Abstract
Protein–lipid interactions regulate many membrane protein functions. Using a multi–scale approach which combines coarse–grained and atomistic molecular dynamics simulations we have predicted the binding site for the anionic phospholipid phosphatidylinositol 4,5–bisphosphate (PIP2) on the Kir2.2 inwardly–rectifying (Kir) potassium channel. Comparison of the predicted binding site to that observed in the recent PIP2–bound crystal structure of Kir2.2 reveals good agreement between simulation and experiment. In addition to providing insight into the mechanism by which PIP2 binds to Kir2.2, these results help to establish the validity of this multi–scale simulation approach and its future application to examine novel membrane protein/lipid interactions in the increasing number of high–resolution membrane protein structures that are now available.
Lipid interactions with membrane proteins are an important component of many cell signaling pathways. The anionic lipid phosphatidylinositol 4,5-bisphosphate (PIP2) plays a major role in such processes by acting as a secondary messenger, localizing proteins to membranes, and by regulating the activity of many different classes of ion channels1. One of the effects of PIP2 is the direct activation of inwardly–rectifying (Kir) potassium channels which modulate cellular electrical activity, and whose dysfunction underlies a wide range of inherited channelopathies2, 3. The binding site for PIP2 in Kir channels was previously predicted using both conventional docking methods and simulation–based approaches4-6. However, these previous studies all employed homology models based upon the structure of a chimera between a eukaryotic and prokaryotic Kir channel in which several of the residues proposed to interact with PIP2 were not fully resolved (PDB id 2QKS). Furthermore, this chimeric Kir/KirBac channel exhibits a complex functional interaction with PIP2 and it is known that PIP2 inhibits prokaryotic Kir channels7, 8. Consequently the accuracy of the binding site predicted in these original simulations remains uncertain.
In this study, we have now taken advantage of several recently solved X–ray crystal structures of eukaryotic Kir channels to evaluate the binding site for PIP2 in the Kir2.2 channel using a multi–scale molecular dynamics simulation approach5. This method employed coarse–grained (CG) simulations to predict initial PIP2–binding events which were then refined by subsequent atomistic (AT) simulations9-13. In order to validate this prediction, the simulations were then compared to the crystal structures of Kir2.2 with either PIP2 (PDB id 3SPI), or with the anionic lipid phosphatidic acid (PPA) bound (PDB id 3SPC). An apo–state structure without any bound lipid has also been solved (PDB id 3JYC)14, 15 and these different structures suggest an activation mechanism in which PIP2 binding induces an upward translation and engagement of the C–terminal domain (CTD) with the transmembrane domain (TMD)14.
To explore the influence of these different initial conformations on these predictions as well as possible effects of the lipid bilayer, bound phospholipids were removed from the 3SPI and 3SPC Kir2.2 structures and both structures then used as input for multi–scale simulations of PIP2 binding (Figure 1). To assess PIP2 binding to a Kir2.2 apo-state, we also performed CG simulations using the 3JYC structure as input (Figure S1 of the Supporting Information). In addition to comparing our predicted binding sites with the PIP2-bound 3SPI crystal structure, we also performed atomistic simulations of this PIP2-bound structure. These reference simulations provide a more valid comparison with the multi–scale simulations, because it is then possible to directly compare simulations of both proteins embedded in a phospholipid bilayer at room temperature, rather than comparing a membrane–bound simulation with a static X–ray structure in the absence of a membrane.
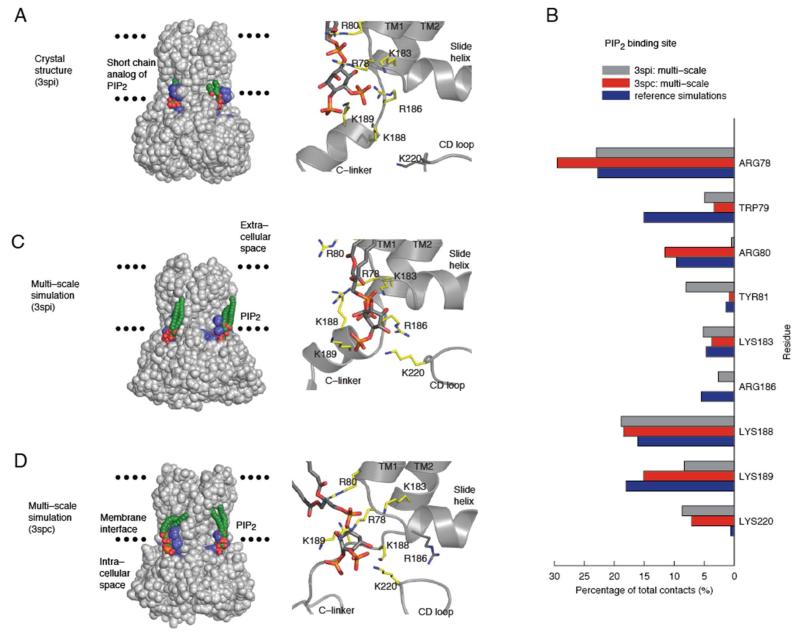
Figure 1.
PIP2 binding in Kir2.2 multi–scale simulations. A) Crystal structure with PIP2 bound. For clarity, only two PIP2 molecules are shown, with PIP2–interacting residues in blue (left). Detailed view of the PIP2 binding site (right) as found in the 3SPI structure. B) Multi–scale simulations used either 3SPI or 3SPC as starting structures and combined coarse–grained (CG; 24 × 0.5 μs) and atomistic (AT; 2 × 0.1 μs) simulations. Reference simulations were AT (2 × 0.1 μs) of the PIP2–bound 3SPI crystal structure. Residues whose side chains make more than 5% of the total contacts (≤4Å) with PIP2 head groups are compared between the multi–scale and reference simulations. C, D) The binding site of PIP2 predicted by (C) 3SPI or (D) 3SPC multi–scale simulations. Parameters for PIP2 can be found in the Supporting Information (Figure S4 and Table 1).
Examination of the 3SPI crystal structure shows that residues which make contacts with the PIP2 head group (Figure 1A) are mostly basic, and are either located on the N–terminal end of the first transmembrane helix TM1 (R78, W79, R80), or on the C–linker which connects the TMD with the CTD (K183, R186, K188, K189)14. We found that in the reference simulation of this structure all PIP2–protein contacts were retained and accounted for nearly 90% of the total contacts made by side chains with the PIP2 head groups (Figure 1B). Those residues which made less frequent contacts (~5%) with the PIP2 head groups (K183, R186) were located on the C–linker. The C–linker is helical in the PIP2–bound 3SPI structure, but flexible in other Kir2.2 crystal structures, and may therefore account for the differences observed for K183 and R186 in these simulations. However, the reference simulations appear to confirm the stability of the 3SPI structure and of the PIP2–protein interactions in this structure.
Importantly, we found that the multi–scale simulations which used the protein coordinates from 3SPI as a starting structure predicted the PIP2 head group to bind to the same cluster of basic residues as observed in the crystal structure (Figure 1C). This is in excellent agreement with both the reference simulations of this structure and with earlier predictions for other Kir channels5, 6. Residues on TM1 (R78) and on the C–linker (K183, R186, K188, K189) were found to form the same number of contacts in the 3SPI multi–scale simulations as in the reference simulations (Figure 1B). Residue W79 on TM1 formed fewer contacts in the multi-scale simulations (5%) compared to the reference simulations (15%). The absence of any specific hydrogen bond interactions of W79 with PIP2 in the crystal structure is likely to render the orientation of this side chain more flexible and might be the reason for less frequent contacts between W79 and PIP2 in the multi–scale simulations compared to the reference simulations. Although W79 is suggested to facilitate PIP2 binding by anchoring TM1 at the membrane interface14, less frequent contacts argue against a direct interaction of W79 with PIP2. The two adjacent residues (R80 and Y81) also exhibit differences between the multi–scale and reference simulations but this is most likely to be related to changes in TM2 which occur during the onset of channel gating. Nevertheless, these results demonstrate agreement between the PIP2–bound 3SPI crystal structure, the 3SPI multi–scale simulations and the reference simulations.
In addition to using 3SPI, we also used the 3SPC protein coordinates as an input for the multi–scale predictions. This structure was crystallized with the anionic phospholipid PPA instead of PIP2 and more closely resembles the apo–state structure (3JYC) in which the CTD is detached and not fully engaged with the TMD14. Strikingly, we found that when the ligand–free 3SPC structure was used as a starting point for the multi–scale simulations of PIP2-binding, there was also good agreement between the multi–scale simulations and reference simulations (Figure 1D). Almost all contacts that were observed in the reference simulations were predicted by the 3SPC multi–scale simulations (Figure 1B) and were similar to those using 3SPI as a starting structure. The only major difference was the absence of any interaction between the head group of PIP2 and R186 (Figure 1B). This is probably because in the 3SPC starting structure this ‘C-linker’ is less well ordered and the R186 side chain points in the opposite direction when compared to the PIP2–bound 3SPI structure. Longer simulations may therefore be needed to see if R186 changes its orientation to interact with the PIP2 head group.
Together these results clearly demonstrate that the multi–scale simulation approach can accurately predict the binding site for PIP2 in Kir2.2 even when slightly different input structures are used. The principal advantage of the multi–scale approach compared to conventional docking methods is that both PIP2 and the Kir2.2 channel are flexible. This can therefore provide insights into the molecular mechanisms underlying PIP2 binding, such as conformational changes or an electrostatic contribution (Figure S2 of the Supporting Information), which more static docking approaches lack. Interestingly, during the simulations we observed that the phosphates of the PIP2 head group formed transient hydrogen bonds with K220 on the CD loop. This resulted in a decrease of the Cα–Cα distance between K220 and a highly–conserved aspartate on the slide helix (D76) by 2–4 Å. This motion appears to be due to the CD loop moving upwards towards the slide helix and C–linker during PIP2 binding, similar to the gating mechanism proposed in several other studies16, 17 and which may form part of a general activation mechanism for Kir channels by PIP26. Our results also suggest that PIP2 binding to the C–linker stabilizes its α–helical structure, and may account for the interaction of PIP2 with R186 in the 3SPI crystal structure. Moreover, using the multi–scale approach, the effect of the lipid bilayer composition on PIP2 binding can be studied. Studies on liposomes of controlled lipid composition highlighted an anionic secondary lipid requirement for PIP2-induced Kir channel activation18. We therefore repeated our CG simulations, replacing half of the lipids on the inner leaflet with the anionic phospholipid phosphatidylserine (PS), noting that PIP2 still binds to Kir2.2, while PS, accumulating on either sides of the slide helix, distributes unevenly around Kir2.2 (Figure S3 of the Supporting Information). This suggests a secondary binding site for anionic lipids in Kir channels.
In conclusion, these findings demonstrate good agreement between the PIP2–bound crystal structure, the multi–scale predictions, and the reference simulations and therefore validate this multi–scale approach5, 6 for the prediction of PIP2 binding sites. Consequently, this method may be confidently applied to the prediction of novel PIP2 binding sites as well as the exploration of other lipid/channel interactions. For example, several eukaryotic voltage-gated potassium channels are regulated by PIP2 19, 20, and enough high–resolution crystal structures now exist to enable similar multi-scale simulations of PIP2 binding to these structures. Also, cholesterol has been shown to directly regulate Kir channel function but its precise binding site is unknown21. Many other unrelated ion channels and transporters, as well other classes of membrane proteins, have also been shown to be regulated by lipids22-24. Therefore, this multi–scale approach now provides an important and powerful tool to help study protein–lipid interactions and will become increasingly important with the ever expanding number of high-resolution membrane protein crystal structures now available.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr Prafulla Aryal for most helpful comments on the manuscript.
Funding
This work was supported by the Engineering and Physical Sciences Research Council, Pfizer Neusentis, the Biotechnology and Biological Sciences Research Council, and the Wellcome Trust.
ABBREVIATIONS
- PIP2
phosphatidylinositol 4,5–bisphosphate
- PPA
phosphatidic acid
- PS
phosphatidylserine
- Kir
inwardly rectifying potassium
- KV
voltage–gated potassium
- TMD
transmembrane domain
- CTD
C-terminal domain
- CG
coarse–grained
- AT
atomistic
Footnotes
ASSOCIATED CONTENT
Detailed methods, PIP2 binding to an apo structure (PDB id 3SPI), electrostatic potential mapped on the PIP2 binding site, simulations in a mixed bilayer, RMSF of the PIP2 binding site. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- (1).Suh BC, Hille B. Ann. Rev. Biophys. 2008;37:175–195. doi: 10.1146/annurev.biophys.37.032807.125859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ashcroft FM. Nature. 2006;440:440–447. doi: 10.1038/nature04707. [DOI] [PubMed] [Google Scholar]
- (3).Lopes CM, Zhang H, Rohacs T, Jin T, Yang J, Logothetis DE. Neuron. 2002;34:933–944. doi: 10.1016/s0896-6273(02)00725-0. [DOI] [PubMed] [Google Scholar]
- (4).Haider S, Tarasov AI, Craig TJ, Sansom MS, Ashcroft FM. EMBO J. 2007;26:3749–3759. doi: 10.1038/sj.emboj.7601809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Stansfeld PJ, Hopkinson R, Ashcroft FM, Sansom MS. Biochemistry. 2009;48:10926–10933. doi: 10.1021/bi9013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Meng X-Y, Zhang H-X, Logothetis DE, Cui M. Biophys. J. 2012;102:2049–2059. doi: 10.1016/j.bpj.2012.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Leal-Pinto E, Gómez-Llorente Y, Sundaram S, Tang Q-Y, Ivanova-Nikolova T, Mahajan R, Baki L, Zhang Z, Chavez J, Ubarretxena-Belandia I, Logothetis DE. J. Biol. Chem. 2010;285:39790–39800. doi: 10.1074/jbc.M110.151373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).D’Avanzo N, Cheng WWL, Doyle DA, Nichols CG. J. Biol. Chem. 2010;285:37129–37132. doi: 10.1074/jbc.C110.186692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Stansfeld PJ, Sansom MSP. J. Chem. Theory Comput. 2011;7:1157–1166. doi: 10.1021/ct100569y. [DOI] [PubMed] [Google Scholar]
- (10).Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink S-J. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- (11).Berger O, Edholm O, Jähnig F. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Scott WRP, Hunenberger PH, Tironi IG, Mark AE, Billeter SR, Fennen J, Torda AE, Huber T, Kruger P, Van Gunsteren WF. J. Phys. Chem. A. 1999;103:3596–3607. [Google Scholar]
- (13).Hess B, Kutzner C, Van der Spoel D, Lindahl E. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- (14).Hansen SB, Tao X, MacKinnon R. Nature. 2011;477:495–498. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tao X, Avalos JL, Chen J, MacKinnon R. Science. 2009;326:1668–1674. doi: 10.1126/science.1180310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Whorton MR, MacKinnon R. Cell. 2011;147:199–208. doi: 10.1016/j.cell.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Bavro VN, De Zorzi R, Schmidt MR, Muniz JRC, Zubcevic L, Sansom MSP, Vénien-Bryan C, Tucker SJ. Nat. Struct. Mol. Biol. 2012;19:158–163. doi: 10.1038/nsmb.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Cheng WWL, D’Avanzo N, Doyle DA, Nichols CG. Biophys. J. 2011;100:620–628. doi: 10.1016/j.bpj.2010.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Rodriguez-Menchaca AA, Adney SK, Tang Q-Y, Meng X-Y, Rosenhouse-Dantsker A, Cui M, Logothetis DE. Proc. Natl. Acad. Sci. USA. 2012;109:2399–2408. doi: 10.1073/pnas.1207901109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Kruse M, Hammond GRV, Hille B. J. Gen. Physiol. 2012;140:189–205. doi: 10.1085/jgp.201210806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Rosenhouse-Dantsker A, Logothetis DE, Levitan I. Biophys. J. 2011;100:381–389. doi: 10.1016/j.bpj.2010.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Tucker SJ, Baukrowitz T. J. Gen. Physiol. 2008;131:431–438. doi: 10.1085/jgp.200709936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Coskun U, Simons K. Structure. 2011;19:1543–1548. doi: 10.1016/j.str.2011.10.010. [DOI] [PubMed] [Google Scholar]
- (24).Aponte-Santamaría C, Briones R, Schenk AD, Walz T, De Groot BL. Proc. Natl. Acad. Sci. USA. 2012;109:9887–9892. doi: 10.1073/pnas.1121054109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.