Abstract
This study demonstrates the requirement of Asp-380 and Asp-386 in the βDELSEED-motif of E. coli ATP synthase for peptide binding and inhibition. We studied the inhibition profiles of wild-type and mutant E. coli ATP synthase in presence of c-terminal amide bound melittin and melittin related peptide. Melittin and melittin related peptide inhibited wild-type ATPase almost completely while only partial inhibition was observed in single mutations with replacement of Asp to Ala, Gln, or Arg. Additionally, very little or no inhibition occurred among double mutants βD380A/βD386A, βD380Q/βD386Q, or βD380R/βD386R signifying that removal of one Asp residue allows limited peptide binding. Partial or substantial loss of oxidative phosphorylation among double mutants demonstrates the functional requirement of βD380 and βD386 Asp residues. Moreover, abrogation of wild-type E. coli cell growth and normal growth of mutant cells in presence of peptides provides strong evidence for the requirement of βDELSEED-motif Asp residues for peptide binding. It is concluded that while presence of one Asp residue may allow partial peptide binding, both Asp residues, βD380 and βD386, are essential for proper peptide binding and inhibition of ATP synthase.
Keywords: F1Fo ATP synthase; E. coli, ATPase; Peptide; Enzyme inhibition; βDELSEED
Introduction
The central role of ATP synthase as a biological nanomotor to generate or hydrolyze ATP, the universal energy currency is well established [1–3]. Lately, the presence of ATP synthase on the cell surface and its involvement in multiple cell functions has become known. A flow of protons through the Fo sector (ab2c10–14) causes rotation of the γ-subunit, which results in conformational changes of the α/β-subunits of the F1 sector (α3β3γδε). This sequence causes either hydrolysis or synthesis of ATP by oxidative phosphorylation or photophosphorylation [4–7]. The direction of the proton gradient across the membrane determines whether ATP synthase operates in a clockwise or anticlockwise direction to either synthesize ATP or hydrolyze ATP. Detailed catalytic and motor function properties of ATP synthase are described in the following references [8–12].
The importance of ATP synthase to human health is revealed by its involvement in many diseases conditions, such as Alzheimer’s, Parkinson’s, Leigh syndrome, neuropathy, Batten’s disease, obesity, hypertension, and the class of severely debilitating diseases known collectively as mitochondrial myopathies ([13] and reference therein). It is a possible molecular therapeutic drug target for such diseases as cancer, tuberculosis, and microbial infections. One potential strategy for the above diseases is interference with ATP synthase by natural or synthetic inhibitors that deprive the cells of required energy, leading to cell death [13–18]. Thus, a better understanding of this enzyme may impact such diseases specifically, and medicine and biology more generally.
A variety of natural and synthetic molecules, including peptides, are known to have antimicrobial, antitumor, antiviral, and antifungal properties [19–22]. Several positively charged peptides with or without a c-terminal amide group have been shown to bind and inhibit wild type ATP synthase [23]. Mutagenic studies of thermophilic Bacillus PS3 suggested that the βDELSEED-motif that consists of residues 380–386 is the site for an intrinsic inhibitor because replacement of D or E residues by A resulted in a loss of inhibition [24–26]. Among known inhibitory peptides a majority possess antimicrobial activity and are thus known as antimicrobial peptides (AMPs). AMPs are found extensively among microbes, plants, invertebrates, and vertebrates. They show potent activity against gram-positive and gram-negative bacteria, fungi, parasites, and viruses and are known to play an important role in vertebrate innate immunity [27].
AMPs were first described in insects as an inducible system of protection against bacterial infection [27–30]. They have been isolated from microbes, plants, invertebrates, and vertebrates, and have been shown to exhibit inhibitory activity against bacteria, fungi, and enveloped viruses [31]. A large number of AMPs are also known to have selective anticancer activity [32]. AMPs have been shown to have a neutralizing effect on bacterial endotoxins [33–35] and multiple additional inhibitory properties with unclear modes of action [36, 37]. At present there are 2427 entries in the Antimicrobial Peptide Database (APD) [38] (http://aps.unmc.edu/AP/main.php) with 82% identified as having antibacterial activity, 37% with antifungal activity, 7% with anticancer activity, and 6% with antiviral activity. The mean number of residues per peptide in the APD is 32 [38].
Antimicrobial peptides have been used in several clinical trials [17, 22, 39, 40]. Multidrug resistant bacterial pathogens necessitate new antimicrobials treatment options for infections, particularly in neonates and children [22]. Cationic AMPs have potential as antimicrobial drugs, especially against multidrug-resistant microbes such as methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE) and multidrug resistant Pseudomonas aeruginosa [41–43]. Cationic AMPs have also been shown to influence the immune system of mammals including humans, where they enhance phagocytosis, wound healing, mobilize numerous immune cells, and up or down regulate the production of cytokines and chemokines in a variety of cell types [44]. Cancer cells exhibit numerous membrane protein targets that inhibitors could bind as possible therapeutic molecules [45].
In previous study we found that melittin, melittin related peptide, and several structurally similar peptides inhibit E. coli ATP synthase. These positively charged amphipathic peptides were assumed to bind at βDELSEED-motif of ATP synthase (Fig. 1) [23]. In order for ATP synthase to be used as a peptide drug target it is of paramount importance to characterize the peptide binding site on the enzyme. Therefore, we embarked on the mutagenic analysis of the proposed peptide binding site (βDELSEED-motif). Proper understanding of the nature of the peptide binding pocket may facilitate structural modifications of peptides for use as antimicrobial and anti-cancer agents. Moreover, inhibitory studies of wild-type and mutant E. coli ATP synthase with peptides may divulge valuable information on structural and functional relationships and could provide a basis for the development of new therapies. Based on mutagenic analysis we present direct evidence of antimicrobial peptides binding at the βDELSEED-motif of ATP synthase using membrane bound E. coli F1Fo-ATP synthase preparations.
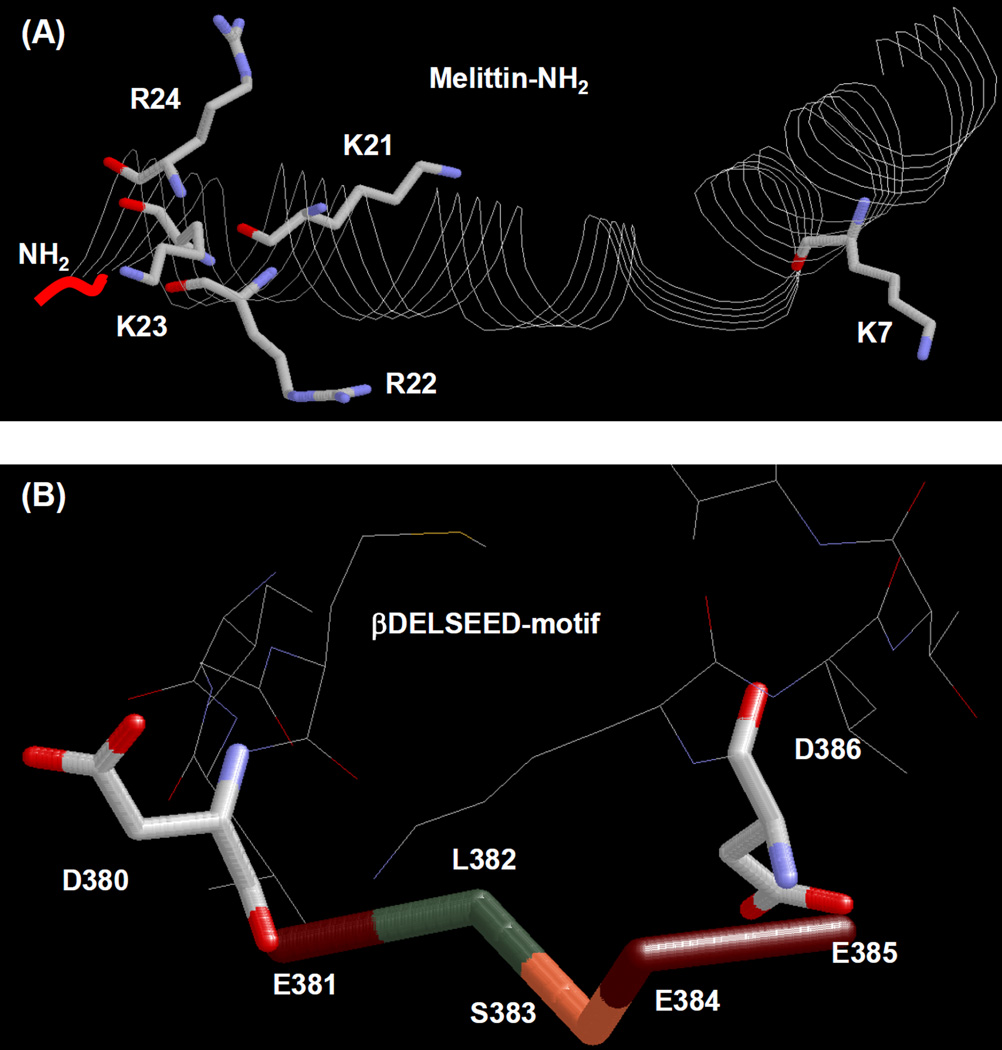
Fig. 1. X-ray structures of melittin-amide and βDELSEED-motif of mitochondrial F1-ATPase.
(A) the 26-residue long structure of the α-helical honey bee (Apis mellifera) venom peptide melittin, was generated by using PDB file 2MLT [86]. C-terminal amide group (NH2) along with five positively charged residues is identified. PDB file 1H8E was used for mitochondrial ATP synthase showing βDELSEED residues [65]. RasMol molecular visualization software was used to generate these figures [87].
Materials and methods
Construction of wild type and mutant E. coli strains
Wild type E. coli strain pBWU13.4/DK8 was used in all experiments [46]. Mutant strains were produced by Stratagene QuikChange Lightning Site-Directed Mutagenesis Kit from Agilent Technologies (catalog #210519-12). Template for PCR introduced base substitutions was M13mp18 template that contained the Hind III-XbaI fragment from pSN6. The pSN6 plasmid carries the βY331W mutation from plasmid pSWM4 [47] introduced on a SacI-EagI fragment into pBWU13.4 [46] and has all the ATP synthase genes. The mutagenic oligonucleotides for single mutants were: βD380A, CCTGGGTATGGCTGAACTGTCTG; βD380R, CCTGGGTATGCGTGAACTGTCTTG; βD380Q, CCTGGGTATGCAGGAACTGTCTG; and βD386A, GTCTGAAGAAGCCAAACTGGTGG; βD386R, CTGTCTGAAGAACGCAAACTGGTGG; βD386Q, CTGTCTGAAGAACAGAAACTGGTGG, where bold underlined bases identify the mutation. DNA sequencing was performed to confirm the presence of mutations and absence of undesired sequence changes. Mutations were transferred to pSN6 on SacI-EagI fragment, generating the new plasmids pZA31 (βD380A/βY331W), pZA32 (βD380R/βY331W), pZA33 (βD380Q/βY331W), pZA53 (βD386A/βY331W), pZA54 (βD386R/βY331W), and pZA55 (βD386Q/βY331W). Plasmids with double mutants pZA62 (βD380A/βD386A), pZA63 (βD380R/βD386R), and pZA64 (βD380Q/βD386Q), were generated by combining pZA31, pZA32, pZA33, and pZA53, pZA54, and pZA55. All plasmids were transformed into E. coli DK8 competent cells that did not express ATP synthase [48]. New mutant strains contained the βY331W Trp mutation which does not affect function, but is used for the measurement of nucleotide binding parameters [47] and was included for possible future use.
Measurement of growth yield in limiting glucose medium; preparation of E. coli membranes; assay of ATPase activity of membranes
ATP synthesis by oxidative phosphorylation was measured by growth on succinate plates (a nonfermentable carbon source). Oxidative and substrate level phosphorylation were measured on limiting glucose (3–5 mM glucose) [49]. ATP synthase bound to cell membranes were prepared as described by Senior et al. [50] using three washes of the initial membrane pellets obtained by French Press and centrifugation. The first wash was a buffer consisting of 50 mM TES, pH 7.0, 15% glycerol, 40 mM 6-aminohexonic acid, and 5 mM p-aminobenzamidine. The two final washes used a buffer of 5 mM TES, pH 7.0, 15% glycerol, 40 mM 6-aminohexonic acid, 5 mM p-aminobenzamidine, 0.5 mM DTT, and 0.5 mM EDTA. Membranes were washed twice more before use by resuspension and ultracentrifugation in 50mM TrisSO4, pH 8.0, 2.5 mM MgSO4.
ATPase activities were measured in 1 ml assay buffer (ATPase cocktail) containing 10 mM NaATP, 4 mM MgCl2, 50 mM TrisSO4, and pH 8.5 at 37 °C. Reactions were started by addition of 1 ml assay buffer to membrane bound F1Fo ATP synthase (membranes) and stopped by addition of 1 ml 10% SDS. Pi release was assayed as in Taussky and Shorr [51]. For both wild-type and mutant membranes 20 – 30 µg of protein was used for the ATPase assay with reaction times from 5–10 min. All reactions were shown to be linear with time and protein concentration. Protein subunit composition and purity were checked by 10% acrylamide SDS-gel electrophoresis and immunoblotting with rabbit polyclonal anti-F1-α and anti-F1-β antibodies [52, 53].
Peptides
Melittin related peptide-amide (MRP-amide) and Melittin-amide were purchased from Biomatik (http://www.biomatik.com). All peptides were determined to have greater than 95% purity by HPLC. Lyophilized powder was stored at −20°C upon receipt and resuspended in deionized water for use as needed.
Inhibition of ATPase Activity by Melittin-amide and MRP-amide
Both wild-type and mutant membrane bound F1Fo ATP synthase were preincubated with different concentrations of peptides for 60 minutes at room temperature (RT) in 50 mM TrisSO4 at pH 8.0. 1 ml ATPase cocktail was used to measure the enzyme activity. The reaction was stopped by the addition of 1 ml SDS to a final concentration of 3.3% (v/v). The addition of an equal volume of Taussky and Shorr reagent gave rise to a blue color that was spectrophotometrically assayed at OD700 [51]. Inhibitory exponential decay and piecewise curves were generated with Sigma plot 10.0. The range of absolute specific activity for membrane bound F1Fo was 11–16 µmol/min/mg at 30 °C for different preparations. Relative ATPase activity was calculated from the absolute values of wild-type in absence of peptides taken as 100%.
Reversal of peptide inhibited ATPase activity
Reversal of inhibition was assayed by dilution of the membrane enzyme. Membranes were first reacted with the maximal inhibitory concentration of peptides for 1 hour at room temperature. These concentrations were based on the maximal inhibition of the ATP synthase (see Fig. 2 and Fig. 3). 50 mM TrisSO4 pH 8.0 buffer was then added to bring the peptide concentrations to non-inhibitory levels, with an additional 1 hour incubation at room temperature before ATPase assay.
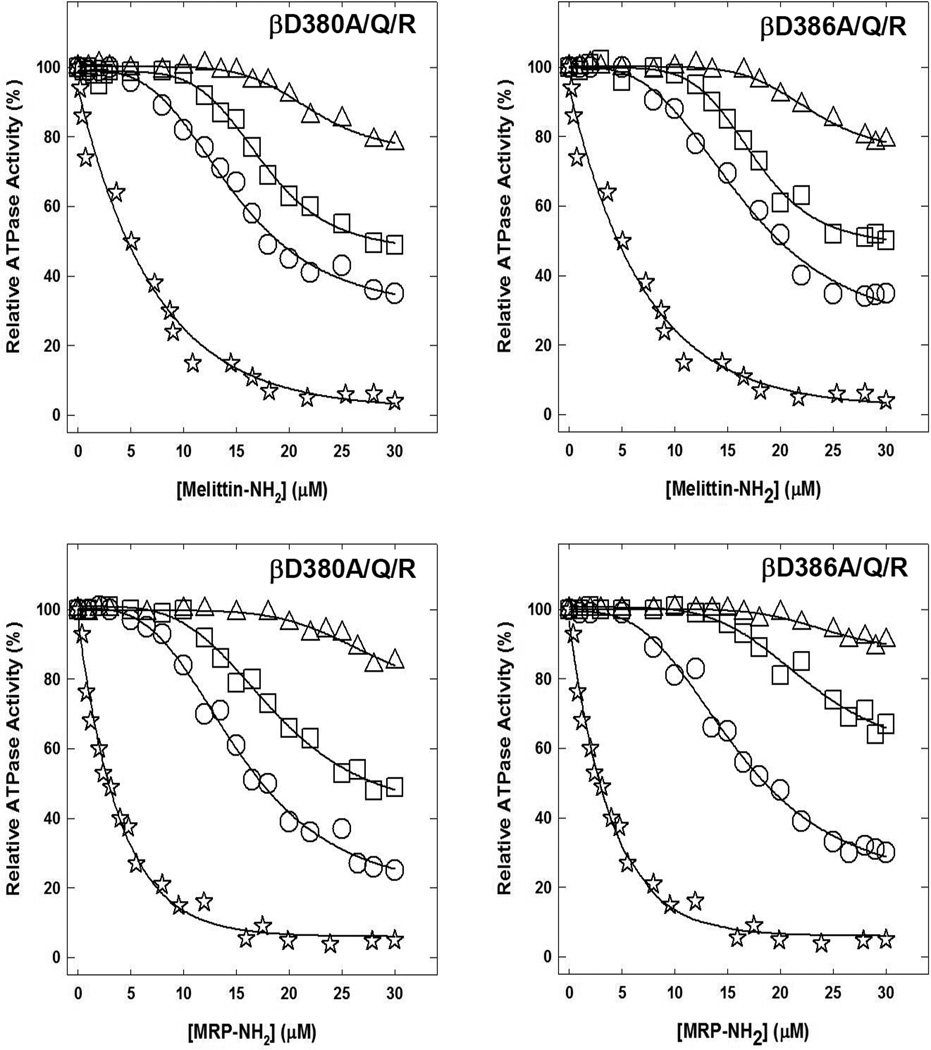
Fig. 2. Inhibition of membrane bound wild-type and mutant ATP synthase by melittin-amide and MRP-amide.
Membranes were preincubated for 60 min at room temperature with varied concentrations of melittin-amide or MRP-amide, then aliquots were added to 1 ml of assay buffer and ATPase activity determined. Materials and Methods section contains the detailed procedure. Symbols used are:, ✰,wild-type; ○, βD380A or βD386A; □, βD380Q or βD386Q; △, βD380R or βD386R. Each data point represents an average of three to four experiments, using 2–3 independent membrane preparations of each mutant. Results agreed within ± 5%.
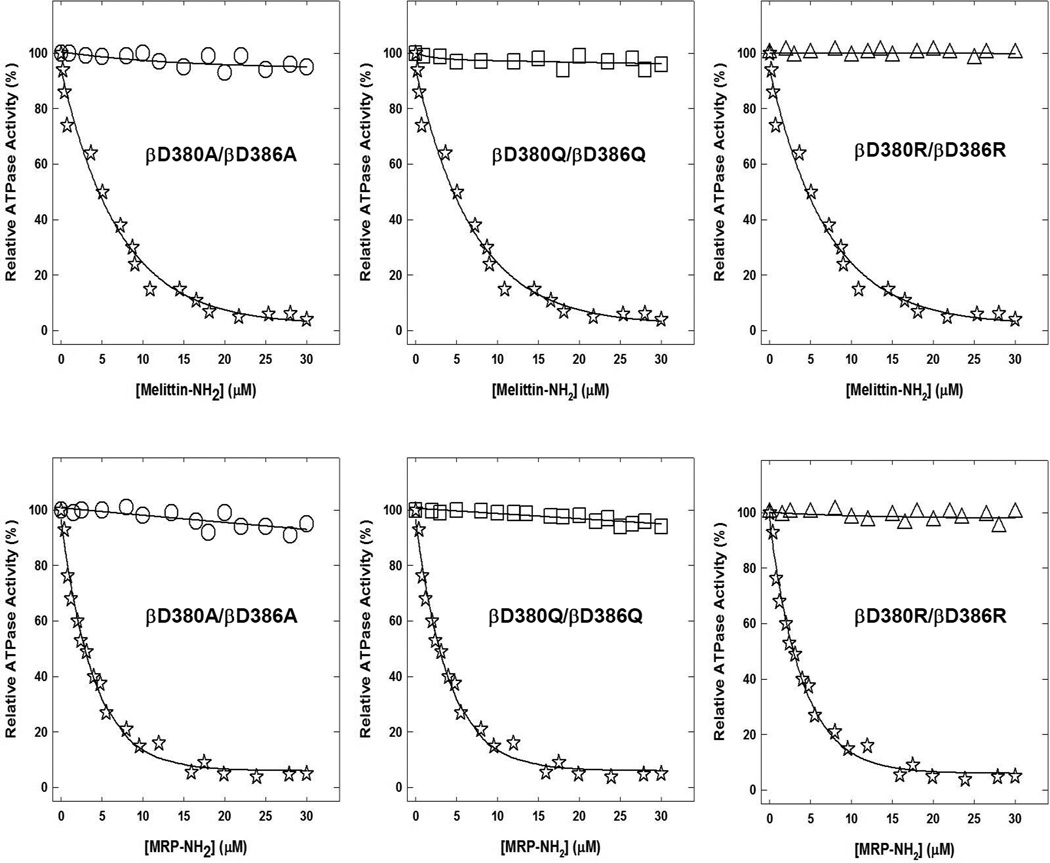
Fig. 3. Melittin-amide and MRP-amide induced inhibition of membrane bound F1Fo ATP synthase of wild-type and double mutants.
Membranes were preincubated for 60 min at room temperature with varied concentration of melittin-amide or MRP-amide, then aliquots added to 1 ml of assay buffer and ATPase activity determined. For experimental details see materials and methods section. Symbols used are:, ✰,wild-type; ○, βD380A/βD386A; □, βD380Q/βD386Q; △, βD380R/βD386R. Each data point represents an average of three to four experiments, using two to three independent membrane preparations of each mutant. Results agreed within ± 5%.
Results
Properties of single βD380A/Q/R, βD386A/Q/R and double βD380A/Q/R-βD386A/Q/R mutants of E. coli ATP synthase
The significance of βDELSEED motif residues βAsp-380 and βAsp-386 in peptide binding were investigated using a series of mutations. βAsp-380 and βAsp-386 were changed to Ala, Gln, and Arg individually, or as double mutants, βD380A/βD386A, βD380Q/βD386Q, or βD380R/βD386R. These two negatively charged residues were chosen to confirm their proposed role in the binding of cationic peptides as well as to determine the peptide induced inhibition mechanism. Asp→Ala removes the charge and mass, Asp→Gln, removes the charge while preserving the volume occupied by side chain by adding little mass, and Asp→Arg adds positive charge and adds some mass too. Double mutations were used to evaluate the overall role of Asp residues in peptide binding.
Duplicate or triplicate preparations of the mutant membranes gave consistently near normal specific activity. Expected mutant ATPase activities also confirm that mutant membranes contained the same amount of α and β subunits as wild-type. Previous work established that inhibition profiles of mutant and wild-type ATP synthase can be assayed using either membrane preparations or purified F1 with comparable results [54–60]. Coomassie Blue-stained SDS-gel electrophoresis, immunoblotting and densitometry of mutant and wild-type membranes (with purified wild-type F1 as reference) established the purity and integrity of F1-α and F1-β subunits [52, 53].
Partial and tardy inhibition of membrane bound F1Fo ATP synthase mutants βD380A/Q/R or βD386A/Q/R by melittin-amide and MRP-amide
Fig. 2 shows the melittin-amide and MRP-amide induced inhibitory profiles of single βD380A/Q/R and βD386A/Q/R mutants. All six single mutants showed decreased inhibition. The melittin-amide induced maximal inhibition in the βD380 and βD386 for replacement of Asp → Ala was ~65%, for Asp → Gln was ~50%, and for Asp → Arg was ~20%. MRP-amide induced inhibition in the βD380 and βD386 for replacement of Asp → Ala was ~75% and 70%, for Asp → Gln was ~50% and 35%, and for Asp → Arg was ~15% and 10% respectively. Multiple previous studies have shown varied maximal inhibition of ATP synthase where mutant or wild-type ATP synthase was partially or incompletely inhibited by natural or synthetic inhibitors such as dietary polyphenols, peptides, NBD-Cl, NaN3, AlCl3 or ScCl3 [23, 54, 60–64].
To affirm that all mutants were maximally inhibited by peptides, we incubated each mutant membrane bound F1Fo preparation with melittin-amide (25µM) and MRP-amide (20µM) with maximal inhibitory concentrations for one hour as in Fig. 2 and Fig. 3. This was followed by a secondary pulse of the same inhibitory peptide concentrations and continued incubation for an additional hour before ATPase assay. It was found that little or no additional inhibition occurred. This shows that peptides fully reacted at the peptide binding site, which resulted in maximal inhibition, and yet residual activity could be retained by mutant enzymes.
Trivial or no inhibition of membrane bound F1Fo ATP synthase double mutants βD380A/βD386A, βD380Q/βD386Q, or βD380R/βD386R by melittin-amide or MRP-amide
The maximal inhibition of βD380A/βD386A or βD380Q/βD386Q double mutant in the presence of melittin-amide or MRP-amide was ~5%, while double mutant β380R/β386R was not inhibited at all (Fig. 3).
Reversal of ATPase activity of membrane bound mutant enzymes from peptide inhibition
Membrane bound mutant enzymes were inhibited with the maximum inhibitory concentrations of peptides for one hour at RT as in Fig. 2 and Fig. 3. Samples were then diluted to a non-inhibitory concentration by adding TrisSO4 pH 8.0 buffer, and ATPase activity was then measured. The inhibitory action by both melittin-amide and MRP-amide was found to be fully reversible in all membrane bound mutant enzymes used in this study.
Effect of peptides on the growth of mutant E. coli cells on limiting glucose and succinate plates
Mutant E. coli strains were grown on succinate plates, limiting glucose, or LB media in the presence or absence of peptides to study their effect on ATP synthesis (Table 1 and Table 2). Table 1 shows the results of oxidative phosphorylation in vivo by growth both on succinate-containing plates and in limiting glucose medium, in absence of peptides. Mutants showed somewhat less than or near normal growth on succinate. Growth on limiting glucose was reduced for βD386R and double mutations while growth for the βD380A/Q/R mutations did not show much effect. Essentially, mutations did not seriously impair ATP synthesis in vivo. Contrary to wild-type (positive control) which showed almost no growth in presence of melittin-amide, or MRP-amide, mutant and null strains (negative control) grew normally.
TABLE 1.
Effect of βD380 and βD386 mutations on cell growth and ATPase activity
| Mutationa | Growthb On succinate |
Growth yield in limiting glucose (%) |
ATPase Activityc µmol/min/mg |
|---|---|---|---|
| Wild-type | ++++ | 100 | 16 |
| Null | − | 46 | 0 |
| βY331W | ++++ | 96 | 15 |
| βD380A | ++++ | 91 | 15 |
| βD380Q | ++++ | 97 | 15 |
| βD380R | ++++ | 89 | 14 |
| βD386A | +++ | 80 | 14 |
| βD386Q | +++ | 84 | 13 |
| βD386R | +++ | 74 | 13 |
| βD380A/βD386A | +++ | 78 | 13 |
| βD380Q/βD386Q | +++ | 77 | 13 |
| βD380R/βD386R | +++ | 69 | 12 |
Wild-type, pBWU13.4/DK8; Null, pUC118/DK8. All mutants were expressed with the βY331W mutation also present, which does not significantly affect growth. Data are means of four to six experiments each.
Growth on succinate plates after 3 days estimated by eye. ++++, heavy growth; +++, lesser growth; −, no growth.
Measured at 30°C and expressed as µmol ATP hydrolyzed/min/mg membrane protein. Each individual experimental point is itself the mean of duplicate assay tubes. Data are derived from two separate membrane preparations. Results from separate membrane preparations were in excellent agreement.
TABLE 2.
Growth of mutant E. coli strains in presence of melittin-amide and MRP-amide
| Mutationa | Growthb On succinate plates (Melittin-amide/MRP-amide) |
Growth yield in limiting glucosec (Melittin-amide/MRP-amide) (%) |
|---|---|---|
| Wild-type | ±/± | 20/25 |
| Null | −/− | 43/46 |
| βY331W | ±/± | 22/24 |
| βD380A | +++/+++ | 89/92 |
| βD380Q | +++/+++ | 90/88 |
| βD380R | +++/+++ | 82/83 |
| βD386A | +++/+++ | 79/77 |
| βD386Q | +++/+++ | 74/72 |
| βD386R | +++/+++ | 70/69 |
| βD380A/βD386A | +++/+++ | 75/70 |
| βD380Q/βD386Q | +++/+++ | 72/75 |
| βD380R/βD386R | ++/++ | 66/70 |
Wild-type, pBWU13.4/DK8; Null, pUC118/DK8. Data are means of four to six experiments each.
Growth on succinate plates after 3 days estimated by eye. ++++, normal growth; +++, less growth; ++, lesser growth; ±, insignificant growth; −, no growth.
Growth on limiting glucose is based on the growth of wild type in absence of peptides and used as 100%. Each individual experimental point is itself the mean of duplicate assay tubes. Data are derived from two-three separate membrane preparations. Results from separate membrane preparations were in excellent agreement.
Discussion
Our goal in this study was to understand the role of the two negatively charged residues, Asp-380 and Asp-386 of the conserved βDELSEED-motif, in peptide binding and inhibition of ATP synthase by peptides. X-ray crystallographic structure of ATP synthase [65] shows the βAsp-380 and βAsp-386 residues forming a perfect cavity by flanking the corners of DELSEED-motif (Fig. 1B). Binding of cationic peptides is thought to be the basis for peptide induced inhibition of ATP synthase. Thus proper knowledge of the amino acids involved in peptide binding is essential for the further development of peptide based cell growth inhibition.
Several peptides including melittin and MRP were found to have variable ATPase inhibitory activity [23–25]. It was shown that both melittin and MRP strongly inhibited the ATPase activity. Moreover, the presence of an amide group at the c-terminal resulted in additional ~20% inhibition. Many peptides show multifunctional effects, i.e. have antimicrobial and antitumor properties, as well as regulatory effects through ATPase inhibition [66]. For example, mitochondrial ATPase inhibitors PSK and SK84 peptides were shown to have potent bactericidal and anticancer properties [67, 68]. Partial or complete inhibition of ATPase has also been observed in the presence of natural or structurally modified polyphenol compounds. Similar inhibition of ATPase activity has also been linked to the reduced growth of bacterial or tumor cells [61–63, 69, 70].
Our results show that single or double mutants βD380A, βD380Q, βD380R, βD386A, βD386Q, βD386R, βD380A/βD386A, βD380Q/βD386Q, or βD380R/βD386R did not affect subunit assembly or structural integrity of membrane bound F1Fo ATP synthase. All mutants were found to have corresponding components of F1-α and β subunits as compared to wild type. Single mutants βD380A, βD380Q, or βD380R showed insignificant inhibitory effects on oxidative phosphorylation as judged by growth on succinate or limiting glucose medium, and did not cause loss of ATPase activity. However, the βD386A, βD386Q, βD386R, βD380A/βD386A, βD380Q/βD386Q, or βD380R/βD386R mutations showed some loss of oxidative phosphorylation, but insignificant change in ATPase activity (Table 1). Another important result is the observed growth patterns of mutant E. coli cells in the presence of melittin-amide and MRP-amide (Table 2). It was found that while both melittin-amide and MRP-amide abrogated growth of wild type E. coli cells, the growth of mutant cells was not much affected. All mutants used grew in either the presence or absence of peptides (Table 1 and Table 2).
Some loss of growth in double mutants βD380A/βD386A, βD380Q/βD386Q, or βD380R/βD386R, without reduction in ATPase activity could be attributed to the possibility is that peptide binding parameters are different during ATP synthesis than during ATP hydrolysis. It is also possible that, in vivo, a modicum of activity that may not be detected by our in vitro assays is sufficient to preserve some synthase function and cell viability. Similar results from past studies have been observed for αPhe-291 in Pi binding [59]. Our null strain (pUC118/DK8) will usually grow 40–50% relative to wild-type (pBWU13.4/DK). This is because the null strain uses only glycolysis to generate ATP whereas the wild-type uses both glycolysis and oxidative phosphorylation.
As expected, inhibitory profiles of single mutants βD380A, βD380Q, βD380R, or βD386A, βD386Q, βD386R showed incremental residual activity with Ala, Gln, and Arg replacements respectively (Fig. 2). Insertion of a positive charge at βD380R apparently repelled cationic peptides, and resulted in only 20% and 15% inhibition (80% and 85% residual activity) in the presence of melittin-amide and MRP-amide respectively. Insertion of Arg at βD386R prevented inhibition by 80% and 90% (20% and 10% inhibition) in presence of melittin-amide and MRP-amide. Changing Asp to Gln in βD380Q and βD386Q resulted in inhibition of 50% to 35% inhibition. Asp to Ala mutations (βD380A and βD386A) caused 65% to 75% inhibition. While double mutants βD380A/βD386A or βD380Q/βD386Q where slightly inhibited (~5%), no inhibition occurred for βD380R/βD386R in the presence of either melittin-amide or MRP-amide (Fig. 3). This inhibitory trend of double mutants is consistent with peptide induced inhibition of single mutants.
Presence of positively charged Arg at βDELSEED seemingly repels positively charged peptides and thus prevents binding and inhibition. Introduction of Gln at βDELSEED reduces the electrostatic attraction of Asp for cationic peptides and results in partial inhibition. Replacement of Asp with Ala removes both the charge and mass of Asp. Although this mutation removes the negative charge attraction for cationic peptides, it also decreases steric hindrance of the Asp side chain. Thus the observed partial inhibition of Ala mutations. In summary all of the mutations used in this study were refractory to melittinamide and MRP-amide induced inhibition to variable degrees, supporting the hypothesis that of βAsp-380 and βAsp-386 are essential for peptide binding.
Previous work suggested that βDELSEED was involved in a coupling between catalysis and rotation. It was shown that the Bacillus PS3 ATP synthase mutants generated by the deletion of 3–7 amino acids within the βDELSEED-motif retained the catalytic activity, and in some cases the activity was found to be higher than the wild-type enzyme [71]. Deleting 7–14 amino acids from or near to this motif suggested that a minimum length is critical for its role in the coupling between catalysis and rotation [72]. Similarly, the βD380 and βD386 mutants used in this study, which did not change the length of polypeptide chain, did not reduce ATPase activity.
The process of inhibition was found to be completely reversible. Membrane bound F1Fo regained activity once brought back to non-inhibitory peptide concentrations by dilution with TrisSO4 buffer. Reversibility indicates non-covalent binding of peptides to the mutant enzymes, as was previously shown in wild-type [23].
Peptides are potential sources of compounds with useful pharmacological properties and medical utility in antimicrobial [73, 74] and anticancer applications [75]. Several mechanisms have been hypothesized for the activity of AMPs, including a membrane permeabilization and cell death model proposed for magainins [76, 77] and a non-pore-dependent cytolytic activity by dermaseptins that causes membrane bilayer miscellization and disintegration [78]. Peptides are known to affect Gram-negative and Gram-positive bacteria, fungi, enveloped viruses, eukaryotic parasites, and cancer cells [75, 79]. Apoptosis via a mitochondrial pathway using ATP synthase as a molecular drug target for a variety of natural and synthetic inhibitors has been previously demonstrated [80–83]. Synergistic effects with AMPs among different α-helical peptides have also been observed [84] and may be a reason for the large number of different isoforms found in several amphibian species [85]. Such observations suggest that the evaluation of potential ATP synthase inhibitory activity by AMPs may be enhanced by combinatorial studies. We conclude that further characterization of the peptide binding βDELSEED-motif, is a promising avenue for understanding ATP synthase as a potential drug target for peptides.
Acknowledgements
This work was supported by the National Institutes of Health Grant GM085771 to ZA.
Abbreviations used
- MRP
melittin related peptide
- AMP
anti-microbial peptide
- membrane
membrane bound F1Fo ATP synthase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Senior AE. J Biol Chem. 2012;287:30049–30062. doi: 10.1074/jbc.X112.402313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad Z, Cox JL. ScientificWorldJournal. 2014;2014:567398. doi: 10.1155/2014/567398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wada Y, Sambongi Y, Futai M. Biochim Biophys Acta. 2000;1459:499–505. doi: 10.1016/s0005-2728(00)00189-4. [DOI] [PubMed] [Google Scholar]
- 4.Senior AE, Nadanaciva S, Weber J. Biochim Biophys Acta. 2002;1553:188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- 5.Abrahams JP, Leslie AG, Lutter R, Walker JE. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 6.Itoh H, Takahashi A, Adachi K, Noji H, Yasuda R, Yoshida M, Kinosita K. Nature. 2004;427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 7.Diez M, Zimmermann B, Borsch M, Konig M, Schweinberger E, Steigmiller S, Reuter R, Felekyan S, Kudryavtsev V, Seidel CA, Graber P. Nat Struct Mol Biol. 2004;11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- 8.Noji H, Yoshida M. J. Biol. Chem. 2001;276:1665–1668. doi: 10.1074/jbc.R000021200. [DOI] [PubMed] [Google Scholar]
- 9.Weber J, Senior AE. FEBS Lett. 2003;545:61–70. doi: 10.1016/s0014-5793(03)00394-6. [DOI] [PubMed] [Google Scholar]
- 10.Martin JL, Ishmukhametov R, Hornung T, Ahmad Z, Frasch WD. Proc Natl Acad Sci U S A. 2014;111:3715–3720. doi: 10.1073/pnas.1317784111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad Z, Okafor F, Laughlin TF. J Amino Acids. 2011;2011:785741. doi: 10.4061/2011/785741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad Z, Senior AE. J Bioenerg Biomembr. 2005;37:437–440. doi: 10.1007/s10863-005-9486-8. [DOI] [PubMed] [Google Scholar]
- 13.Hong S, Pedersen PL. Microbiol. Mol. Biol. Rev. 2008;72:590–641. doi: 10.1128/MMBR.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen PL. J. Bioenerg. Biomembr. 2007;39:1–12. doi: 10.1007/s10863-007-9070-5. [DOI] [PubMed] [Google Scholar]
- 15.Schagger H, Ohm TG. Eur J Biochem. 1995;227:916–921. doi: 10.1111/j.1432-1033.1995.tb20219.x. [DOI] [PubMed] [Google Scholar]
- 16.Ahmad Z, Laughlin TF. Curr Med Chem. 2010;17:2822–2836. doi: 10.2174/092986710791859270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmad Z, Okafor F, Azim S, Laughlin TF. Curr Med Chem. 2013;20:1956–1973. doi: 10.2174/0929867311320150003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nesci S, Ventrella V, Trombetti F, Pirini M, Pagliarani A. Med Hypotheses. 2014;83:160–165. doi: 10.1016/j.mehy.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Wu D, Gao Y, Qi Y, Chen L, Ma Y, Li Y. Cancer Lett. 2014;351:13–22. doi: 10.1016/j.canlet.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Reygaert WC. Front Microbiol. 2014;5:434. doi: 10.3389/fmicb.2014.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosseinzadeh H, Mehri S, Abolhassani MM, Ramezani M, Sahebkar A, Abnous K. Daru. 2013;21:25. doi: 10.1186/2008-2231-21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashby M, Petkova A, Hilpert K. Curr Opin Infect Dis. 2014;27:258–267. doi: 10.1097/QCO.0000000000000057. [DOI] [PubMed] [Google Scholar]
- 23.Laughlin TF, Ahmad Z. Int J Biol Macromol. 2010;46:367–374. doi: 10.1016/j.ijbiomac.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gledhill JR, Walker JE. Biochem J. 2005;386:591–598. doi: 10.1042/BJ20041513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bullough DA, Ceccarelli EA, Roise D, Allison WS. Biochim Biophys Acta. 1989;975:377–383. doi: 10.1016/s0005-2728(89)80346-9. [DOI] [PubMed] [Google Scholar]
- 26.Kato-Yamada Y, Bald D, Koike M, Motohashi K, Hisabori T, Yoshida M. J Biol Chem. 1999;274:33991–33994. doi: 10.1074/jbc.274.48.33991. [DOI] [PubMed] [Google Scholar]
- 27.Jakubke H-D, Sewald N. Peptides from A to Z : a concise encyclopedia. Weinheim: Wiley-VCH; 2008. [Google Scholar]
- 28.Hultmark D, Steiner H, Rasmuson T, Boman HG. Eur J Biochem. 1980;106:7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x. [DOI] [PubMed] [Google Scholar]
- 29.Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. Nature. 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 30.Boman HG, Nilsson I, Rasmuson B. Nature. 1972;237:232–235. doi: 10.1038/237232a0. [DOI] [PubMed] [Google Scholar]
- 31.Powers JP, Hancock RE. Peptides. 2003;24:1681–1691. doi: 10.1016/j.peptides.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 32.Hoskin DW, Ramamoorthy A. Biochim Biophys Acta. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenfeld Y, Papo N, Shai Y. J Biol Chem. 2006;281:1636–1643. doi: 10.1074/jbc.M504327200. [DOI] [PubMed] [Google Scholar]
- 34.Martin GS, Mannino DM, Eaton S, Moss M. N Engl J Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 35.Mangoni ML, Shai Y. Biochim Biophys Acta. 2009;1788:1610–1619. doi: 10.1016/j.bbamem.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 36.da Rocha Pitta MG, Galdino SL. Curr Protein Pept Sci. 2010;11:236–247. doi: 10.2174/138920310791112066. [DOI] [PubMed] [Google Scholar]
- 37.Reddy KV, Yedery RD, Aranha C. Int J Antimicrob Agents. 2004;24:536–547. doi: 10.1016/j.ijantimicag.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Wang G. Nucleic Acids Res. 2004;32:D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butler MS, Cooper MA. J Antibiot (Tokyo) 2011;64:413–425. doi: 10.1038/ja.2011.44. [DOI] [PubMed] [Google Scholar]
- 40.Hancock RE. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 41.Cai Y, Chai D, Wang R, Liang B, Bai N. J Antimicrob Chemother. 2012;67:1607–1615. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- 42.Saravolatz LD, Pawlak J, Johnson L, Bonilla H, Saravolatz LD, 2nd, Fakih MG, Fugelli A, Olsen WM. Antimicrob Agents Chemother. 2012;56:4478–4482. doi: 10.1128/AAC.00194-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uccelletti D, Zanni E, Marcellini L, Palleschi C, Barra D, Mangoni ML. Antimicrob Agents Chemother. 2010;54:3853–3860. doi: 10.1128/AAC.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang D, Biragyn A, Kwak LW, Oppenheim JJ. Trends Immunol. 2002;23:291–296. doi: 10.1016/s1471-4906(02)02246-9. [DOI] [PubMed] [Google Scholar]
- 45.Shadidi M, Sioud M. Drug Resist Updat. 2003;6:363–371. doi: 10.1016/j.drup.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Ketchum CJ, Al-Shawi MK, Nakamoto RK. Biochem J. 1998;330(Pt 2):707–712. doi: 10.1042/bj3300707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber J, Wilke-Mounts S, Lee RS, Grell E, Senior AE. J Biol Chem. 1993;268:20126–20133. [PubMed] [Google Scholar]
- 48.Klionsky DJ, Brusilow WS, Simoni RD. J Bacteriol. 1984;160:1055–1060. doi: 10.1128/jb.160.3.1055-1060.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Senior AE, Latchney LR, Ferguson AM, Wise JG. Arch Biochem Biophys. 1984;228:49–53. doi: 10.1016/0003-9861(84)90045-6. [DOI] [PubMed] [Google Scholar]
- 50.Senior AE, Langman L, Cox GB, Gibson F. Biochem J. 1983;210:395–403. doi: 10.1042/bj2100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taussky HH, Shorr E. J. Biol. Chem. 1953;202:675–685. [PubMed] [Google Scholar]
- 52.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 53.Rao R, Perlin DS, Senior AE. Arch Biochem Biophys. 1987;255:309–315. doi: 10.1016/0003-9861(87)90398-5. [DOI] [PubMed] [Google Scholar]
- 54.Ahmad Z, Senior AE. J Biol Chem. 2004;279:31505–31513. doi: 10.1074/jbc.M404621200. [DOI] [PubMed] [Google Scholar]
- 55.Ahmad Z, Senior AE. J Biol Chem. 2004;279:46057–46064. doi: 10.1074/jbc.M407608200. [DOI] [PubMed] [Google Scholar]
- 56.Ahmad Z, Senior AE. J Biol Chem. 2005;280:27981–27989. doi: 10.1074/jbc.M503955200. [DOI] [PubMed] [Google Scholar]
- 57.Ahmad Z, Senior AE. FEBS Lett. 2005;579:523–528. doi: 10.1016/j.febslet.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 58.Ahmad Z, Senior AE. FEBS Lett. 2006;580:517–520. doi: 10.1016/j.febslet.2005.12.057. [DOI] [PubMed] [Google Scholar]
- 59.Brudecki LE, Grindstaff JJ, Ahmad Z. Arch Biochem Biophys. 2008;471:168–175. doi: 10.1016/j.abb.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 60.Li W, Brudecki LE, Senior AE, Ahmad Z. J Biol Chem. 2009;284:10747–10754. doi: 10.1074/jbc.M809209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahmad Z, Ahmad M, Okafor F, Jones J, Abunameh A, Cheniya RP, Kady IO. Int J Biol Macromol. 2012;50:476–486. doi: 10.1016/j.ijbiomac.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dadi PK, Ahmad M, Ahmad Z. Int J Biol Macromol. 2009;45:72–79. doi: 10.1016/j.ijbiomac.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 63.Chinnam N, Dadi PK, Sabri SA, Ahmad M, Kabir MA, Ahmad Z. Int J Biol Macromol. 2010;46:478–486. doi: 10.1016/j.ijbiomac.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahmad Z, Winjobi M, Kabir MA. Biochemistry. 2014;53:7376–7385. doi: 10.1021/bi5013063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menz RI, Walker JE, Leslie AG. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- 66.Lee SR, Kurata S, Natori S. FEBS Lett. 1995;368:485–487. doi: 10.1016/0014-5793(95)00717-n. [DOI] [PubMed] [Google Scholar]
- 67.Lu J, Chen ZW, Wu Y, Zhang M, Ding JP, Cederlund E, Jornvall H, Bergman T. Biochem Biophys Res Commun. 2014;446:519–522. doi: 10.1016/j.bbrc.2014.02.138. [DOI] [PubMed] [Google Scholar]
- 68.Lu J, Chen ZW. Peptides. 2010;31:44–50. doi: 10.1016/j.peptides.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 69.Gledhill JR, Montgomery MG, Leslie AG, Walker JE. Proc Natl Acad Sci U S A. 2007;104:13632–13637. doi: 10.1073/pnas.0706290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sekiya M, Chiba E, Satoh M, Yamakoshi H, Iwabuchi Y, Futai M, Nakanishi-Matsui M. Int J Biol Macromol. 2014;70:241–245. doi: 10.1016/j.ijbiomac.2014.06.055. [DOI] [PubMed] [Google Scholar]
- 71.Mnatsakanyan N, Krishnakumar AM, Suzuki T, Weber J. J Biol Chem. 2009;284:11336–11345. doi: 10.1074/jbc.M900374200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mnatsakanyan N, Kemboi SK, Salas J, Weber J. J Biol Chem. 2011;286:29788–29796. doi: 10.1074/jbc.M111.254730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conlon JM, Al-Ghaferi N, Abraham B, Leprince J. Methods. 2007;42:349–357. doi: 10.1016/j.ymeth.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 74.Rinaldi AC. Curr Opin Chem Biol. 2002;6:799–804. doi: 10.1016/s1367-5931(02)00401-5. [DOI] [PubMed] [Google Scholar]
- 75.Papo N, Shai Y. Cell Mol Life Sci. 2005;62:784–790. doi: 10.1007/s00018-005-4560-2. [DOI] [PubMed] [Google Scholar]
- 76.Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 77.Matsuzaki K, Nakamura A, Murase O, Sugishita K, Fujii N, Miyajima K. Biochemistry. 1997;36:2104–2111. doi: 10.1021/bi961870p. [DOI] [PubMed] [Google Scholar]
- 78.Oren Z, Shai Y. Biopolymers. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 79.Hancock RE. Lancet Infect Dis. 2001;1:156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 80.Pervaiz S. FASEB J. 2003;17:1975–1985. doi: 10.1096/fj.03-0168rev. [DOI] [PubMed] [Google Scholar]
- 81.Clement MV, Hirpara JL, Chawdhury SH, Pervaiz S. Blood. 1998;92:996–1002. [PubMed] [Google Scholar]
- 82.Mills KI, Woodgate LJ, Gilkes AF, Walsh V, Sweeney MC, Brown G, Burnett AK. Biochem Biophys Res Commun. 1999;263:294–300. doi: 10.1006/bbrc.1999.1356. [DOI] [PubMed] [Google Scholar]
- 83.Johnson KM, Cleary J, Fierke CA, Opipari AW, Jr, Glick GD. ACS Chem Biol. 2006;1:304–308. doi: 10.1021/cb600143j. [DOI] [PubMed] [Google Scholar]
- 84.Bowie JH, Separovic F, Tyler MJ. Peptides. 2012;37:174–188. doi: 10.1016/j.peptides.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 85.Mor A, Hani K, Nicolas P. J Biol Chem. 1994;269:31635–31641. [PubMed] [Google Scholar]
- 86.Anderson D, Terwilliger TC, Wickner W, Eisenberg D. J Biol Chem. 1980;255:2578–2582. [PubMed] [Google Scholar]
- 87.Sayle RA, Milner-White EJ. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]