Abstract
Rotaxanes, pseudorotaxanes and catenanes are supramolecular complexes with potential use in nanomachinery, molecular computing and single-molecule studies. Here we constructed a protein rotaxane in which a polypeptide thread is encircled by a Cytolysin A (ClyA) nanopore and capped by two proteins stoppers. The rotaxane could be switched between two states. At low negative applied potentials (<−50 mV) one of the protein stoppers resided inside the nanopore indefinitely. Under this configuration the rotaxane prevents the diffusion of protein molecules across the lipid bilayer and provides a useful platform for single-molecule analysis. High negative applied potentials (−100 mV) dismantled the interlocked rotaxane system by the forceful translocation of the protein stopper, allowing new proteins to be trapped inside or transported across the nanopore. The observed voltage threshold for the translocation of the protein stopper through the nanopore related well to the biphasic voltage dependence of the residence time measured for the freely diffusing protein stopper. We propose a model in which molecules translocate through a nanopore when the average dwell time decreases with the applied potential.
Keywords: ClyA nanopore, protein translocation, rotaxane, voltage dependent residence time, DHFR
Introduction
The bottom-up assembly of nanoscale elements into more complex molecular structures has potential applications in nanotechnology. DNA has been used to build a variety of nanoscale structures including rotaxanes,1-3 catenenes,4 reconfigurable three-dimensional objects,5 biomimetic systems6-8 and chemically fuelled molecular motors.9-11 Proteins possess larger chemical diversity than nucleic acids and are, therefore, attractive building elements in nanotechnology. However, the inability of predicting the three-dimensional structure of proteins strongly limited the use of proteins as building blocks for de novo structures. To our knowledge, only one artificial protein catenane has been described in the literature.12 One strategy to use proteins in nanotechnology is to combine proteins with a known structure. Biological nanopores are suitable building elements for this task because they self-assemble on biological membranes into a well-defined structure. Furthermore, when reconstituted in lipid bilayers, an external potential induces electrophoretic and electroosmotic forces that can be used to control the assembly of supramolecular structures. Using this strategy, single-molecule DNA-nanopore rotaxanes have been formed.13, 14 Such systems, however, showed limitations as they were formed exclusively with avidin as an external protein stopper and DNA as internal thread.
An additional advantage of using nanopores is that the ionic flux through individual nanopores provides means to identify single-molecules lodged inside the nanopore. For example, pseudo-rotaxanes formed by a DNA thread capped by a DNA processing enzyme are used in nanopore and protein sequencing applications.15-23 Nanopores might also be used to study single proteins. Ionic currents through nanopores are very sensitive to the environment of the nanopore and small differences between protein homodimers24 or isomeric protein-DNA interactions25 can be observed by specific changes to the nanopore conductance. Recently we have shown that folded proteins might be trapped transiently inside the lumen of the Cytolysin A (ClyA) nanopore without the need of complex immobilization strategies or covalent chemistry.25-28 Remarkably, ionic current recordings could monitor the binding of ligands to internalised proteins, indicating that an enzymatic activity can be translated into an electrical signal.28 Conveniently, ClyA could be isolated in different oligomeric states, providing nanopores with identical chemical composition but different pore dimensions.27 One limitation of this method, however, is that the trapping of proteins inside ClyA nanopores is voltage dependent and not always possible if the proteins are small (less than ~20 kDa), have an elongated shape (e.g. green fluorescent proteins) or are highly negatively charged.
In analogy to the mechanism of action of anthrax29 or diphtheria toxins,30 transmembrane proteins might also be engineered to transport molecules into the cytosol of target cells.31-34 We have shown that ClyA nanopores can be engineered to selectively internalize proteins into the nanopore interior.25-28 Therefore, if proteins translocate through the nanopore under physiological conditions of ionic strength and applied potential, ClyA might be used to convey therapeutic proteins across biological membranes. Proving the translocation of proteins across nanopores, however, is challenging. Previously, the translocation of bovine serum albumin (BSA) through a ~18 nm silicon nitride nanopore35 or the translocation of transport receptors through a ~40 nm nuclear pore complex mimic36 was shown by fluorescence spectroscopy following the collection of the protein translocated across the nanopore after ~50 hours recording.35 Due to the fragility of the lipid bilayer, however, this approach is not applicable to biological nanopores. More recently, the unfolded translocation of a recombinant protein covalently linked to an oligonucleotide through the aerolysin nanopore was shown by performing a PCR amplification of the DNA adducts from the translocated protein-DNA fraction.37 This approach, however, can only be applied to protein-DNA hybrids.
Here we assemble a supramolecular protein structure where two protein stoppers lock a polypeptide thread inside a biological nanopore. The rotaxane was formed at low-applied potentials and dismantled at high-applied potentials, proving that the crossing of the protein stopper through the ClyA nanopore can be controlled by the applied potential. A voltage threshold for protein translocation related well to the biphasic voltage dependence of the residence time of the un-locked protein stopper, revealing that the voltage dependent dwell time of a molecule relates with good approximation to its translocation probability through the nanopore.
Results
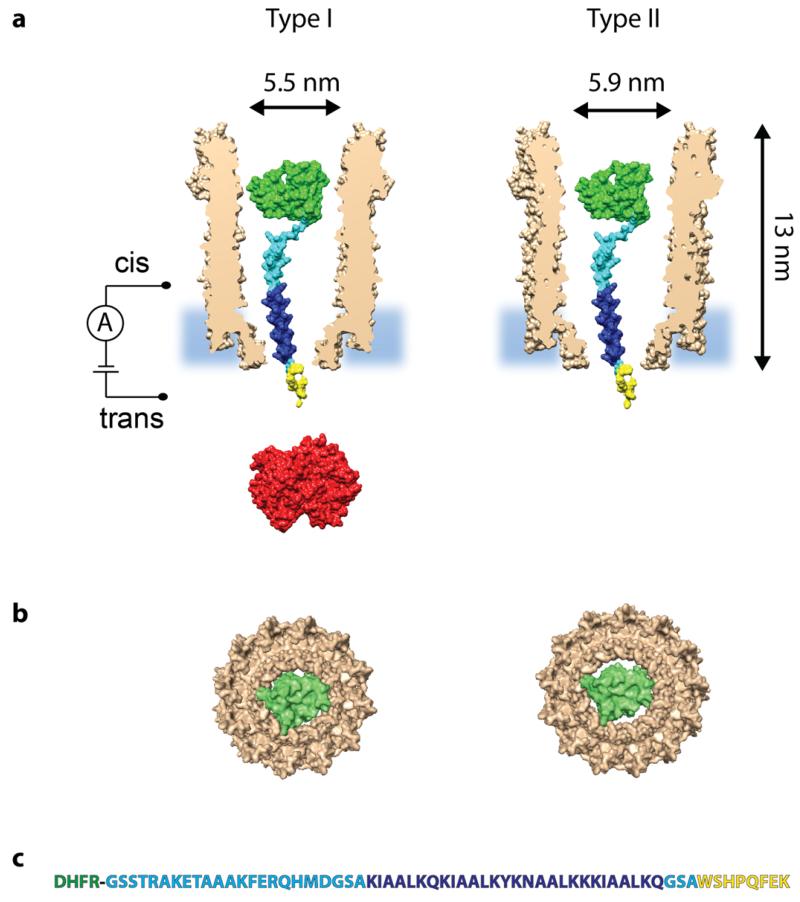
A rotaxane is formed when a macrocycle encircles a thread capped by two stoppers. We used a ClyA nanopore as macrocycle and a cysteine-free (C85→A C152→S) dihydrofolate reductase from E. coli (DHFR, Mw = 18 kDa, Figure 1) as a first protein stopper. The C-terminus of DHFR was genetically extended with a polypeptide terminating with a Strep-Tag (DHFRST, Figure 1). The 62 amino acids thread (Strep-tag) is long enough to protrude through the transmembrane region of the nanopore and to connect with the second rotaxane stopper Strep-Tactin-HRP (an engineered Streptavidin that binds the Strep-Tag with high affinity complexed with horseradish peroxidase) added to the opposite side of the lipid bilayer (Figure S1). Strep-Tactin-HRP is too large to translocate through the ClyA nanopore.
Figure 1. A single DHFRST protein captured inside Type I ClyA-AS and Type II ClyA-CS.
a, Surface representation of a Type I ClyA-AS and a Type II ClyA-CS nanopore (brown, shown as cross section) containing a single E. coli DHFR (green, PDB_ID 1RH3) extended with a polypeptide linker (cyan), a positively charged threading tag (dark blue) and a Strep-Tag (yellow). Strep-Tactin is shown in red. The dimensions of the ClyA nanopore are indicated considering the Van der Waals radii of the atoms.27 b, Bottom view of a single DHFR molecule inside Type I ClyA (left) and Type II ClyA (right), showing the tight fit in dimensions between DHFR and the transmembrane part of Type I ClyA, while in Type II ClyA nanopores DHFR is expected to experience less steric hindrance upon translocation. Both the DHFR and the ClyA nanopore are shown as surface representations. c, Sequence of the DHFR (green) fusion construct, with the polypeptide linker shown in cyan, the positively charged threading tag in dark blue and the Strep-Tag in yellow.
We tested the interaction of DHFRST with two ClyA nanopore types: Type I ClyA-AS and Type II ClyA-CS nanopores. ClyA-AS (C87A/L99Q/E103G/F166Y/I203V/C285S/K294R/H307Y) and ClyA-CS (L99Q/E103G/F166Y/C285S/K294R) are engineered versions of ClyA from Salmonella Typhi, selected for their favourable properties in planar lipid bilayers, with Type I most likely corresponding to the dodecameric and Type II to the tridecameric version of the nanopore (Figure 1).27
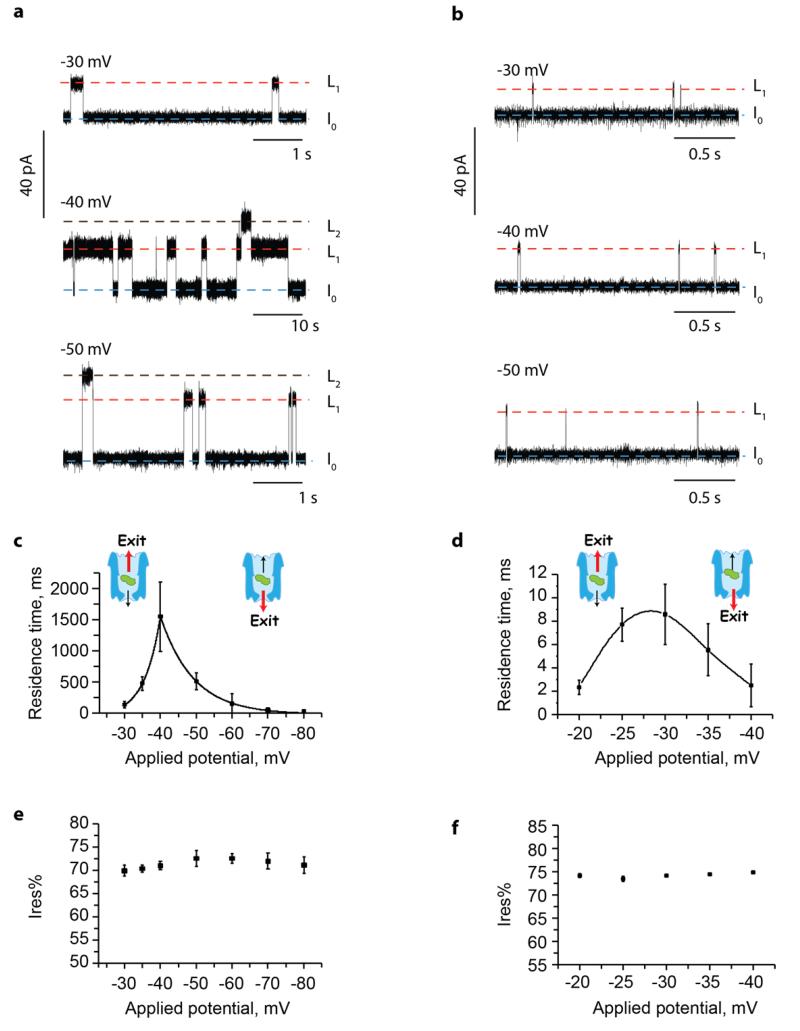
In 150 mM NaCl, 15 mM Tris HCl pH 7.5, the addition of ~100 nM of DHFRST to the cis compartment of Type I ClyA-AS and Type II ClyA-CS induced current blockades only at negative applied potentials. Since DHFRST carries a weak negative charge (theoretical pI=6.7), these data indicate that the main driving force that promotes the entry of the protein into the cis vestibule of ClyA is the electroosmotic flow.26, 38-41 For Type I ClyA-AS, in the voltage regime between −30 mV and −80 mV, the observed current blockades were transient and showed two different residual current levels (L1 and L2, Figure 2a). At −40 mV, L1 had a residual current (Ires%), defined as the percent ratio between the blocked and open pore currents, of 70.5±2.6 % (n=25 individual DHFRST blockades, N=5 single ClyA-AS pores) and L2 displayed Ires% equal to 49.2±6.4 % (n=25 individual DHFRST blockades, N=5 single ClyA-AS pores). The vast majority of DHFRST blockades only displayed L1 current levels (Figure S2). Occasionally long (e.g. more than several seconds) L2 blockades were observed, in which case ramping to positive applied potentials restored the open-pore current. At potentials below −40 mV L2 blockades almost entirely disappeared.
Figure 2. DHFRST blockades to Type I ClyA-AS and Type II ClyA-CS.
a,b, Typical current blockades induced by DHFRST (~100 nM, added in cis) to Type I ClyA-AS (a) and Type II ClyA-CS (b) nanopores at various negative applied potentials. The open-pore current IO is indicated by a blue dashed line. L1 and L2 current levels are indicated by red and black dashed lines, respectively. c,d, Voltage dependencies of the DHFRST blockade dwell times for Type I ClyA-AS (c) and Type II ClyA-CS (d) nanopores. The fitted lines are an exponential fit for Type I ClyA-AS and a spline trend line for Type II ClyA-CS. A likely explanation to the data is that at applied potentials lower than the dwell time maximum proteins preferentially exit the nanopore from the cis entrance, while at potentials higher than the dwell time maximum proteins exit the nanopore from the narrower trans exit. The red arrow in the molecular schemes indicates the preferred direction of protein exit. e,f, Voltage dependencies of the Ires% values of the DHFRST blockades for Type I ClyA-AS (e) and Type II ClyA-CS (f) nanopores. All current traces were collected in 150 mM NaCl, 15 mM Tris HCl pH 7.5, at 28°C, by applying a Bessel-low pass filter with a 2 kHz cut-off and sampled at 10 kHz. Errors are shown as standard deviations.
DHFRST blockades to Type II ClyA-CS were shorter than to Type I ClyA-AS (~15-fold at −30 mV, ~80-fold at −35 mV and ~600-fold at −40 mV, Figure 2a,b) and mainly comprised of L1 current levels (L1 = 74.0±1.1 % at −30 mV, n=25, N=4, Figure 2b). DHFRST also rarely dwelled on a current level with lower residual current. However, the dwell time at this current level was too short to allow reliable determination of the Ires% values (Figure S3).
The blockades’ average dwell time to Type I ClyA-AS first increased to a maximum value at approximately −40 mV (dwell time = 1549±556 ms, n=1300, N=6) and then rapidly decreased (Figure 2c). The dwell times of DHFRST blockades to Type II ClyA-CS also initially increased and then decreased with the applied potential, although less sharply. The dwell times to Type II ClyA-CS showed a peak maximum around −30 mV (dwell time = 8.6±2.6 ms, n=550, N=7, Figure 2d). Remarkably, increasing the diameter of the nanopore by just 0.4 nm decreased the maximum average dwell time by about three orders of magnitude. For both nanopore types, the Ires% remained constant over the investigated applied potential range (Figure 2e,f), suggesting that the protein remained folded in the range of applied potentials considered.
A likely explanation for the observed voltage dependences, as previously reported in the literature for other moleules,42-46 is that at potentials lower than the dwell time maximum (−40 mV for Type I ClyA-AS and −30 mV for Type II ClyA-CS) the majority of DHFRST proteins enter and exit the nanopore from the cis side, while at potentials above the dwell time maximum the majority of proteins translocate through the nanopore (Figure 2c,d). Following this interpretation, a rotaxane with DHFR as an internal stopper, the Strep-tag as the tread and Strep-Tactin-HRP as the second external stopper (Figure S1) will be assembled at low-applied potentials, where DHFRST enters but does not translocate the nanopore. The rotaxane can then be disassembled at high-applied potentials where DHFRST translocates through the nanopore.
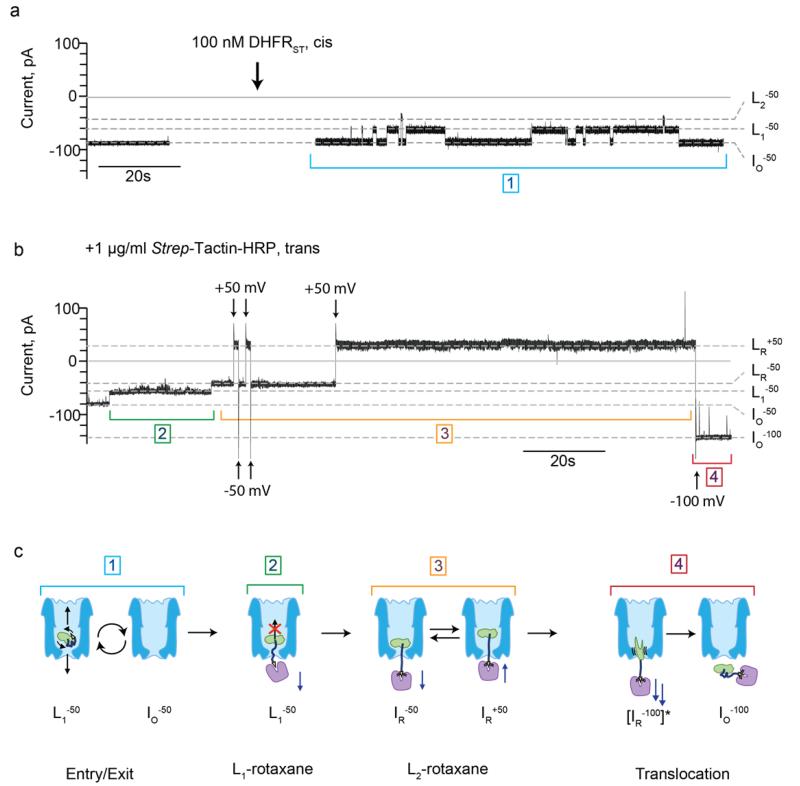
At −50 mV, the addition of DHFRST (~100 nM, cis) to Type I ClyA-AS induced transient blockades that showed a mean dwell time of 510±134 ms (n=1400, N=5, Figure 3a,c step 1) and mainly showed a L1 (Ires% = 71.5±1.4 %, n=30, N=7) current level, although occasional visits to L2 (Ires% = 52.2±3.1 %, n=30, N=7) were also observed. After Strep-Tactin-HRP was added (1μl/ml, trans) ~40-50 % of the transient blockades turned into long-lasting L1 events (Ires% L1−50 = 71.0±1.2 %, n=46, N=6), suggesting that the interaction between DHFRST and Strep-Tactin-HRP prevented the exit of the DHFR stopper from the nanopore. Upon switching the potential to +50 mV, however, the open-pore current (IO+50) was restored (Figure S4a), indicating that the interaction between Strep-Tactin and the Strep-Tag was not strong enough to retain DHFRST inside the ClyA nanopore at positive applied potentials.
Figure 3. DHFRST rotaxane formation at −50 mV and DHFRST translocation at −100 mV.
a, Current trace of a single Type I ClyA-AS nanopore at −50 mV before (left) and after (right) the addition of ~100 nM DHFRST to the cis compartment. b, The subsequent addition of 1 μg/ml Strep-Tactin-HRP to the trans compartment induced the formation of a protein rotaxane. c, Schematic representations of the different steps of the current trace displayed in (a,b). Step 1: Entry and exit of DHFRST. The electroosmotic flow promotes the entry of DHFRST inside the nanopore from the cis side. The protein then exits the nanopore from either the cis or trans side. Step 2: L1-rotaxane formation. After the addition of Strep-Tactin-HRP to the trans compartment, the dwell time of the L1 blockades drastically increases, indicating that Strep-Tactin-HRP interacted with the Strep-Tag of DHFRST. Under this configuration, reversal of the potential induces dismantling of the rotaxane (Figure S4a). Step 3: L2-rotaxane. Occasionally, a long-living L1 blockade switches to a long-living L2 blockade. Under this configuration, the rotaxane displays a high stability at positive applied potentials. Step 4: DHFRST translocation. At −100 mV the rotaxane is dismantled by translocation of the rotaxane complex through ClyA. The ClyA nanopore is depicted in blue, DHFR in green, the positively charged polypeptide tag in dark blue, the Strep-Tag in yellow, and Strep-Tactin in purple. The grey dashed lines represent the open-pore currents at −50 mV (IO−50) and −100 mV (IO−100), the L1 current at −50 mV (L1−50), the current level after rotaxane formation at −50 mV (LR−50) and +50 mV (LR+50). The current trace was collected in 150 mM NaCl, 15 mM Tris HCl pH 7.5, at 28°C, by applying a Bessel-low pass filter with a 2 kHz cut-off and sampled at 10 kHz.
Interestingly, in ~30 % of the observed long-lasting L1 events (“L1 rotaxane”, Figure 3b,c, step 2), L1 converted to a long-lasting L2 current level (LR−50, Ires% = 50.6±2.2 %, n=44, N=6, Figure 3b,c, step 3, “L2-rotaxane”), which showed a slightly lower Ires% value than the L2 states observed in the absence of Strep-Tactin-HRP in the trans solution. Upon switching the potential to +50 mV, a permanent blocked current level was observed (LR+50, Ires%=43.7±4.9 %, n=55, N=6), indicating that Strep-Tactin-HRP did not release from the Strep-Tag of DHFRST (Figure 3b,c, step 3, Figure S4b, Figure S5). A blocked current level was also observed at +100 mV (Figure S6), indicating that the protein rotaxane is stable over the whole applied positive potential range. The subsequent switching of the potential to −100 mV restored the Type I ClyA-AS open-pore current (IO−100), indicating that at this potential DHFRST translocated through the nanopore (Figure 3b,c, step 4, Figure S4b, Figure S5), thereby dismantling the rotaxane. The addition of 10 μM of biotin to the trans compartment prevented the formation of rotaxanes, indicating that our observations were indeed the result of the specific interaction between the Strep-Tag of DHFRST and Strep-Tactin-HRP (Figure S7).
Rotaxanes could also be formed at lower applied potentials (e.g. at −35 mV, Figure S8). However, working at this potential was impractical because the conversion of the long-lasting L1 blockades (L1−35) to the stable rotaxane current level (LR−35) could only be rarely observed, probably because the driving force at lower applied potentials was not strong enough to ensure full threading of the Strep-tag through the transmembrane part of ClyA-AS. Rotaxanes could not be formed using Type II ClyA-CS nanopores, presumably because the dwell time of DHFRST inside the larger Type II nanopore is too short for the Strep-Tag to interact with Strep-Tactin-HRP.
Discussion
In this work we described a strategy to assemble a nanopore-based all-protein rotaxane. Compared with previously described nanopore rotaxanes built with a DNA molecule as thread and a biotin-binding molecule as external stopper, our design allowed the incorporation of a generic protein stopper inside the lumen of the nanopore. This configuration might have applications in single-molecule sensing or enzymology studies. Further, at high-negative applied potentials the rotaxane was dismantled by the forced transport of the protein stopper through the nanopore, indicating, therefore, that the protein stopper could translocate through the nanopore, albeit at high-applied potentials.
Proving the translocation of a molecule across a nanopore is important in many nanopore applications, but generally challenging to accomplish especially using ionic current measurements alone. Here, we observed that the residence time of un-locked DHFRST inside ClyA nanopores first increased and then decreased with the applied potential. A possible explanation for the observed voltage dependencies of DHFRST, as previously postulated in the literature for other molecules,42-46 is that at lower applied potentials the electroosmotic flow (SI Discussion) is not strong enough to keep the protein inside the nanopore lumen and random thermal fluctuations are likely to promote the exit of the protein from the entry side (the cis side). Increasing the applied potential, however, increases the electroosmotic flow, which then might become strong enough to force the translocation of the protein through the narrow transmembrane region of ClyA. In this view, a potential threshold might exist for the translocation of DHFRST across ClyA nanopores. At potentials lower than the threshold (<−40 mV for Type I ClyA-AS and <−30 mV for Type II ClyA-CS) proteins enter and exit the nanopore from the cis side of the lipid bilayer (cis exit), while at potentials above the threshold proteins can translocate through the nanopore. Therefore, the dwell time maximum represents a regime in which cis exit and translocation have equal probabilities.
The protein rotaxane described in this work provided experimental evidence supporting this interpretation. If the biphasic voltage dependence of DHFRST is related to the existence of a threshold potential for protein translocation, a rotaxane would be formed at applied potentials below the threshold (<−40 mV) and dismantled at potentials higher than the threshold (>−40 mV). To our delight, we found that the assembly and dismantling of the protein rotaxane was voltage dependent, providing therefore evidence for a potential threshold for protein translocation. However, we also found that the rotaxane was formed more efficiently and could not be dismantled at potentials above the dwell time maximum of free DHFRST. A likely explanation is that the threshold for protein translocation differs for free proteins and proteins locked in a rotaxane configuration. Although we cannot exclude a possible effect of the Strep-Tactin-HRP on the translocation process (e.g. by changing the electroosmotic flow through the nanopore), it is likely that a specific orientation of DHFRST inside ClyA is important to allow the translocation of DHFR across the nanopore. Free DHFRST has a higher rotational and translational freedom than the immobilized DHFRST, and can therefore efficiently sample the translocation configuration at lower applied potentials. By contrast, when DHFRST is locked in a rotaxane configuration a higher driving force might be required to sample a configuration inside the nanopore that allows protein translocation.
Conclusions
In this work we constructed a protein rotaxane in which a polypeptide chain is encircled by a ClyA nanopore and capped by a DHFR stopper on the cis side and a Strep-Tactin-HRP stopper on the trans side. Rotaxane architectures might find applications as switches in molecular electronics or might be used as supramolecular protein systems for nanomachining applications. In this work we formed a rotaxane to prove the translocation of proteins across the ClyA nanopore. We showed that the rotaxane was assembled at low-applied potentials (V <−50 mV) and dismantled at high-applied potentials (−100 mV), indicating that a voltage threshold exists for the translocation of the DHFR stopper across the ClyA nanopore. Although the immobilization is likely to affect the dynamics of protein translocation, this observation related well with the biphasic voltage dependence observed for the dwell times of free DHFR. Our results provide experimental evidence to suggest that a decrease in the residence time with the applied potential can be used as an indication for the translocation of molecules across a nanopore.
Supplementary Material
References
- 1.Ackermann D, Schmidt TL, Hannam JS, Purohit CS, Heckel A, Famulok M. Nat Nanotechnol. 2010;5(6):436–42. doi: 10.1038/nnano.2010.65. [DOI] [PubMed] [Google Scholar]
- 2.Lohmann F, Ackermann D, Famulok M. J Am Chem Soc. 2012;134(29):11884–7. doi: 10.1021/ja3042096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohmann F, Weigandt J, Valero J, Famulok M. Angew Chem Int Ed Engl. 2014;53(39):10372–6. doi: 10.1002/anie.201405447. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt TL, Heckel A. Nano Lett. 2011;11(4):1739–42. doi: 10.1021/nl200303m. [DOI] [PubMed] [Google Scholar]
- 5.Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J. Nature. 2009;459(7243):73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 6.Bell NA, Engst CR, Ablay M, Divitini G, Ducati C, Liedl T, Keyser UF. Nano Lett. 2012;12(1):512–7. doi: 10.1021/nl204098n. [DOI] [PubMed] [Google Scholar]
- 7.Burns JR, Stulz E, Howorka S. Nano Lett. 2013;13(6):2351–6. doi: 10.1021/nl304147f. [DOI] [PubMed] [Google Scholar]
- 8.Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC. Science. 2012;338(6109):932–6. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin J-S, Pierce NA. Journal of the American Chemical Society. 2004;126(35):10834–10835. doi: 10.1021/ja047543j. [DOI] [PubMed] [Google Scholar]
- 10.Yin P, Yan H, Daniell XG, Turberfield AJ, Reif JH. Angewandte Chemie International Edition. 2004;43(37):4906–4911. doi: 10.1002/anie.200460522. [DOI] [PubMed] [Google Scholar]
- 11.Tian Y, He Y, Chen Y, Yin P, Mao C. Angewandte Chemie International Edition. 2005;44(28):4355–4358. doi: 10.1002/anie.200500703. [DOI] [PubMed] [Google Scholar]
- 12.Blankenship JW, Dawson PE. Protein Sci. 2007;16(7):1249–56. doi: 10.1110/ps.062673207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Quesada J, Saghatelian A, Cheley S, Bayley H, Ghadiri MR. Angewandte Chemie-International Edition. 2004;43(23):3063–7. doi: 10.1002/anie.200453907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franceschini L, Soskine M, Biesemans A, Maglia G. Nature Communications. 2013;4:2415. doi: 10.1038/ncomms3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri MR. Angewandte Chemie-International Edition. 2005;44(9):1401–4. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cockroft SL, Chu J, Amorin M, Ghadiri MR. Journal of the American Chemical Society. 2008;130(3):818–20. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(19):7702–7. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoddart D, Maglia G, Mikhailova E, Heron AJ, Bayley H. Angewandte Chemie-International Edition. 2010;49(3):556–559. doi: 10.1002/anie.200905483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franceschini L, Mikhailova E, Bayley H, Maglia G. Chem Commun (Camb) 2012;48(10):1520–2. doi: 10.1039/c1cc16124e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Phys Rev Lett. 2000;85(14):3057–60. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 21.Nivala J, Marks DB, Akeson M. Nature Biotechnology. 2013;31(3):247–50. doi: 10.1038/nbt.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benner S, Chen RJ, Wilson NA, Abu-Shumays R, Hurt N, Lieberman KR, Deamer DW, Dunbar WB, Akeson M. Nat Nanotechnol. 2007;2(11):718–24. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Larrea D, Bayley H. Nature Nanotechnology. 2013;8(4):288–95. doi: 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nir I, Huttner D, Meller A. Biophys J. 2015;108(9):2340–9. doi: 10.1016/j.bpj.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Meervelt V, Soskine M, Maglia G. ACS nano. 2014;8(12):12826–35. doi: 10.1021/nn506077e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soskine M, Biesemans A, Moeyaert B, Cheley S, Bayley H, Maglia G. Nano Letters. 2012;12(9):4895–900. doi: 10.1021/nl3024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soskine M, Biesemans A, De Maeyer M, Maglia G. J Am Chem Soc. 2013;135(36):13456–63. doi: 10.1021/ja4053398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soskine M, Biesemans A, Maglia G. J Am Chem Soc. 2015;137(17):5793–7. doi: 10.1021/jacs.5b01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S, Moayeri M, Leppla SH. Trends Microbiol. 2014;22(6):317–25. doi: 10.1016/j.tim.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collier RJ. Bacteriol Rev. 1975;39(1):54–85. doi: 10.1128/br.39.1.54-85.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arora N, Klimpel KR, Singh Y, Leppla SH. The Journal of biological chemistry. 1992;267(22):15542–8. [PubMed] [Google Scholar]
- 32.Arora N, Leppla SH. Infect Immun. 1994;62(11):4955–61. doi: 10.1128/iai.62.11.4955-4961.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu H, Leppla SH. PLoS One. 2009;4(11):e7946. doi: 10.1371/journal.pone.0007946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arora N, Williamson LC, Leppla SH, Halpern JL. The Journal of biological chemistry. 1994;269(42):26165–71. [PubMed] [Google Scholar]
- 35.Fologea D, Ledden B, McNabb DS, Li J. Appl Phys Lett. 2007;91(5):539011–539013. doi: 10.1063/1.2767206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowalczyk SW, Kapinos L, Blosser TR, Magalhaes T, van Nies P, Lim RY, Dekker C. Nat Nanotechnol. 2011;6(7):433–8. doi: 10.1038/nnano.2011.88. [DOI] [PubMed] [Google Scholar]
- 37.Pastoriza-Gallego M, Breton MF, Discala F, Auvray L, Betton JM, Pelta J. ACS nano. 2014;8(11):11350–60. doi: 10.1021/nn5042398. [DOI] [PubMed] [Google Scholar]
- 38.Gu LQ, Cheley S, Bayley H. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15498–503. doi: 10.1073/pnas.2531778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong CTA, Muthukumar M. Journal of Chemical Physics. 2007;126(16) doi: 10.1063/1.2723088. [DOI] [PubMed] [Google Scholar]
- 40.Firnkes M, Pedone D, Knezevic J, Doblinger M, Rant U. Nano letters. 2010;10(6):2162–2167. doi: 10.1021/nl100861c. [DOI] [PubMed] [Google Scholar]
- 41.Muthukumar M. Protein Pept Lett. 2014;21(3):209–16. doi: 10.2174/09298665113209990079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stefureac RI, Trivedi D, Marziali A, Lee JS. J Phys Condens Matter. 2010;22(45):454133. doi: 10.1088/0953-8984/22/45/454133. [DOI] [PubMed] [Google Scholar]
- 43.Rincon-Restrepo M, Mikhailova E, Bayley H, Maglia G. Nano Letters. 2011;11(2):746–50. doi: 10.1021/nl1038874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Nat Nanotechnol. 2009;4(4):265–70. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 45.Cracknell JA, Japrung D, Bayley H. Nano Letters. 2013;13(6):2500–5. doi: 10.1021/nl400560r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wanunu M, Sutin J, McNally B, Chow A, Meller A. Biophysical Journal. 2008;95(10):4716–25. doi: 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.