycin-resistant colonies following phage infection. Two isolates of each were confirmed by PCR to be lysogenized with φPOC-J13Δstx. Polymerase chain reaction also determined that φPOC-J13Δstx had inserted into S1742 or a homologous gene in BS1038, BS1058, and BS1061. It is interesting to note that the BS1040 φPOC-J13Δstx lysogens did not produce a positive PCR product for insertion into S1742. Analysis of the sequence from the assembled BS1040 genome indicated that it did not encode S1742 or the adjacent genes, suggesting that the phage inserted into a different locus in BS1040.
Because none of the 7 S sonnei isolates from Haitian patients were stx positive, we wanted to investigate whether the S sonnei isolate (BS1058) could be stably lysogenized with wild-type φPOC-J13 and produce toxin. Two colonies of BS1058 lysogenized with φPOC-J13 were isolated and confirmed by PCR analysis to contain the stx genes flanked by phage sequence, as well as insertion into S1742. The 2 BS1058 φPOC-J13 lysogens also produced and released toxin in a Vero cell cytotoxicity assay.
Lastly, because of the increasing emergence of antibiotic resistance in clinical isolates of Shigella, we wanted to determine the antibiotic resistance profiles of all 8 of the isolates characterized. Overall, the antibiotic resistance profiles were unremarkable. The 7 S flexneri isolates were resistant to tetracyline and chloramphenicol, and only the S sonnei isolate (BS1058) showed sensitivity. All 8 of the isolates were sensitive to ciprofloxacin.
DISCUSSION
Our previous reports identifying Stx-producing Shigella species in international travelers led to the hypothesis that emergence of these strains is related to travel to Haiti and the Dominican Republic. This study reinforces that hypothesis by confirming the presence of stx-encoding Shigella in stool samples from Haitian patients. Although there were a low number of isolates over the study period, approximately 29% of Shigella isolated between 2012 and 2014 from the 5 clinical sites were stx positive. Moreover, all of the stx-encoding Shigella species identified harbored a phage identical to φPOC-J13. This suggests that φPOC-J13 is responsible for the horizontal spread of stx genes to diverse Shigella serogroups and serotypes in Haiti.
Although the majority of stx-positive Shigella isolates carrying φPOC-J13 that we have characterized are S flexneri 2a [8, 9], clinical isolates of S flexneri Y and S dysenteriae 4 harboring φPOC-J13 have been reported [8]. In addition, we have identified φPOC-J13 as the carrier of the stx genes from a previous report [6] of 4 isolates of stx-encoding S dysenteriae 4 associated with travel to Hispaniola (our unpublished data) as well as Shigella boydii 19 isolates from travelers returning to the United States from that region (our unpublished data). Although the limited species range identified for φPOC-J13 thus far suggests that the phage is specific to certain Shigella serogroups and serotypes, in this study we were able to lysogenize stx-negative clinical isolates of S flexneri 3a, S flexneri 6, and S sonnei from Haitian patients with φPOC-J13. This finding demonstrates that the host range of φPOC-J13 is more extensive than previously recognized. It is possible that as φPOC-J13 spreads to additional Shigella species, Stx-producing Shigella may become more prevalent in Haiti.
An outbreak of ciprofloxacin-resistant S sonnei that recently occurred in the United States was associated with international travel, including patients who had traveled to the Dominican Republic or Haiti [13]. Although none of the isolates characterized in our study were ciprofloxacin-resistant, the association of these Stx-producing Shigella species with international travel [8, 9] highlight their potential for global spread. More importantly, because ciprofloxacin is the current recommendation for treatment of Shigella infections for persons 18 years of age and older, the emergence of these stx-producing strains of Shigella should raise some alarms. Treatment of STEC infections with ciprofloxacin may have significant adverse clinical consequences because it can induce the lysogenic lambdoid phage that encodes the toxin and increase toxin production [14]. We have shown that φPOC-J13 can also be induced by treatment with ciprofloxacin with concomitant increase in Stx production (our unpublished data). Given the concerns noted above with ciprofloxacin resistance, combined with the risk that ciprofloxacin-sensitive strains may be carrying Stx (raising the possibility of phage and toxin induction with ciprofloxacin therapy), it may be wise to limit empiric use of ciprofloxacin in Shigella-infected patients who have a recent history of travel to Hispaniola.
CONCLUSIONS
Infection with bacteria that produce Stx can lead to more serious disease including the development of HUS, as found in approximately 10% of cases [15]. Although none of the isolates we have identified so far resulted in HUS, the potential exists for these species to cause more severe disease. The prevalence of Stx-producing Shigella should continue to be monitored to better understand the extent of the emergence in Hispaniola, the spread to foreign travelers, and the clinical consequences of infection with Stx-producing Shigella.
Acknowledgments
We thank the participating clinics in Haiti and Benoit St. Hilare for technical support. We also thank Stephen Darnell for technical assistance and Chris Grimm for assistance with antibiotic resistance determination.
Disclaimer. The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the Uniformed Services University of the Health Sciences.
Financial support. This work was supported by grants R01AI24656 and R01AI097405 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (to A. T. M. and J. G. M., respectively). This work was also supported in part by Department of Defense grant C0654_12_UN (to A. A.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Butler T. Haemolytic uraemic syndrome during shigellosis. Trans R Soc Trop Med Hyg 2012; 106:395–9. [DOI] [PubMed] [Google Scholar]
- 2.McDonough MA, Butterton JR. Spontaneous tandem amplification and deletion of the Shiga toxin operon in Shigella dysenteriae 1. Mol Microbiol 1999; 34:1058–69. [DOI] [PubMed] [Google Scholar]
- 3.Unkmeir A, Schmidt H. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect Immun 2000; 68:4856–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt H. Shiga-toxin-converting bacteriophages. Res Microbiol 2001; 152:687–95. [DOI] [PubMed] [Google Scholar]
- 5.Beutin L, Strauch E, Fischer I. Isolation of Shigella sonnei lysogenic for a bacteriophage encoding gene for production of Shiga toxin. Lancet 1999; 353:1498. [DOI] [PubMed] [Google Scholar]
- 6.Gupta SK, Strockbine N, Omondi M et al. . Emergence of Shiga toxin 1 genes within Shigella dysenteriae type 4 isolates from travelers returning from the Island of Hispanola. Am J Trop Med Hyg 2007; 76:1163–5. [PubMed] [Google Scholar]
- 7.Nyholm O, Lienemann T, Halkilahti J et al. . Characterization of Shigella sonnei isolate carrying Shiga toxin 2-producing gene. Emerg Infect Dis 2015; 21:891–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray MD, Lacher DW, Leonard SR et al. . Prevalence of Stx-producing Shigella species isolated from French Travelers returning from the Caribbean: an emerging pathogen with international implications. Clin Microbiol Infect 2015; 21:765.e9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray MD, Lampel KA, Strockbine NA et al. . Clinical isolates of Shiga toxin 1a-producing Shigella flexneri with an epidemiological link to recent travel to Hispaniola. Emerg Infect Dis 2014; 20:1669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livio S, Strockbine NA, Panchalingam S et al. . Shigella isolates from the global enteric multicenter study inform vaccine development. Clin Infect Dis 2014; 59:933–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark CS, Maurelli AT. Shigella flexneri inhibits staurosporine-induced apoptosis in epithelial cells. Infect Immun 2007; 75:2531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lech K, Brent R. 2001. Plating lambda phage to generate plaques, Chapter 1, unit 1.11. In Ausubel FM, Brent R, Moore DD, Seidman JG, Smith JA, Struhl K (ed), Current protocols in molecular biology. Wiley, New York, NY. [DOI] [PubMed] [Google Scholar]
- 13.De Lappe N, O'Connor J, Garvey P et al. . Ciprofloxacin-resistant Shigella sonnei associated with travel to India. Emerg Infect Dis 2015; 21:894–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, McDaniel AD, Wolf LE et al. . Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J Infect Dis 2000; 181:664–70. [DOI] [PubMed] [Google Scholar]
- 15.Walker CL, Applegate JA, Black RE. Haemolytic-uraemic syndrome as a sequela of diarrhoeal disease. J Health Popul Nutr 2012; 30:257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]