Abstract
Membrane protein structural biology has made tremendous advances over the last decade but there are still many challenges associated with crystallization, data collection and structure determination. Two independent groups, Axford et al. [(2015), Acta Cryst. D71, 1228–1237] and Huang et al. [(2015), Acta Cryst. D71, 1238–1256], have published methods that make a major contribution to addressing these challenges.
Keywords: commentary, membrane proteins, in meso crystallization, in meso data collection, in situ crystallization, in situ data collection
Membrane protein structural biology has made tremendous advances over the last decade as indicated by the exponential growth in the number of structures that have been published (http://blanco.biomol.uci.edu/mpstruc/). These advances are a result of many factors (Bill et al., 2011 ▸), including improvements in membrane protein overexpression, stabilization of proteins using antibodies or thermostabilizing mutations, and the enhancement of crystallization technologies such as crystallization in lipidic cubic phase (LCP, in meso crystallization). However, there are still many challenges associated with membrane protein crystallization, data collection and structure determination. Major problems often arise because membrane proteins frequently form tiny crystals, which either cannot be improved in size or which can be improved in size, but, as a consequence, lose diffraction quality. In addition, crystal handling, such as mounting the crystals and soaking in cryoprotectants, is often the reason for the loss of diffraction quality through mechanical shear-induced microlesions. This is particularly true for membrane protein crystals, which are often very fragile because of their high solvent content and being very thin in one dimension. In this issue of Acta Cryst. D, two independent groups, Axford et al. (2015 ▸) and Huang et al. (2015 ▸), have published methods that make a major contribution to addressing these problems, which will facilitate high-resolution data-collection of fragile crystals.
In the methodology demonstrated by Axford et al. (2015 ▸), a standard in situ 96-well sitting-drop crystallization plate was used to crystallize TehA from Haemophilus influenzae in a final volume of 200 nl. The plate was left for several days until crystals grew to their maximum size (up to 75 µm in the largest dimension). Instead of harvesting and mounting the crystals, the team mounted the entire plate on the beamline (in this case I24 at the Diamond Light Source; Fig. 1 ▸) and standard procedures were then used for data collection from the membrane protein crystals, i.e. each crystal was centered in the beam and wedges of data were collected at room temperature. This avoided simultaneously two major potential problems, namely crystal handling and cryocooling. Wedges of data were collected from multiple different crystals (30–50 images per wedge, 0.2° rotation each), thus reducing the effects of radiation damage, and then they were merged to obtain a data set at 2.3 Å resolution (90% complete). A direct comparison was performed between the structure of TehA determined from a cryocooled crystal at 1.5 Å resolution (98% complete, one crystal) and from the room temperature plate collection strategy (56 crystals); only minor changes were observed in flexible regions such as loop regions. This study therefore provides a proof of principle that membrane protein structures can be determined at a synchrotron using in situ room temperature data collection strategies.
Figure 1.
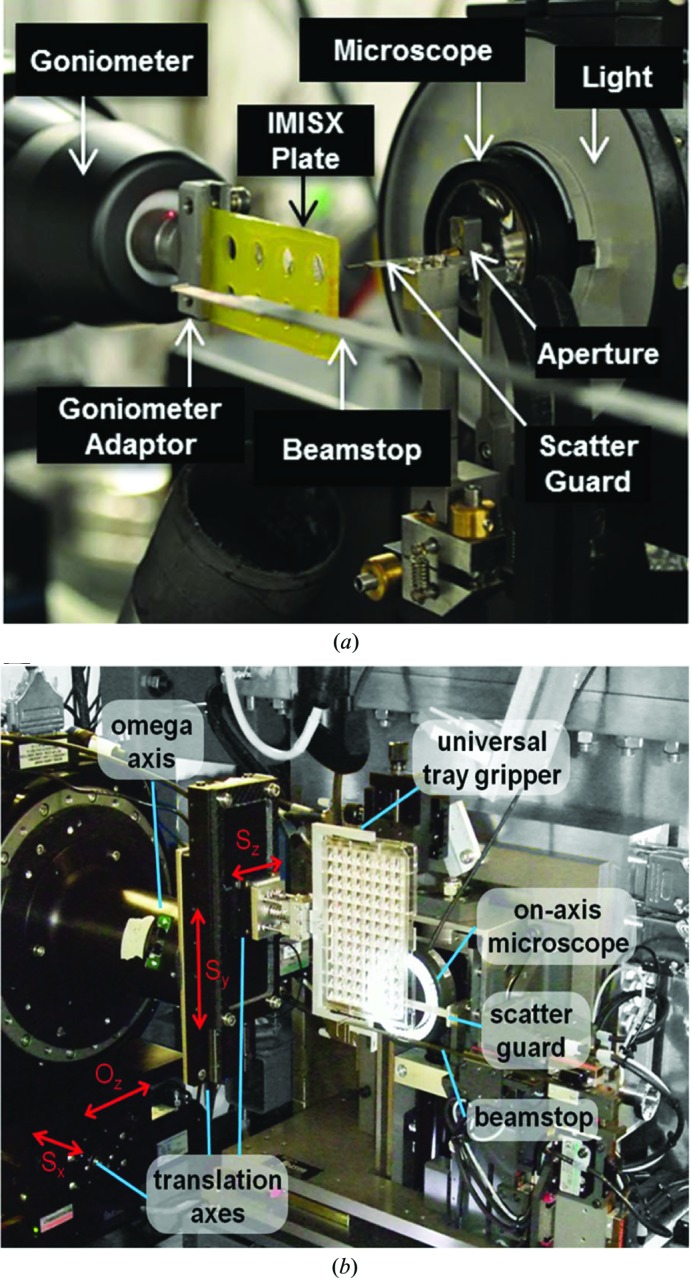
Two solutions for collecting in situ diffraction data from crystallization plates. (a) The set-up at SLS for IMISX (Huang et al., 2015 ▸). (b) The set-up at Diamond for data collection from 96-well plates (Axford et al., 2015 ▸).
Huang et al. (2015 ▸) took the in situ approach one step further and showed the applicability of room temperature data collection for in meso/in situ crystallization (IMISX) and its use for high-throughput crystallography of membrane proteins crystallized in meso using the LCP technology. This study used two different membrane proteins (the β-barrel AlgE and the α-helical protein PepTSt) and lysozyme for their demonstration of proof-of-principle. The major difficulties associated with using the LCP crystallization method include the high viscosity of the LCP, the temperature sensitivity of the lipid phase and the extreme difficulties encountered mounting crystals, because the normal set-up has the crystallization matrix sandwiched between two glass plates. These issues have been entirely circumvented by the development of a thin plastic film of a cyclic olefin copolymer (COC) to act as the support to sandwich the LCP matrix during crystallogenesis. As plastics allow more water to evaporate through their surfaces than glass, each of the crystallization set-ups was also sealed between glass plates to ensure reproducibility of crystallization and to provide additional protection to the plastic films. As the COC support does not affect X-ray diffraction, in situ data collection could then be used. The IMISX set-ups were shipped in temperature-controlled Styrofoam boxes to the Swiss Light Source (beamline PX II, X10SA) and mounted onto the beamline (Fig. 1 ▸). Data were collected both with IMISX and a cryocooled crystal of the same samples to compare methodologies. In the case of AlgE, the structure was refined to 2.8 Å resolution (94% complete; 244 crystals) from the data collected at room temperature in situ, which compared well with the structure determined from a single cryocooled crystal (2.9 Å resolution, 96% complete). The structure of PepTSt was more challenging from in situ data collection, requiring data from 572 crystals that yielded a structure to 2.8 Å resolution (100% complete), compared with the data collection from a single cryocooled crystal (2.3 Å resolution, 99% complete). Thus, the IMISX methodology was well validated for the structure determination of membrane proteins and is particularly impressive given the small size of the crystals used, which were often only about 10–20 µm in the largest dimension.
The two in situ high-throughput methodologies open up new perspectives in X-ray crystallography of membrane proteins and will provide a more rapid route to structure determination where the crystals are too small or fragile to mount, or where radiation sensitivity requires data collection from hundreds of crystals. In situ data collection will save considerable time, both from not having to improve crystal size, which could take months, and also during data collection where bespoke software and imaging allows rapid collection of diffraction data. In situ data collection therefore provides an excellent alternative to data collection at the X-ray free-electron laser, which cannot currently provide sufficient time for users.
References
- Axford, D. et al. (2012). Acta Cryst. D68, 592–600. [DOI] [PMC free article] [PubMed]
- Axford, D., Foadi, J., Hu, N.-J., Choudhury, H.G., Iwata, S., Beis, K., Evans, G. & Alguel, Y. (2015). Acta Cryst. D71, 10.1107/S139900471500423X. [DOI] [PMC free article] [PubMed]
- Bill, R.M., Henderson, P. J., Iwata, S., Kunji, E.R., Michel, H., Neutze, R., Newstead, S., Poolman, B.Tate, C.G. & Vogel, H. (2011). Nat. Biotechnol. 29 335–340. [DOI] [PubMed]
- Huang, C.-Y., Olieric, V., Ma, P., Panepucci, E., Diederichs, K., Wang, M. & Caffrey, M. (2015). Acta Cryst. D71, 10.1107/S1399004715005210. [DOI] [PMC free article] [PubMed]


