Abstract
Loss of NOTCH signaling in postnatal murine joints results in osteoarthritis (OA), indicating a requirement for NOTCH during joint cartilage maintenance. Unexpectedly, NOTCH components are significantly up-regulated in human and murine post-traumatic OA, suggesting either a reparative or pathological role for NOTCH activation in OA. Here we investigated the potential dual role for NOTCH in joint maintenance and OA by generating two mouse models overexpressing the NOTCH1 intracellular domain within postnatal joint cartilage; one with sustained NOTCH activation that likely resembles pathological NOTCH signaling and one with transient NOTCH activation that more closely reflects physiological NOTCH signaling. Sustained NOTCH signaling in joint cartilage leads to an early and progressive OA pathology, while on the contrary, transient NOTCH activation enhances cartilage matrix synthesis and promotes joint maintenance under normal physiological conditions. Using RNA-seq, immunohistochemical, and biochemical approaches we identified several novel targets potentially responsible for NOTCH-mediated cartilage degradation, fibrosis, and OA progression, including components of the IL6/STAT3 and ERK/p38 MAPK pathways; factors that may also contribute to post-traumatic OA development. Collectively, these data demonstrate a dual role for the NOTCH pathway in joint cartilage and identify important downstream NOTCH effectors as potential targets for disease modifying osteoarthritis drugs (DMOADs).
Keywords: Osteoarthritis, articular cartilage, NOTCH signaling, IL6, STAT3, ERK, p38
Introduction
Osteoarthritis (OA) is the most common joint disorder observed worldwide and a major cause of disability that carries an extremely high socioeconomic burden (1). In the United States alone, nearly 25% of the population is expected to have physician diagnosed OA by 2030 (2), which is estimated to cost between $100 and $200 billion annually (3, 4). OA is characterized clinically by fibrosis and degradation of the joint cartilages, osteophyte formation, subchondral bone sclerosis, and synovial tissue hyperplasia (5–7). Most of these disease outcomes are dictated by abnormal chondrocyte differentiation coupled with an imbalance in cartilage extracellular matrix (ECM) turnover. Under physiological conditions, articular and joint chondrocytes maintain a dynamic balance between synthesis and degradation of ECM components. Normal joint cartilages are largely composed of an ECM rich in Type II Collagen (COL2A1) and the proteoglycan Aggrecan (ACAN), which in the context of OA are degraded by specific collagenases (MMP13, MMP9, MMP3, etc.) and aggrecanases (ADAMTS4, ADAMTS5, etc.) (8–10). OA disease progression can also be exacerbated by inflammatory factors, such as Interleukins 1β and 6 (IL1β and IL6) and tumor necrosis factor-α (TNFα) (11–13), which both suppress matrix synthesis and promote matrix degradation. While several factors have been implicated in the pathogenesis of OA, the genetic pathways that precisely regulate the catabolic or anabolic cartilage response or the balance of these processes are just beginning to be understood (5, 14, 15). Understanding the molecular mechanisms that regulate cartilage anabolism and catabolism to maintain joint cartilage homeostasis will be extremely important in developing future disease modifying osteoarthritis drugs (DMOADs).
Recently, the NOTCH pathway was identified as a potential regulator of both catabolic and anabolic molecules in the cartilage ECM during development (16–18). In mammals, NOTCH signaling is primarily initiated when a NOTCH ligand interacts with one of the NOTCH receptors, leading to a series of receptor cleavages that ultimately releases the NOTCH intracellular domain (NICD) into the cytoplasm. The NICD then translocates to the nucleus and activates downstream target gene expression, including genes of the Hes/Hey families, via interactions with the RBPjκ-MAML transcriptional complex (19–21). NOTCH pathway components are expressed in both the developing growth plate cartilage and adult articular cartilage (22, 23), suggesting a functional role for NOTCH in regulating both cartilage development and homeostasis. We recently discovered that loss of RBPjκ-dependent NOTCH signaling in all joint tissues, as well as, postnatal joint cartilages results in an early and progressive OA-like pathology (24), indicating a requisite role for NOTCH in articular cartilage and joint maintenance. Interestingly, recent studies have also demonstrated that the NOTCH pathway is highly activated in mouse and human joint tissues during post-traumatic OA (25–27), and that temporary suppression of NOTCH signaling in murine joints leads to delayed OA progression (25). These data collectively suggest that physiological NOTCH signaling within joint tissues is essential for joint maintenance, however, when NOTCH signaling is abnormally activated such as during post-traumatic OA, temporary inhibition of the NOTCH pathway or its downstream effectors may provide a means for altering the progression of post-traumatic OA. Since the NOTCH pathway is just beginning to be understood in the context of OA and joint cartilage maintenance, identifying potential downstream effectors that may also serve as better drug targets will be crucial in both our understanding of the disease and the development of future therapeutics.
To assess the potential dual role for NOTCH signaling in joint cartilages and to determine whether the amplitude, duration, and/or frequency of NOTCH activation influences cartilage physiology or pathology, we generated mouse models overexpressing the NICD1 within postnatal joint cartilages in a sustained or transient manner. We further investigated the underlying mechanisms and downstream targets of NOTCH signaling during OA development and cartilage degeneration.
Results
Sustained Activation of NOTCH1 Signaling in Postnatal Chondrocytes Results in a Progressive OA-like Pathology
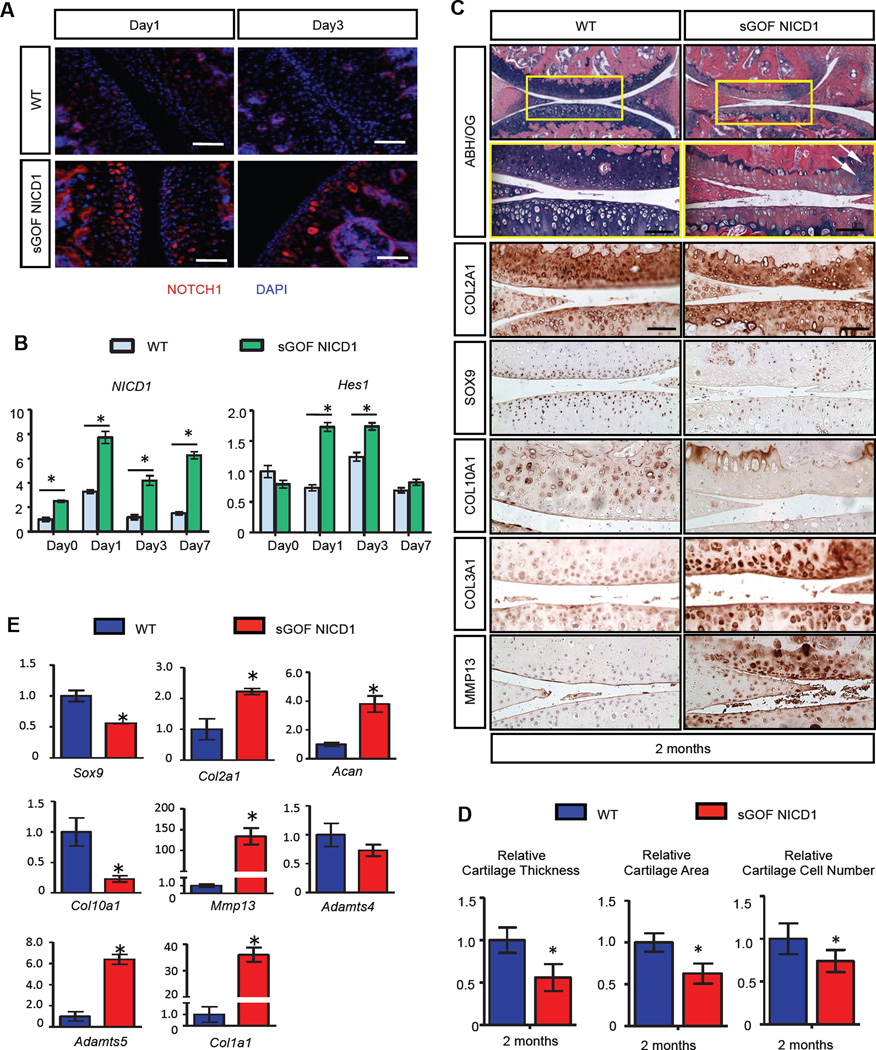
To determine whether NOTCH activation in OA was a reparative response or contributed to the pathology, we generated a NOTCH gain-of-function (GOF) genetic mouse model using the tetracycline-on (Tet-On) system in combination with the Cre-recombinase system: Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+ (fig. S1A). Only in the presence of the reverse tetracycline transactivator (rtTA) and tetracycline (or its commercially available alternative doxycycline), the tetO promoter can drive over-expression of NICD1 within cartilages. Therefore, we can control the robustness and duration of NOTCH signaling by adjusting the dose and frequency of doxycycline (DOX) administration. We first generated and characterized the NOTCH1 over-expression profile in a sustained NOTCH GOF model using high doses and frequencies of DOX injections. Knee joints of Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+ mutants (sGOF NICD1) and Rosa-rtTAf/+ littermate controls (WT) were injected with a single high dose of DOX (100µg/g body weight) and harvested at day 1 and 3 post injection. Immunofluorescence (IF) analyses revealed that NOTCH1 was over-expressed in all zones of the articular cartilage in sGOF NICD1 mice at day1 post injection, and maintained high levels of expression at day 3 following DOX delivery, suggesting sustained NOTCH1 activation (Fig. 1A). To confirm these results, RNA was isolated from articular chondrocytes of 1-month old WT and sGOF NICD1 mice at day 0, 1, 3 and 7 following a single high dose DOX injection. Real-time qPCR revealed that NICD1 expression peaked at day 1 following DOX injection (8-fold induced), and maintained a high level of expression at days 3 and 7 (4-fold and 6-fold induced respectively) (Fig. 1B). Expression of the NOTCH target gene, Hes1, was also elevated at day 1 and 3 (~1.6-fold induced at both time points), although expression returned to basal levels by day 7 (Fig. 1B). Therefore, we injected 1-month old sGOF NICD1 mutant and control mice with high doses and high frequencies of DOX (100µg/g body weight, three times/week) to obtain sustained NOTCH activation (fig. S1B), and harvested knee joints at 2-months of age. Histological analyses revealed hallmark features of an OA-like pathology in sGOF NICD1 mutants including: 1) joint cartilage fibrosis and degeneration, 2) meniscus degradation, and 3) synovial tissue hyperplasia (Fig. 1C). Alcian blue, hematoxylin, and orange-g (ABH/OG) staining demonstrated a loss of proteoglycan content in the articular cartilage of sGOF NICD1 knee sections with the development of chondrocyte clusters in regions retaining some stain, a hallmark of early OA. Severe synovial hyperplasia was also observed in sGOF NICD1 mutants. Additionally, the growth plates often collapsed in sGOF NICD1 mutants, which lead to an altered architecture of the subchondral bone. Histomophometry performed on 2-month-old ABH/OG stained knee sections established that articular cartilage thickness and area were significantly reduced (by ~30% and ~40%, respectively) in sGOF NICD1 mutants, and the number of chondrocytes was dramatically decreased by approximately 50% (Fig. 1D). TUNEL staining revealed a near 60% increase in TUNEL-positive cells on sGOF NICD1 mutant knee sections, especially in the regions where chondrocytes formed cell clusters (fig. S2A). These data suggest that sustained NOTCH1 activation in joint cartilage leads to early and severe cartilage loss, potentially mediated by the induction of apoptosis in articular chondrocytes.
Figure 1. Sustained activation of NOTCH1 signaling in postnatal chondrocytes results in a progressive OA-like pathology.
(A) IF and (B) real-time qPCR analyses revealed sustained Notch signaling activation after high dose (100 µg/g body weight) of DOX administration. (Scale bars, 50µm.) Gene expression analyses were performed for NICD1 and Hes1 in triplicate. All samples were normalized to Beta-actin and then normalized to the controls. Bars represent means ± SD. “*” denotes P<0.05, two-tailed Student’s t test. (C) ABH/OG staining and IHC analyses of COL2A1, SOX9, COL10A1, COL3A1 and MMP13 on 2-month-old WT and sGOF NICD1 mutant knee sections. High magnification of a centralized domain of articular cartilage is shown in a yellow box. White arrows indicate cell clusters. (Scale bars, 50µm) (D) Histomorphometry analyses of articular cartilage thickness, area, and chondrocyte number performed on 2-month-old WT and sGOF NICD1 mutant knee sections. All results were normalized to WT control. Bars represent means ± SD. “*” denotes P<0.05, two-tailed Student’s t test. (E) Real-time qPCR comparing gene expression in the articular chondrocytes isolated from 2-month-old WT and sGOF NICD1 mutant mice. Gene expression analyses were performed for Sox9, Col2a1, Acan, Col10a1, Mmp13, Adamts4, Adamts5, and Col1a1 in triplicate. All samples were normalized to Beta-actin and then normalized to the controls. Bars represent means ± SD. “*” denotes P<0.05, two-tailed Student’s t test. n>3 biological repeats for all experiments.
To identify specific changes in the expression of ECM-related molecules we performed IHC for COL2A1, COL10A1, SOX9, COL3A1, and MMP13 on 2-month-old sGOF NICD1 mutant and control knee sections (Fig. 1C). IHC analysis demonstrated that fibrotic cartilage regions exhibited enhanced COL3A1 expression, suggesting chondrocyte de-differentiation and fibrosis. Expression of COL2A1 was moderately reduced in the articular cartilage in sGOF NICD1 mutant sections, while COL10A1 expression was increased in the articular cartilage deep zone. Notably, SOX9 was largely absent from the superficial zones of sGOF NICD1 mutant articular cartilage. In regions with severe cartilage degeneration and fibrosis, SOX9 was completely absent. Consistent with articular cartilage loss, we detected a striking increase in the catabolic marker, MMP13, within areas of cartilage degeneration. Real-time qPCR was performed on RNA isolated from articular chondrocytes of 2-month-old sGOF NICD1 mutant and control mice (Fig. 1E). Gene expression analyses demonstrated that expression of the anabolic gene Sox9 was dramatically decreased by 50% in sGOF NICD1 mutant mice, whereas the expression of the catabolic genes Mmp13 and Adamts5 were significantly increased (by 132- and 6-fold respectively). Consistent with the IHC analyses, the de-differentiation marker Col1a1 was dramatically up-regulated (~35-fold) in sGOF NICD1 mutants. Moderate up-regulation of Col2a1 (~2-fold) and Acan (~4-fold) was observed in sGOF NICD1 mutants, however the expression of Col10a1 was 70% down-regulated, suggesting abnormal cartilage ECM synthesis. Collectively, these data revealed that sGOF NICD1 mutants exhibit altered ECM synthesis, increased cartilage degradation, and cartilage fibrosis, consistent with a progressive OA-like pathology. These phenotypes were largely replicated at 2- and 8-months of age in another sustained GOF NICD1 genetic mouse model that uses tamoxifen (1mg/10g body weight for 5 days at 1-month of age) to drive lower levels of cartilage-specific sustained NICD1 expression from the Rosa26 locus that is induced by the AcanCreERT2 driver (NICD1AcanTM)(fig. S3), further highlighting that enhanced NOTCH signaling specifically within chondrocytes is the driving force for OA development in these mice.
Transient Activation of NOTCH1 Signaling in Postnatal Chondrocytes Results in Increased Cartilage ECM Synthesis and Joint Maintenance
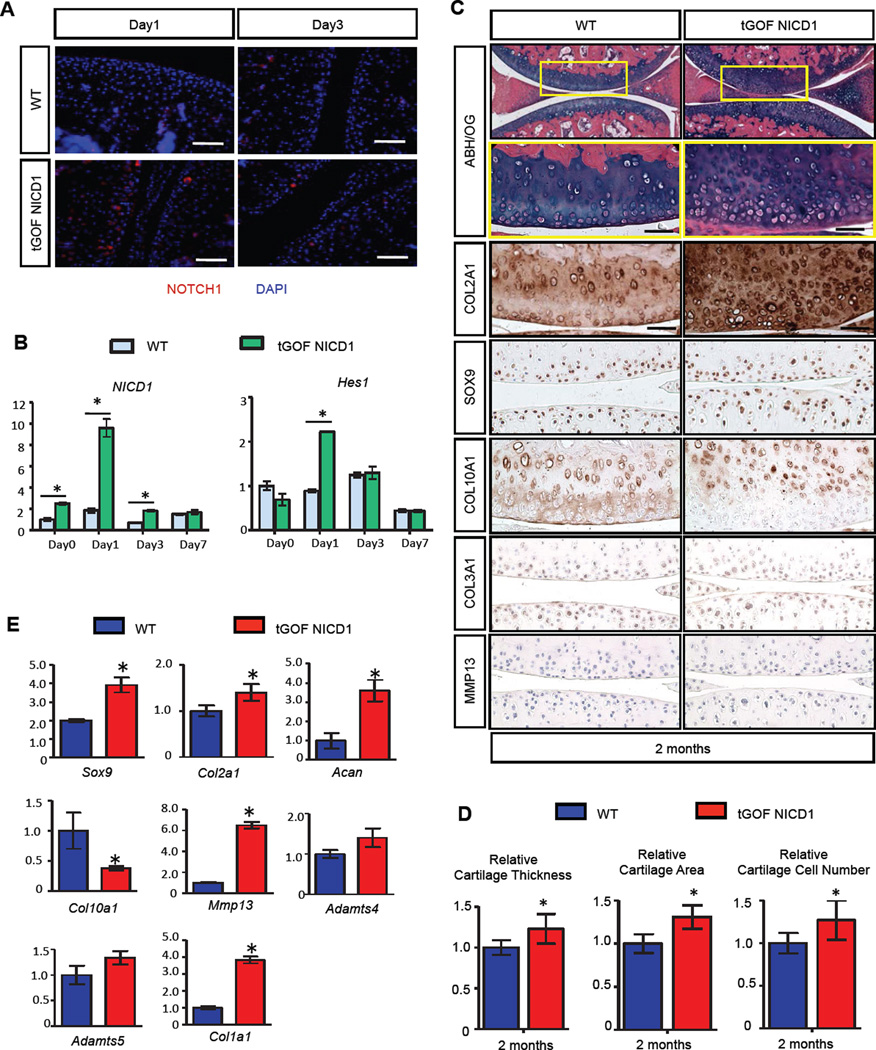
Since sustained NOTCH activation at low or high levels results in an early and progressive OA-like pathology, we set out to determine whether transient NOTCH activation in joint cartilages leads to a different bioactivity by more closely mimicking physiologic Notch signaling in immature cartilage during development. To generate a transient NOTCH GOF model, we injected the Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+ mutant mice (tGOF NICD1) and Rosa-rtTAf/+ littermate controls (WT) with low dose and low frequency of DOX (1µg/g body weight, once/week for 1 month) at 1-month of age (fig. S1B), and harvested knee joints a 2- and 4-months of age. IF and real-time qPCR analyses performed on 1-month-old tGOF NICD1 and control mice confirmed that a single low dose of DOX administration can only transiently activate NOTCH1 signaling for 1 to 3 days in sparse chondrocytes of the articular cartilage (Fig. 2A, B). Joint integrity analyses demonstrated that, by 2-months of age, tGOF NICD1 mutant mice that received a single DOX injection per week exhibited normal knee joint architecture with increased cartilage ECM synthesis (Fig. 2C). Histomophometry analyses revealed that articular cartilage thickness and area were significantly increased by 23% and 31% respectively in tGOF NICD1 mutants as compared to controls (Fig. 2D). Notably, the number of articular chondrocytes was also increased by 27% (Fig. 2D). By 4-months of age, tGOF NICD1 mutant mice exhibited the continuation of joint cartilage maintenance, with trends for increased cartilage area and chondrocyte number, and a 13% increase in articular cartilage thickness as compared to controls (fig. S4A, B). TUNEL staining revealed no significant change of apoptosis in tGOF NICD1 mutants at both 2- and 4-months of age (fig. S2B, C).
Figure 2. Transient activation of NOTCH1 signaling in postnatal chondrocytes results in increased cartilage ECM synthesis and joint maintenance.
(A) IF and (B) real-time qPCR analyses revealed transient Notch signaling activation after low dose (1 µg/g body weight) of DOX administration. (Scale bars, 50µm.) Gene expression analyses were performed for NICD1 and Hes1 in triplicate. All samples were normalized to Beta-actin and then normalized to the controls. Bars represent means ± SD. “*” denotes P<0.05, two-tailed Student’s t test. (C) ABH/OG staining and IHC analyses of COL2A1, SOX9, COL10A1, COL3A1 and MMP13 on 2-month-old WT and tGOF NICD1 mutant knee sections. High magnification of a centralized domain of articular cartilage is shown in a yellow box. (Scale bars, 50µm) (D) Histomorphometry analyses of articular cartilage thickness, area, and chondrocyte number performed on 2-month-old WT and tGOF NICD1 mutant knee sections. All results were normalized to WT control. Bars represent means ± SD. “*” denotes P<0.05, two-tailed Student’s t test. (E) Real-time qPCR comparing gene expression in the articular chondrocytes isolated from 2-month-old WT and tGOF NICD1 mutant mice. Gene expression analyses were performed for Sox9, Col2a1, Acan, Col10a1, Mmp13, Adamts4, Adamts5, and Col1a1 in triplicate. All samples were normalized to Beta-actin and then normalized to the controls. Bars represent means ± SD. “*” denotes P<0.05, two-tailed Student’s t test. n>3 biological repeats for all experiments.
To identify specific changes in the expression of ECM-related molecules, we performed IHC for COL2A1, COL10A1, SOX9, COL3A1, and MMP13 on tGOF NICD1 mutant and control knee sections at 2- and 4-months of age (Fig. 2C, fig. S4A). By 2-months of age, COL2A1 expression was significantly enhanced in the articular cartilage of tGOF NICD1 mutant knee sections. The number of chondrocytes expressing SOX9 was also increased in the superficial and tangential zones of tGOF NICD1 mutant knee sections, while mild up-regulation of COL10A1 expression was observed strictly in the deep zone. The increased expression of COL2A1 and SOX9 can also be observed in 4-month old tGOF NICD1 mutant mice. COL3A1 and MMP13 were almost non-detectable in both tGOF NICD1 mutants and controls at 2- and 4-months of age. RNA was isolated from articular chondrocytes of 2- and 4-month-old tGOF NICD1 mutant and control mice to access the changes of ECM-related genes (Fig. 2E and fig. S4C). Real-time qPCR analyses demonstrated that by the age of 2-months, expression of Sox9, Col2a1 and Acan were significantly up-regulated (by 2-fold, 40% and 3-fold respectively) in tGOF NICD1 mutant mice, while Col10a1 was decreased by ~60%. Moderate up-regulation of Col1a1 and Mmp13 was also observed at this time point (by 3.5 and 6.2 fold respectively). No significant change was observed in Adamts4 and Adamts5 expression. Interestingly, the anabolic effect observed in 2-month old tGOF NICD1 mutants was maintained until 4-months of age, which was 2 months after the last DOX administration. Gene expression analyses revealed increased Sox9, Col2a1 and Acan in tGOF NICD1 mutants (by 4-fold, 3.2-fold and 3-fold respectively), while the previously elevated expression of Col1a1 and Mmp13 were reduced by 50%. Collectively, these data were consistent with the IHC analyses and demonstrated that transient activation of NOTCH1 signaling in adult joint cartilages enhanced articular cartilage ECM synthesis and promoted joint maintenance for at least two months following the last DOX administration.
To investigate whether transient NOTCH1 activation can protect joint integrity against trauma-induced OA, we performed a meniscal-ligamentous injury (MLI) surgery on 8-week-old tGOF NICD1 mutant and control mice followed by 4 low doses of DOX injections (1µg/g body weight, once/week). Knee joints were harvested 8-weeks after surgery and joint integrity was analyzed by ABH/OG stained histology (fig. S4D). Accelerated OA progression was observed in tGOF NICD1 mutants, including joint cartilage degeneration and clefting, meniscus degradation, osteophyte formation, and severe synovial tissue expansion (fig. S4D). These data suggest that transient Notch activation can only promote cartilage and joint maintenance under physiological conditions, but in pathological situations, such as the trauma-induced inflammatory environment, even transient Notch activation accelerates OA development and leads to rapid joint degradation potentially due to synergistic effects with other pro-inflammatory factors within the injured joint environment.
Large-scale Temporal Gene Expression Profiling Reveals Potential NOTCH1 Target Genes Responsible for Cartilage Fibrosis and Degradation
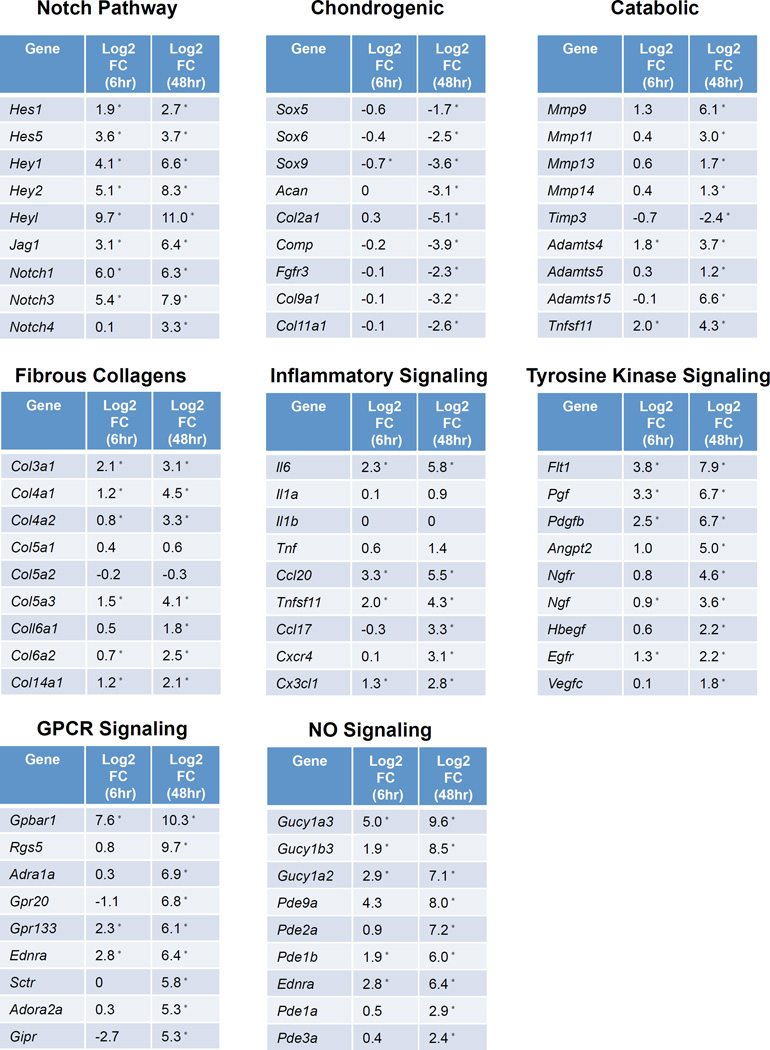
We have demonstrated that NOTCH signaling is a critical regulator of cartilage and joint maintenance under physiological conditions, and sustained activation of NOTCH in postnatal cartilages leads to a progressive OA-like pathology. However, the downstream NOTCH targets responsible for OA development remain unclear. To address this issue, we developed an in vitro sustained NOTCH GOF model using costal chondrocytes isolated from Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+ mutant and control mice and performed RNA-sequencing (RNA-seq) analyses at 6 hours and 48 hours following DOX administration in culture. These data revealed that NOTCH activation induced 451 genes at 6 hours and 1249 genes at 48 hours, including NOTCH pathway genes (HeyL, Notch3, Jag1, etc.), cartilage degradation enzymes (Mmp13, Mmp9, Adamts4, Adamts5, etc.), fibrous collagens (Col3a1, Col4a1, Col5a3, etc.), growth factors (Pdgfb, Tgfb1, Tgfb2, etc.), and specific cytokines/chemokines (Il6, Ccl20, Ccl17, etc.), while other inflammatory factors often associated with OA remained relatively low (Il1a, Il1b, Tnf). NOTCH activation also down-regulated the expression of 75 genes at 6 hours and 867 genes at 48 hours, including cartilage-related collagens (Col2a1, Col9a1, Col11a1, etc.) and chondrogenic genes (Sox9, Sox5, Sox6, Acan, Comp, etc.). In particular, we highlight here the profiles of NOTCH target genes, chondrogenic genes, catabolic genes, fibrous collagens, and inflammatory factors that are likely associated with the NOTCH induced OA progression in vivo (Fig. 3). The majority of chondrogenic genes were dramatically down-regulated, whereas the expression of Mmp and Adamts family genes were significantly up-regulated at 48 hours following DOX treatment. Interestingly, the expression of collagens went in two separate directions: the expression of genes encoding cartilage-related collagens such as Col2a1, Col9a1, Col9a2, Col9a3, Col11a1, Col11a2 were significantly repressed, while the expression of genes encoding fibrous collagens such as Col3a1, Col4a1, Col4a2, Col5a3, Col6a2 and Col14a1 were all dramatically induced. These data identified novel NOTCH regulated genes in chondrocyte cultures, and demonstrated that sustained NOTCH signaling suppresses the chondrogenic phenotype and promotes cartilage fibrosis and degradation, implicating the NOTCH pathway as a critical regulator of the pathogenesis of OA. We also found the expression of some inflammatory factors moderately up-regulated, such as several Interleukin and Tnf related genes. In particular, the expression of Il6 was 5- and 54-fold up-regulated at 6 hours and 48 hours, which was one of the most robustly responsive genes following DOX treatment in sGOF NICD1 chondrocytes. In addition, pathway analysis identified several clustered genes related to tyrosine kinase signaling, G protein coupled receptor (GPCR) signaling, and Nitric Oxide (NO) signaling pathways as being significantly up-regulated, many of which have been implicated in OA and/or cartilage catabolism (Fig. 3).
Figure 3. Large-scale temporal gene expression profiling reveals potential NOTCH1 target genes responsible for cartilage fibrosis and degradation.
P2 costal chondrocyte cultures isolated from control and Col2a1Cre; tetO-NICD1f/+; Rosa-rtTAf/+ mutant pups were treated with 10µg/ml DOX for 6 or 48 hours. RNA was collected for RNA-seq experiments. Each group contained three biological repeats. Log2(Fold Change) (Log2FC) of some genes of interest are shown, including Notch pathway genes, typical chondrogenic and catabolic genes, fibrous collagens, as well as genes from inflammatory signaling, tyrosine kinase signaling, GPCR signaling, and NO signaling. “*” denotes statistical significance with P<0.05 after Benjamini-Hochberg correction for multiple-testing (Cufflinks version 2.0.2).
Sustained NOTCH1 Signaling Activates the IL6/STAT-3 Pathway in OA Cartilages
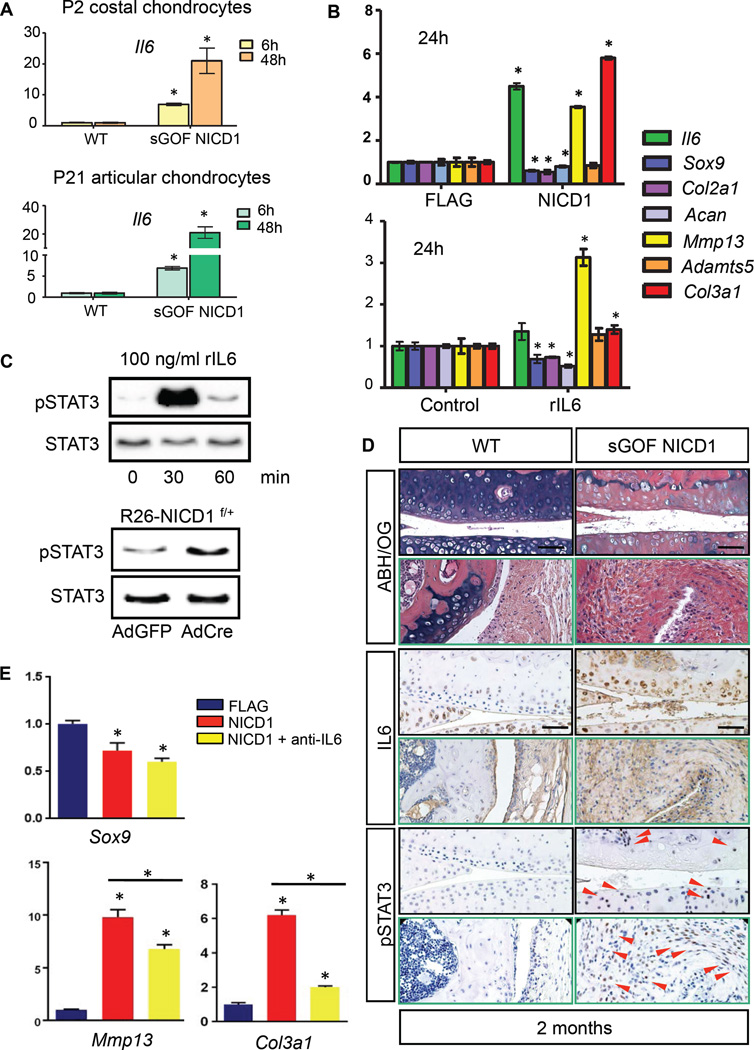
IL6 is a pro-inflammatory cytokine produced by various types of cells and is associated with human OA (28). It is modestly expressed in normal articular cartilage, but significantly over-expressed in OA synovial membrane, cartilage and synovial fluid, which stimulates not only the production of the cartilage-degrading proteases, but also suppresses the expression of anabolic genes such as Sox9, Col2a1 and Acan (7, 9, 11). IL6 signaling is initiated by ligand binding to a membrane bound IL6 receptor (IL6R) or a soluble form of IL6R (sIL6R) and the co-receptor gp130, which ultimately leads to activation of JAK kinase and signals propagated via the ERK MAPK pathway and the STAT1/STAT3 pathway (29–32). To better understand the role of IL6 following NOTCH activation, we first confirmed the RNA-seq results with real-time qPCR performed on P2 costal chondrocytes, P21 articular chondrocytes, and the chondrogenic cell line ATDC5 (Fig. 4A,B). Dramatic up-regulation of Il6 was observed in all cell models following NOTCH activation.
Figure 4. Sustained NOTCH1 signaling activates the IL6/STAT3 pathway in cultured cells and OA cartilages.
(A) Real-time qPCR analyses for Il6 were performed on RNA collected from P2 costal chondrocyte cultures or P21 articular chondrocyte cultures isolated from WT and sGOF NICD1 mutant pups treated with 10µg/ml DOX for 6 or 48 hours in triplicate. All samples were normalized to Beta-actin and then normalized to the controls. Bars represent means ± SD. “*” denotes P<0.05, two-tailed Student’s t test. (B) Real-time qPCR comparing gene expression in ATDC5 cells transfected with FLAG control or NICD1 over-expression vectors for 24 hours, and gene expression in ATDC5 cells treated with control diluents or recombinant IL6 proteins (rIL6, 1ng/ml) for 24 hours. Gene expression analyses were performed for Il6, Sox9, Col2a1, Acan, Mmp13, Adamts5, and Col3a1 in triplicate. All samples were normalized to Beta-actin and then normalized to the controls. Bars represent means ± SD. “*” denotes P<0.05, two-tailed Student’s t test. (C) Western blot analyses for STAT3 and phosphorylated STAT3 (pSTAT3) in proteins extracted from ATDC5 cells treated with rIL6 (100 ng/ml) for 0, 30, and 60 min, and in proteins extracted from control AdGFP infected or AdCre infected R26-NICD1f/+ primary chondrocytes. Results are representative of three independent experiments. (D) ABH/OG and IHC analyses of IL6 and pSTAT3 on 2-month-old WT and sGOF NICD1 mutant knee sections. (Scale bars, 50µm.) Red arrowheads indicate pSTAT3 positive cells. (E) Real-time qPCR comparing gene expression in ATDC5 cells transfected with FLAG control, NICD1 over-expression vectors, and NICD1 over-expression vectors plus IL6 neutralizing antibody (anti-IL6) for 24 hours. Gene expression analyses were performed for Sox9, Mmp13, and Col3a1 in triplicate. All samples were normalized to Beta-actin and then normalized to the controls. Bars represent means ± SD. “*” denotes P<0.05, one-way ANOVA, followed by Bonferroni method.
To better understand IL6 regulation of ECM-related and OA-associated factors, we utilized an in vitro culture model of ATDC5 chondrogenic cells. Differentiated ATDC5 cells were transformed with NICD1 over-expression vectors or treated with recombinant IL6 proteins (rIL6) for 24 hours, and RNA was harvested for real-time qPCR analyses. Gene expression analyses revealed that rIL6 treatment suppressed Sox9, Cola2a1 and Acan expression, and induced Mmp13 and Col3a1 expression, consistent with gene changes observed under sustained NOTCH activation (Fig. 5B). Since STAT3 is an important downstream mediator of IL6 signaling and is the most abundant Stat molecule expressed in cartilage, we assessed the capacity of IL6 to induce an activated form of the STAT3 protein (phosphorylated STAT3, pSTAT3) in chondrogenic ATDC5 cells. Western blot analysis revealed that rIL-6 treatment rapidly activated STAT3 in an acute manner, evidenced by a rapid increase in its phosphorylation at the residue Tyr705. Based on the facts that sustained NICD1 signals stimulate Il6 expression and that IL-6 can activate STAT3 in chondrogenic cells, we predicted that STAT3 signaling would be elevated in NICD1 GOF chondrocytes. Indeed, Western blot analysis showed a significant increase in the level of pSTAT3 48 hours following NOTCH activation.
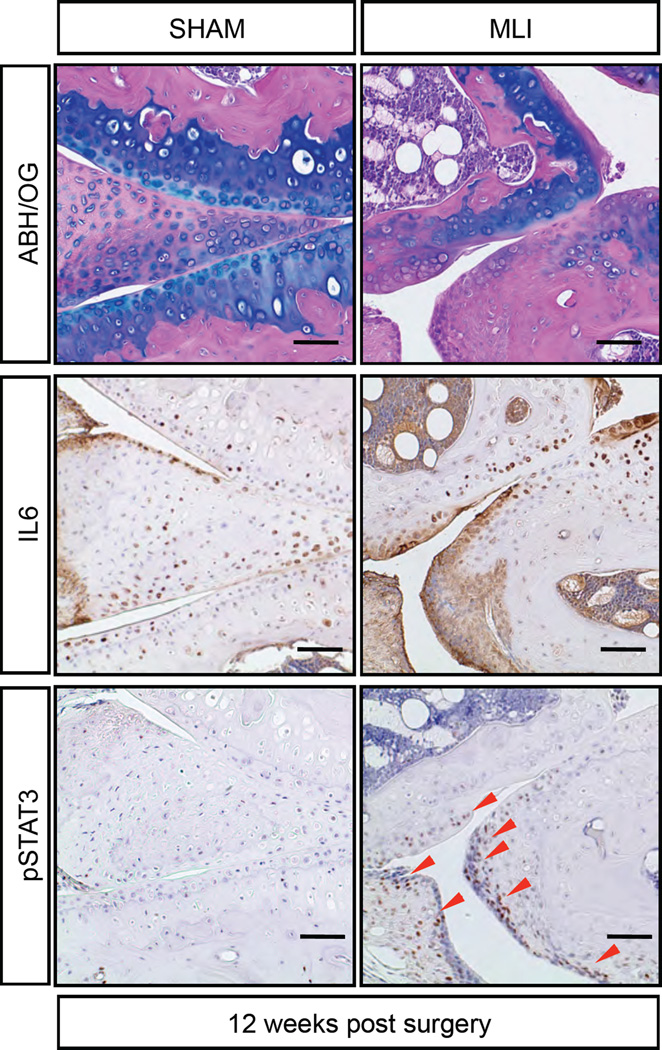
Figure 5. IL6/STAT3 pathway is activated in a trauma-induced OA model.
ABH/OG staining and IHC analyses of IL6 and pSTAT3 on sham or MLI knee sections of 5-month-old WT mice that were 12 weeks post surgery. (Scale bars, 50µm.) Red arrowheads indicate pSTAT3 positive cells. n>5 biological repeats.
To determine whether the NOTCH-induced IL6/STAT3 signaling occurred in vivo within the joints of our NOTCH GOF models (sGOF NICD1 and NICD1AcanTM), we performed IHC analyses for IL6 and p STAT3 on joint sections from 2- and 8-month old mutant and control mice. A low level of IL6 was expressed in normal articular cartilage and synovial tissues of WT animals, but was abundantly over-expressed in the cartilage ECM and hyperplastic synovium of sustained NOTCH signaling mutants, especially in regions of severe fibrosis (Fig. 4D and fig. S3D). To confirm that increased IL6 was associated with enhanced STAT3 activation, we examined the expression of pSTAT3 in both NOTCH GOF models. IHC revealed that no pSTAT3 signal was detected in WT knee joints, nevertheless, significant up-regulation of pSTAT3 was detected in the fibrotic cartilages, degraded menisci, and hyperplastic synovial tissues in both sGOF NICD1 and NICD1AcanTM mutants (Fig. 4D and fig. S3D).
To further demonstrate whether the effect observed under NOTCH activation was IL6 dependent, we pre-treated differentiated ATDC5 cells with IL6 neutralizing antibodies for 24 hours, and transformed the cells with FLAG control or NICD1 over-expression vectors. Real-time qPCR revealed that IL6 neutralization did not alleviate the NOTCH-mediated suppression of Sox9, but partially reduced the NOTCH-mediated induction of Mmp13. Interestingly, neutralization of IL6 almost completely eliminated the NOTCH-mediated induction of Col3a1. These data suggest that NOTCH-mediated regulation of Sox9 is IL6-independent, while NOTCH regulation of Mmp13 and Col3a1 occur in an IL6-dependent manner (Fig. 4E). Collectively, these results suggest that prolonged NOTCH activity induces the expression of IL6 in OA cartilage, which may directly result in joint cartilage degeneration and fibrosis via IL6/STAT-3 signaling activities.
We also confirmed IL6/STAT3 activation in a surgical-induced OA model, where 2-month-old WT mice were administered a meniscal-ligamentous injury (MLI) surgery that produces a progressive OA-like phenotype from 4 to 20 weeks post injury (33, 34). At 12-weeks post surgery, OA-like pathologies can be observed in MLI induced mice, including joint cartilage degeneration and fibrosis, osteophyte formation, and synovial hyperplasia (Fig. 5). IHC demonstrated enhanced IL6 expression in OA tissues, especially the fibrotic and degenerative cartilages and synovium, and pSTAT3 was detected in IL6 positive regions (Fig. 5). These results suggest that injury-induced joint cartilage fibrosis and degeneration may also occur via IL6/STAT3 signaling pathway changes, a mechanism that may be shared with NOTCH-induced OA.
ERK and p38 MAPK Pathways Likely Contribute to the NOTCH1-induced OA-related Phenotypes
In addition to the STAT3 regulation, the ERK and p38 MAPK pathways are known to be important mediators of pathological IL6 signaling as well as numerous other cytokines and growth factors. To test whether IL6 can activate these two pathways in chondrogenic cells, we examined the effects of rIL6 treatments on the activation and phosphorylation of ERK and p38 MAPK in ATDC5 cells. Western blot analysis showed that phospho-ERK1/2 and phospho-p38 levels increased dramatically within 30 minutes of rIL6 treatment, and then rapidly declined, indicating that IL6 can simultaneously activate both ERK and p38 MAPK pathways in ATDC5 cells (Fig. 6A). In line with the NOTCH-induced up-regulation of IL6 in costal chondrocytes, phosphorylated (active) forms of ERK and p38 were also significantly increased in these NICD GOF primary cells (Fig. 6A). Similar to our pSTAT3 analyses, IHC performed on articular cartilage sections from sGOF NICD1 mice further confirmed elevated levels of phospho-p38 in vivo (Fig. 6B). Unfortunately, available phospho-ERK1/2 antibodies were not compatible with IHCs performed on our paraffin sections of adult knee joints. Therefore, further in vivo studies may be needed to elucidate the potential p38 and ERK1/2 contributions to NOTCH-induced OA-like phenotypes.
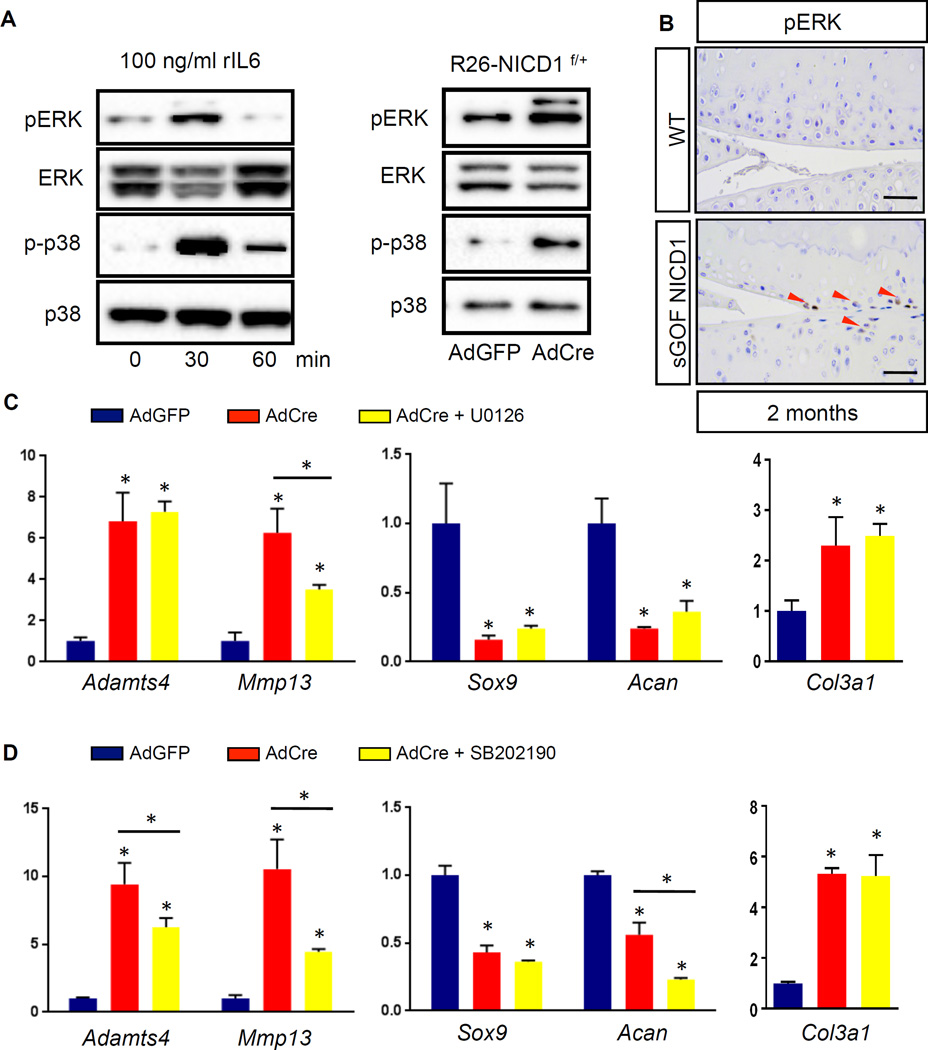
Figure 6. Sustained NOTCH1 signaling activates the ERK/p38 cascade and leads to selected effects on the expression of catabolic, chondrogenic, and fibrous genes.
(A) Western blot analyses for ERK, phosphorylated ERK (pERK), p38, and phosphorylated p38 (p-p38) in proteins extracted from ATDC5 cells treated with rIL6 (100 ng/ml) for 0, 30, and 60 min, and in proteins extracted from control AdGFP infected or AdCre infected R26-NICD1f/+ primary chondrocytes. Results are representative of three independent experiments. (B) IHC analyses of pERK on 2-month-old WT and sGOF NICD1 mutant knee sections. (Scale bars, 50µm.) Red arrowheads indicate pERK positive cells. (C, D) Real-time qPCR comparing gene expression in RNA extracted from control AdGFP infected and AdCre infected R26-NICD1f/+ primary chondrocytes. (C) AdGFP/AdCre infected plus ERK inhibitor U0126, and (D) AdGFP/AdCre infected plus p38 inhibitor SB202190. Gene expression analyses were performed for Adamts4, Mmp13, Sox9, Acan, and Col3a1 in triplicate. All samples were normalized to Beta-actin and then normalized to the controls. Bars represent means ± SD. “*” denotes P<0.05, one-way ANOVA, followed by Bonferroni method.
To determine whether ERK and p38 MAPK contributed to the OA-associated phenotypes caused by sustained NICD1 signals in vitro, we evaluated the effects of ERK inhibitor U0126 and p38 MAPK inhibitor SB202190 on the expression of anabolic (Sox9, Acan), catabolic (Adamts5, Mmp13), and fibrotic (Col3a1) genes. Real-time qPCR revealed that both U0126 and SB202190 treatment had no significant impact on the NOTCH-mediated up-regulation of Col3a1 or suppression of Sox9 (Fig. 6C,D). While U0126 treatment had no effect on the NOTCH-mediated suppression of Acan (Fig. 6C), SB202190 treatment exacerbated the NOTCH-mediated suppression of Acan expression (Fig. 6D). Interestingly, both U0126 and SB202190 treatment significantly alleviated NOTCH-induced Mmp13 expression (Fig. 6C,D). Moreover, SB202190 treatment also partially rescued NOTCH-mediated up-regulation of Adamts4 (Fig. 6D). Collectively, these data suggest that elevated ERK and p38 MAPK at least partially contributes to the NOTCH-mediated up-regulation in cartilage catabolic activity, although interference with p38 MAPK may induce further down-regulation of some cartilage anabolic factors. Furthermore, inhibition of either ERK or p38 MAPK does not affect NOTCH-induced cartilage fibrosis.
Alternative Signaling Pathways Activated by Sustained NOTCH1 Signaling in Chondrocytes
In addition to inflammatory signaling, gene and pathway analysis of our RNA-seq data revealed that multiple alternative signaling pathways were significantly up-regulated in sGOF NICD1 chondrocytes. We highlight here the top 3 significant pathways in our analyses that have also been previously reported to be relevant in cartilage biology and arthritis-related pathologies. These include tyrosine kinase signaling, G protein coupled receptor (GPCR) signaling, and Nitric Oxide (NO) signaling pathways (Fig. 3).
Many genes related to tyrosine kinase signaling are dramatically induced by sustained NOTCH1 signaling in chondrocytes (Fig. 3). Several of them, such as Pgf, Flt1, and Ngf, have previously been shown to be involved in the pathology of osteoarthritis and cartilage catabolism (35–39). To investigate whether tyrosine kinase signaling contributed to NOTCH1-induced OA-related molecular phenotypes in vitro, we treated chondrocytes with Genistein, a tyrosine kinase inhibitor. qPCR analysis revealed that Genistein treatment did not alleviate the NOTCH1-mediated Sox9 and Acan suppression, but significantly attenuated the NOTCH1 induction of catabolic (Adamts4 and Mmp13) and fibrotic (Col3a1) genes (fig. S5). These data indicate that tyrosine kinase signaling may partially mediate NOTCH1-induced catabolic and fibrotic processes.
Since several GPCR signaling factors were identified in our RNAseq dataset (Fig. 3) and have also been implicated in cartilage biology and joint disease (40, 41), we next tested whether elevated GPCR signaling contributed to the NOTCH1-induced OA-associated molecular phenotypes. We individually inhibited three major downstream pathways of GPCR signaling, Gαs/adenylate cyclase, Gαi, and Gαq/11/PLC, using adenylate cyclase inhibitor SQ-22536 (fig. S6A), Gαiinhibitor Pertussis toxin (PTx) (fig. S6B), and the PLC inhibitor Edelfosine (fig. S6C), respectively. Inhibition of any of these three pathways did not alleviate the NOTCH1-mediated induction of Adamts4 and Mmp13, nor the NOTCH1-mediated suppression of Sox9 and Acan (fig. S6A,B,C). However, both PTx and Edelfosine treatment slightly reduced NOTCH1-mediated induction of Col3a1 (fig. S6B,C), suggesting that GPCR signaling may partially mediate the induction of fibrotic Col3a1 by sustained NOTCH1 signaling.
NO signaling has been strongly implicated in human OA and experimental OA (42–45). Interestingly, a group of genes related to NO signaling, including genes encoding endothelin receptor A, soluble guanylyl cyclase (the only known receptor for nitric oxide), and phosphodiesterase, were remarkably up-regulated by sustained NICD1 signals (Fig. 3). However, endothelin receptor A (Edra) antagonist BQ-123 or soluble guanylyl cyclase (sGC) inhibitor ODQ treatment only slightly alleviated the NOTCH1-mediated induction of Col3a1 (fig. S7A,B). Similarly, phosphodiesterase inhibitor IBMX only modestly attenuated the NOTCH1-mediated suppression of Sox9 and Acan (fig. S7C). In contrast, none of these treatments rescued the NOTCH1-mediated induction of Adamts4 and Mmp13 (fig. S7A,B,C), indicating that NO-related signaling is unlikely to mediate the effect of NOTCH1 activation on the up-regulation of catabolic genes, but may have an impact on the NOTCH-regulated anabolic and fibrotic responses in cartilage.
Discussion
In this study, we have provided the first genetic evidence that sustained versus transient NOTCH activation in postnatal joint cartilages leads to opposing effects on articular cartilage and joint maintenance. This study establishes that sustained NOTCH activation in adult joint cartilage results in a severe, early, and progressive OA-like pathology, while transient NOTCH activation results in increased cartilage ECM synthesis and joint maintenance only under physiological conditions. In vitro and in vivo studies demonstrate the capability of NOTCH signaling to regulate anabolic, catabolic, and fibrotic gene expression, and RNA-seq experiments determined that sustained NOTCH activation suppresses chondrogenic genes but promotes the expression of cartilage-related proteases (MMPs and ADAMTSs), fibrotic collagens, inflammatory factors including Il6, and a host of broad signaling pathways (tyrosine kinase, GPCR, and NO signaling) that are known to affect cartilage biology and potentially contribute to OA. Collectively, these data indicate that NOTCH signaling is a critical pathway that regulates joint cartilage homeostasis and in pathological situations of sustained signaling is involved in joint cartilage degradation and fibrosis, likely via induction of at least the IL6/STAT3, ERK/p38 MAPK pathways, and potentially others. Therefore, an appropriate balance of NOTCH signaling must be achieved in order to maintain normal articular cartilage homeostasis and joint integrity.
NOTCH components are widely expressed in human adult articular cartilage, indicating a potential role for NOTCH signaling in articular cartilage maintenance during adult life (46, 47). Interestingly, the expression of NOTCH signaling molecules is significantly up-regulated in OA cartilage (25–27, 46), indicating a role for NOTCH in OA on-set and progression. We demonstrated here that sustained NOTCH activation in postnatal joint cartilages leads to an early and progressive OA-like pathology, including joint cartilage degradation, fibrosis, and synovial expansion, which is likely due to the dramatic up-regulation of cartilage degradation enzymes and pro-inflammatory factors. It has recently been shown that loss or reductions in cartilage-specific NOTCH signaling are capable of reducing MMP13 expression within murine joints and delaying cartilage degeneration over the short-term (8-weeks) (25). Interestingly, our previous data have demonstrated that although decreased expression of both anabolic and catabolic factors can be observed with short-term NOTCH inhibition in joint cartilages, long-term (6 to 8-months) NOTCH reduction disrupts normal physiology of the cartilage and ultimately results in cartilage degeneration (24). These data collectively suggest that NOTCH signaling may play a complex role in cartilage homeostasis, such that both sustained activation or permanent reduction of NOTCH signaling leads to cartilage degradation and joint failure.
NOTCH signaling is involved in both anabolism and catabolism of cartilage. We and others have previously shown that sustained NICD activation in committed growth plate chondrocytes (17, 48) and articular chondrocytes (49) stimulated the expression of the catabolic factor Mmp13, while permanent NOTCH inhibition within growth plate and primary articular chondrocyte cultures reduced Mmp13 expression (17, 48). Several lines of evidence have demonstrated that sustained NOTCH signaling in mesenchymal progenitors and growth plate chondrocytes suppresses Sox9, Col2a1, and Acan (17, 48, 50, 51). Data presented by Mead and colleagues (18) and our lab (Kohn et al., unpublished data) have indicated that loss of NOTCH signaling leads to inappropriate expression of Sox9 in hypertrophic chondrocytes. Furthermore, we have demonstrated here that sustained over-expression of NICD1 leads to suppression of Sox9, Col2a1, and Acan in adult articular cartilage and chondrogenic cell cultures, while transient over-expression of NICD1 in adult joint cartilages promotes the expression of Sox9, Col2a1 and Acan in vivo. Interestingly, others have demonstrated that short-term or transient NOTCH signaling promotes chondrocyte anabolism by inducing Sox9 in vitro (52), although the mechanisms for this action remain unclear. Collectively, these data suggest that transient or physiological NOTCH signaling in chondrocytes favors a balanced anabolic and catabolic cartilage maintenance response, while sustained or high levels of NOTCH activity elicits a pathological response via the simultaneous suppression of chondrogenic genes and induction of catabolic factors.
All sustained NICD1 GOF models exhibited robust up-regulation of IL6 and pSTAT3 in degenerating and fibrotic regions of joint cartilages and synovial tissues. Prior studies identified a conserved RBPjκ binding site within the promoter of Il6, which overlaps a NF-κB binding site (53). RBPjκ can directly bind this site, and NOTCH signaling can induce Il6 transcription via interactions with the NF-κB pathway (53, 54). Consistent with these findings, we find elevated IL6 production after NOTCH activation in vivo and in vitro. RNA-seq analyses revealed that the expression level of Il6 was 5- and 54-fold up-regulated after 6 and 48 hours of NOTCH1 activation respectively, confirming the concept that Il6 may be a direct NOTCH target gene. An in vitro study performed in chondrocyte cultures has also suggested that neutralization of IL6 cannot modify the NOTCH-mediated suppression of Sox9 and Col2a1, but opposed Mmp13 induction (55). Here we further demonstrated that neutralization of IL6 at least partially inhibits NOTCH-mediated induction of Mmp13 and also abolishes NOTCH-mediated induction of Col3a1. Interestingly, STAT3 activity can be modulated by NOTCH and the NOTCH effectors, HES1 and HES5, in cultured neural cells where both the NICD and the HES proteins were shown to associate and facilitate the complex formation between JAK2 and STAT3 (56). Therefore, these data suggest a potential role for NOTCH and HES proteins in promoting cartilage degeneration in both an IL6-independent and -dependent manner by regulating the phosphorylation and activity of STAT3.
We also determined that sustained NOTCH1 signaling activates the ERK and p38 MAPK pathways in chondrocytes both in vitro and in vivo, which has not been previously reported. ERK and p38 MAPK can function as downstream effectors of IL6 signaling, but are also regulated independent of IL6 via numerous pathways including other inflammatory cytokines and growth factor/receptor pathways (TGFβ/TGFβRs, RTKs and GPCRs). While the mechanism by which NOTCH1 activates ERK and p38 MAPK in chondrocytes may occur via the induction of IL6, we have also demonstrated the ability of NOTCH1 to highly induce other inflammatory cytokines (Ccl20, Ccl17, Cxcr4, etc.), RTK components (Flt1, Pgf, Pdgfb, Ngf, Ngfr, etc.), and GPCR family members (Gpbar1, Rgs5, Gpr20, Ednra, etc.). Several of these factors have been shown to separately activate an ERK and p38 MAPK signaling cascade and have also been implicated in cartilage biology and joint disease (35–41, 57). Attenuation of specific branches of these broad signaling pathways (RTKs and GPCRs), as well as others induced by NOTCH1 in chondrocytes (NO), have demonstrated the complex nature by which NOTCH signaling regulates cartilage anabolism, catabolism, and fibrotic gene regulation. Future studies will be directed at dissecting this complex regulation in order to uncover the precise mechanism(s) involved in NOTCH-induced OA and to identify novel targets for the development of disease modifying osteoarthritis drugs (DMOADs).
Materials and Methods
Mouse strains
Animal studies were approved by the University of Rochester Committee on Animal Resources. All mouse strains, including AcanCreERT2 (58), Rosa-NICD1f/f (59), Col2a1Cre (60), tetO-NICD1 (61), and Rosa-rtTAf/+ (62) have been described previously. Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+ and AcanCreERT2; Rosa-NICD1f/f (NICD1AcanTM) mice were viable and produced in Mendelian ratios. Doxycycline was administrated to Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+ mice and littermate controls with two strategies starting at 1-month of age: 1 µg/g body weight, once/week or 100 µg/g body weight, three times/week. Mice were harvested at 2-months and 4-months of age. Tamoxifen (1 mg/10 g body weight) was administered daily via intraperitoneal injection to all NICD1AcanTM and littermate controls for 5 continuous days starting at 1-month of age. Mice were harvested at 2-months and 8-months of age.
Trauma-induced OA model and surgical procedures
We used a well-established mouse meniscal-ligamentous injury (MLI) model to mimic the clinical situation of trauma-induced OA (34). All experiments were performed according to the protocol approved by the Institutional Animal Care & Use Committee at the University of Rochester Medical Center. MLI surgery was performed on 8-week-old Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+ (tGOF NICD1) mice and littermate controls. Briefly, the bilateral hind limbs were shaved and prepared for aseptic surgery. The right knee joint was exposed and the medial collateral ligament was transected. Then the joint space was opened slightly and the medial meniscus was detached from its anterior-medial tibial attachment using a microsurgical technique. The contralateral knee joint was sham-operated without any ligament transection or meniscus detachment. The skin incision was closed after the surgery. Doxycycline (DOX) was administered to both tGOF NICD1 and control mice via i.p. injection (1 µg/g body weight, once/week) for 4 weeks after MLI surgery. Knee joints were harvested 8 weeks post MLI surgery by the age of 4-months.
Analysis of mouse tissue sections
Knee joints were harvested and fixed in 10% neutral buffered formalin for 3 days, decalcified in Formic Acid Bone Decalcifier (Immunocal, Decal Chemical Corp.) for 7–10 days, paraffin processed, and embedded for sectioning. Tissues were sectioned at 5 µm and stained with ABH/OG. IHC analyses were performed on sections using traditional antigen retrieval and colorimetric development methodologies with the following primary antibodies: SOX9 (Santa Cruz), COL2A1 (Thermo Scientific), COL10A1 (Quartett), COL1A1 (Abcam), COL3A1 (Abcam), MMP-13 (Thermo Scientific), IL6 (Abcam), and phosphorylated STAT3 (Cell Signaling). TUNEL cell death assay was performed on sections using the in situ Cell Death Detection Kit, Fluorescein (Roche) according to the manufacturer’s instructions. IF analysis and β-galactosidase staining was performed on frozen sections. Knee joints were harvested and fixed in 4% paraformaldehyde (PFA) for 2 hours at 4 °C and decalcified with 14% EDTA at 4 °C for 10 days. Tissues were washed in sucrose gradient, embedded with Tissue-TEK OCT medium, snap frozen in liquid nitrogen and sectioned at 10 µm using a Lecia CM 1850 cryotome. The NOTCH1 primary antibody (Santa Cruz) was used for IF analysis. Beta-galactosidase staining was performed as previously described (63).
Murine costal chondrocyte isolation and RNA-seq
Murine costal chondrocytes were isolated as previously described with modifications (15). Briefly, rib cages were dissected from 2-day-old Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+ mutant or control pups with soft tissue removed and plated in 1X PBS. Rib cages were digested with 2mg/ml of Pronase (Roche) in 1X PBS for 1 hour in a 37°C shaking water bath, and then digested in 3mg/ml Collagenase D (Roche) in DMEM high glucose medium (Gibco) for 1 hour at 37°C. The rib cages were transferred into a petri dish and digested in 3mg/ml Collagenase D in DMEM high glucose medium for 4–6 hours. Murine costal chondrocyte cell suspensions were then filtered through 40µm filters, and seeded at a density of 500,000 cells/well in 6-well tissue culture plates in DMEM high glucose medium supplemented with 10% Fetal Bovine Serum (FBS) (Sigma) and 1% Penicillin/Streptomycin (P/S). Both mutant and control cell cultures were treated with 10ug/ml doxycycline (Sigma) for 6 or 48 hours.
RNA was extracted with the RNeasy Mini Kit (Qiagen) and the RNase-Free DNase Set (Qiagen) according to the manufacturer’s instructions. Three biological replicates were performed in both mutant and control groups. 1µg of total RNA from each sample was sent to the Genomics Research Center (GRC) at URMC for mRNA sequencing and data processing. According to the GRC, RNA concentration is determined using a NanoDrop 1000 spectrophotometer and RNA quality is assessed using an Agilent Bioanalyzer (Agilent). The TruSeq RNA Sample Preparation Kit V2 (Illumina) was used for next generation sequencing library construction according to the manufacturer’s protocols. Briefly, mRNA was purified from 100ng total RNA using oligo-dT magnetic beads and fragmented. First-stand cDNA synthesis was performed with random hexamer priming followed by second-strand cDNA synthesis. End repair and 3’adenylation was performed on the double stranded cDNA. Illumina adaptors were ligated to both ends of the cDNA, purified by gel electrophoresis, and amplified using PCR primers specific to the adaptor sequences to generate amplicons of approximately 200–500bp in size. The amplified libraries are hybridized to the Illumina single end flow cell and amplified using the cBot (Illumina) at a concentration of 8 picomoles per lane. Single end reads of 100nt were generated for each sample and aligned to the organism specific reference genome. Raw reads generated from the Illumina HiSeq2500 sequencer were de-multiplexed using configurebcl2fastq.pl version 1.8.3. Low complexity reads and vector contamination were removed using sequence cleaner (“seqclean”) and the NCBI univec database, respectively. The FASTX toolkit (fastq_quality_trimmer) was applied to remove bases with quality scores below Q=13 from the end of each read. Processed reads were then mapped to the UCSC XXX genome build with SHRiMP version 2.2.3 and differential expression analysis was performed using Cufflinks version 2.0.2; specifically, cuffdiff2 and usage of the general transfer format (GTF) annotation file for the given reference genome.
ATDC5 Cell culture and quantitative gene expression analyses
ATDC5 cells (RIKEN BRC) were maintained in DMEM /F-12 (1:1) (Gibco) medium supplemented with 5% FBS and 1% P/S. For ATDC5 cell differentiation studies, ATDC5 cells were cultured with DMEM/F-12 (1:1) medium supplemented with 5% FBS, 1% P/S and 0.1% ITS premix (BD Biosciences). Differentiated ATDC5 chondrogenic cells cultured for 7–14 days were then cultured in serum free DMEM /F-12 (1:1) medium for 6 hours, transfected with FLAG control/NICD1 over-expression vectors using Lipofectamine 2000 (Invitrogen) according to the manufacture’s instruction, or treated with control diluents/recombinant IL6 proteins (R&D System) for 24 hours. For acute signaling studies, differentiated ATDC5 cells were serum starved overnight, and then treated with control diluents or recombinant mouse IL6 protein (R&D System) for indicated time. For experiments using the IL6 neutralizing antibody (Abcam), differentiated ATDC5 cells were pre-treated with the antibody for 24 hours, and then transformed with FLAG control/NICD1 over-expression vectors using Lipofectamine 2000. RNA was isolated using the RNeasy Mini Kit (Qiagen). Complementary DNA synthesis and real-time qPCR were performed as previously described (16). Primer sequences for NICD1, Hes1, Sox9, Col2a1, Acan, Col10a1, Col3a1, Col1a1, Mmp13, Adamts4, Adamts5, and Il6 are available upon request.
Adenovirus infection and inhibitor treatment
For inhibitor experiments, primary costal chondrocytes were isolated from neonatal Rosa-NICD1f/+ pups and cultured in DMEM media with 10% FBS and 1% P/S (referred as growth medium) as described above. Isolated chondrocytes were seeded in 12-well plates at 0.5×106 cells/well. After overnight culture, cells were infected with adenoviruses expressing either green fluorescence protein (Ad-GFP) or Cre (Ad-CRE) at a multiplicity of infection (MOI) of 100 in the presence of 8 ug/ml polybrene. 24 hours after viral infection, cells were cultured either in fresh growth medium (for protein analysis) or in fresh growth medium containing vehicle or indicated inhibitors (for RNA analysis). 48 hours later, cells were harvested for protein or RNA isolation.
ERK1/2 inhibitor U0126 was from Sigma, and used at 10 µM. p38 MAPK inhibitor SB202190 was from Sigma and used at 2 µM. Protein tyrosine kinases inhibitor Genistein was from TOCRIS and used at 60µM. PLC inhibitor Edelfosine was from TOCRIS, and used at 5µM. Adenylate cyclase inhibitor SQ-22536 was from Sigma, and used at 10 µM. Gαiinhibitor Pertussis toxin (PTx) was from TOCRIS and used at 100 ng/ml. Nitric oxide-sensitive guanylyl cyclase inhibitor ODQ was from Sigma, and used at 10 µM. Endothelin receptor A antagonist BQ-123 was from Sigma, and used at 5µM. Phosphodiesterase inhibitor IBMX was from Sigma, and used at 65 µM.
Western blot analyses
For western blot analyses, total proteins were extracted from cells using RIPA buffer [20 mM Tris (pH 8.0), 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate] supplemented with protease and phosphatase inhibitors (Roche). 30 µg protein samples were separated on 10% SDS-polyacrylamide gels and were transferred to PVDF membranes. Immunoblots were then blocked with 5% nonfat milk and incubated overnight with primary antibodies at a final dilution of 1:1000. Antibodies for STAT3, phospho-STAT3 (Tyr705), ERK1/2, phospho-ERK1/2 (Thr202/Tyr204), p38 MAPK, and phospho-p38 MAPK (Thr180/Tyr182) were all purchased from Cell Signaling Technology.
Histomorphometry
Quantitative histomorphometry was performed on ABH/OG stained sections using an OsteoMeasure analysis system (OsteoMetrics). Cartilage thickness was measured from the middle of the femoral and tibial condyles. Cartilage area was traced from both articular cartilage surfaces using the area tool in OsteoMeasure software. 3–5 mice were analyzed in each group and at least 3 slides were examined for each mouse.
Statistical analysis
Statistical analyses were performed using Student’s t-test and one-way ANOVA followed by Bonferroni method where appropriate.
Supplementary Material
Acknowledgements
This work was supported in part by the following United States National Institute of Health grants: R01 grants (AR057022 and AR063071 to MJH), R21 grant (AR059733 to MJH), a P50 Center of Research Translation grant (AR054041 to RJO), and a P30 Core Center grant (AR061307). We thank Dr. Amy E. Kiernan for providing important mouse strains. We would like to gratefully acknowledge the technical expertise and assistance of Sarah Mack, Kathy Maltby, and Ashish Thomas within the Center for Musculoskeletal Research Histology, Biochemistry, and Molecular Imaging Core. We would also like to thank Dr. John Ashton and colleagues within the Genomics Research Center at URMC for RNA sequencing and data processing.
Footnotes
Disclosures
All authors state that they have no conflict of interests.
Supplemental Materials List
Supplemental Figure 1. Development of a NOTCH1 activation model with the combined Tet-On /Cre system.
Supplemental Figure 2. Sustained activation of NOTCH1 signaling in postnatal chondrocytes results in increased chondrocyte apoptosis.
Supplemental Figure 3. Sustained activation of NOTCH1 signaling in postnatal chondrocytes results in a progressive OA-like pathology.
Supplemental Figure 4. Transient activation of NOTCH1 signaling in postnatal chondrocytes results in increased cartilage ECM synthesis and joint maintenance as long as 3 months after DOX injection but does not protect cartilage degradation from MLI surgery.
Supplemental Figure 5. Suppression of tyrosine kinase signaling leads to selected effects on NOTCH-affected genes.
Supplemental Figure 6. Suppression of GPCR signaling leads to selected effects on NOTCH-affected genes.
Supplemental Figure 7. Suppression of NO signaling leads to selected effects on NOTCH-affected genes.
References
- 1.Hunter DJ, Schofield D, Callander E. The individual and socioeconomic impact of osteoarthritis. Nature reviews. Rheumatology. 2014;10:437–441. doi: 10.1038/nrrheum.2014.44. [DOI] [PubMed] [Google Scholar]
- 2.Hootman JM, Helmick CG. Projections of US prevalence of arthritis and associated activity limitations. Arthritis and rheumatism. 2006;54:226–229. doi: 10.1002/art.21562. [DOI] [PubMed] [Google Scholar]
- 3.Bitton R. The economic burden of osteoarthritis. The American journal of managed care. 2009;15:S230–S235. [PubMed] [Google Scholar]
- 4.Kotlarz H, Gunnarsson CL, Fang H, Rizzo JA. Insurer and out-of-pocket costs of osteoarthritis in the US: evidence from national survey data. Arthritis and rheumatism. 2009;60:3546–3553. doi: 10.1002/art.24984. [DOI] [PubMed] [Google Scholar]
- 5.Sandell LJ. Etiology of osteoarthritis: genetics and synovial joint development. Nature reviews. Rheumatology. 2012;8:77–89. doi: 10.1038/nrrheum.2011.199. [DOI] [PubMed] [Google Scholar]
- 6.Bos SD, Slagboom PE, Meulenbelt I. New insights into osteoarthritis: early developmental features of an ageing-related disease. Current opinion in rheumatology. 2008;20:553–559. doi: 10.1097/BOR.0b013e32830aba48. [DOI] [PubMed] [Google Scholar]
- 7.Goldring MB, Goldring SR. Osteoarthritis. Journal of cellular physiology. 2007;213:626–634. doi: 10.1002/jcp.21258. [DOI] [PubMed] [Google Scholar]
- 8.Heinegard D, Saxne T. The role of the cartilage matrix in osteoarthritis. Nature reviews. Rheumatology. 2011;7:50–56. doi: 10.1038/nrrheum.2010.198. [DOI] [PubMed] [Google Scholar]
- 9.Lee AS, Ellman MB, Yan D, Kroin JS, Cole BJ, van Wijnen AJ, Im HJ. A current review of molecular mechanisms regarding osteoarthritis and pain. Gene. 2013;527:440–447. doi: 10.1016/j.gene.2013.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M, Shen J, Jin H, Im HJ, Sandy J, Chen D. Recent progress in understanding molecular mechanisms of cartilage degeneration during osteoarthritis. Annals of the New York Academy of Sciences. 2011;1240:61–69. doi: 10.1111/j.1749-6632.2011.06258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nature reviews. Rheumatology. 2011;7:33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 12.Daheshia M, Yao JQ. The interleukin 1beta pathway in the pathogenesis of osteoarthritis. The Journal of rheumatology. 2008;35:2306–2312. doi: 10.3899/jrheum.080346. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi M, Squires GR, Mousa A, Tanzer M, Zukor DJ, Antoniou J, Feige U, Poole AR. Role of interleukin-1 and tumor necrosis factor alpha in matrix degradation of human osteoarthritic cartilage. Arthritis and rheumatism. 2005;52:128–135. doi: 10.1002/art.20776. [DOI] [PubMed] [Google Scholar]
- 14.Reynard LN, Loughlin J. Insights from human genetic studies into the pathways involved in osteoarthritis. Nature reviews. Rheumatology. 2013;9:573–583. doi: 10.1038/nrrheum.2013.121. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, Huang X, Li L, Huang H, Xu R, Luyten W. Insights on biology and pathology of HIF-1alpha/-2alpha, TGFbeta/BMP, Wnt/beta-catenin, and NF-kappaB pathways in osteoarthritis. Current pharmaceutical design. 2012;18:3293–3312. doi: 10.2174/1381612811209023293. [DOI] [PubMed] [Google Scholar]
- 16.Dong Y, Jesse AM, Kohn A, Gunnell LM, Honjo T, Zuscik MJ, O'Keefe RJ, Hilton MJ. RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development. 2010;137:1461–1471. doi: 10.1242/dev.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohn A, Dong Y, Mirando AJ, Jesse AM, Honjo T, Zuscik MJ, O'Keefe RJ, Hilton MJ. Cartilage-specific RBPjkappa-dependent and -independent Notch signals regulate cartilage and bone development. Development. 2012;139:1198–1212. doi: 10.1242/dev.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mead TJ, Yutzey KE. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14420–14425. doi: 10.1073/pnas.0902306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. Journal of cell science. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 22.Hayes AJ, Dowthwaite GP, Webster SV, Archer CW. The distribution of Notch receptors and their ligands during articular cartilage development. Journal of anatomy. 2003;202:495–502. doi: 10.1046/j.1469-7580.2003.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowthwaite GP, Bishop JC, Redman SN, Khan IM, Rooney P, Evans DJ, Haughton L, Bayram Z, Boyer S, Thomson B, Wolfe MS, Archer CW. The surface of articular cartilage contains a progenitor cell population. Journal of cell science. 2004;117:889–897. doi: 10.1242/jcs.00912. [DOI] [PubMed] [Google Scholar]
- 24.Mirando AJ, Liu Z, Moore T, Lang A, Kohn A, Osinski AM, O'Keefe RJ, Mooney RA, Zuscik MJ, Hilton MJ. RBP-Jkappa-dependent Notch signaling is required for murine articular cartilage and joint maintenance. Arthritis and rheumatism. 2013;65:2623–2633. doi: 10.1002/art.38076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosaka Y, Saito T, Sugita S, Hikata T, Kobayashi H, Fukai A, Taniguchi Y, Hirata M, Akiyama H, Chung UI, Kawaguchi H. Notch signaling in chondrocytes modulates endochondral ossification and osteoarthritis development. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1875–1880. doi: 10.1073/pnas.1207458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahjoub M, Sassi N, Driss M, Laadhar L, Allouche M, Hamdoun M, Romdhane KB, Sellami S, Makni S. Expression patterns of Notch receptors and their ligands in human osteoarthritic and healthy articular cartilage. Tissue & cell. 2012;44:182–194. doi: 10.1016/j.tice.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson C, Brantsing C, Egell S, Lindahl A. Notch1, Jagged1, and HES5 are abundantly expressed in osteoarthritis. Cells, tissues, organs. 2008;188:287–298. doi: 10.1159/000121610. [DOI] [PubMed] [Google Scholar]
- 28.Livshits G, Zhai G, Hart DJ, Kato BS, Wang H, Williams FM, Spector TD. Interleukin-6 is a significant predictor of radiographic knee osteoarthritis: The Chingford Study. Arthritis and rheumatism. 2009;60:2037–2045. doi: 10.1002/art.24598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Reilly S, Ciechomska M, Cant R, Hugle T, van Laar JM. Interleukin-6, its role in fibrosing conditions. Cytokine & growth factor reviews. 2012;23:99–107. doi: 10.1016/j.cytogfr.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Mihara M, Hashizume M, Yoshida H, Suzuki M, Shiina M. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin Sci (Lond) 2012;122:143–159. doi: 10.1042/CS20110340. [DOI] [PubMed] [Google Scholar]
- 31.Hashizume M, Hayakawa N, Mihara M. IL-6 trans-signalling directly induces RANKL on fibroblast-like synovial cells and is involved in RANKL induction by TNF-alpha and IL-17. Rheumatology (Oxford) 2008;47:1635–1640. doi: 10.1093/rheumatology/ken363. [DOI] [PubMed] [Google Scholar]
- 32.Hashizume M, Mihara M. High molecular weight hyaluronic acid inhibits IL-6-induced MMP production from human chondrocytes by up-regulating the ERK inhibitor, MKP-1. Biochemical and biophysical research communications. 2010;403:184–189. doi: 10.1016/j.bbrc.2010.10.135. [DOI] [PubMed] [Google Scholar]
- 33.Kamekura S, Hoshi K, Shimoaka T, Chung U, Chikuda H, Yamada T, Uchida M, Ogata N, Seichi A, Nakamura K, Kawaguchi H. Osteoarthritis development in novel experimental mouse models induced by knee joint instability. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2005;13:632–641. doi: 10.1016/j.joca.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Sampson ER, Beck CA, Ketz J, Canary KL, Hilton MJ, Awad H, Schwarz EM, Chen D, O'Keefe RJ, Rosier RN, Zuscik MJ. Establishment of an index with increased sensitivity for assessing murine arthritis. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2011;29:1145–1151. doi: 10.1002/jor.21368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iannone F, De Bari C, Dell'Accio F, Covelli M, Patella V, Lo Bianco G, Lapadula G. Increased expression of nerve growth factor (NGF) and high affinity NGF receptor (p140 TrkA) in human osteoarthritic chondrocytes. Rheumatology. 2002;41:1413–1418. doi: 10.1093/rheumatology/41.12.1413. [DOI] [PubMed] [Google Scholar]
- 36.Lowin T, Weidler C, Jenei-Lanzl Z, Capellino S, Baerwald CG, Buttgereit F, Straub RH. Relationship between placenta growth factor 1 and vascularization, dehydroepiandrosterone sulfate to dehydroepiandrosterone conversion, or aromatase expression in patients with rheumatoid arthritis and patients with osteoarthritis. Arthritis and rheumatism. 2012;64:1799–1808. doi: 10.1002/art.34338. [DOI] [PubMed] [Google Scholar]
- 37.Pecchi E, Priam S, Gosset M, Pigenet A, Sudre L, Laiguillon MC, Berenbaum F, Houard X. Induction of nerve growth factor expression and release by mechanical and inflammatory stimuli in chondrocytes: possible involvement in osteoarthritis pain. Arthritis research & therapy. 2014;16:R16. doi: 10.1186/ar4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramos YF, den Hollander W, Bovee JV, Bomer N, van der Breggen R, Lakenberg N, Keurentjes JC, Goeman JJ, Slagboom PE, Nelissen RG, Bos SD, Meulenbelt I. Genes involved in the osteoarthritis process identified through genome wide expression analysis in articular cartilage; the RAAK study. PloS one. 2014;9:e103056. doi: 10.1371/journal.pone.0103056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoo SA, Yoon HJ, Kim HS, Chae CB, De Falco S, Cho CS, Kim WU. Role of placenta growth factor and its receptor flt-1 in rheumatoid inflammation: a link between angiogenesis and inflammation. Arthritis and rheumatism. 2009;60:345–354. doi: 10.1002/art.24289. [DOI] [PubMed] [Google Scholar]
- 40.Appleton CT, James CG, Beier F. Regulator of G-protein signaling (RGS) proteins differentially control chondrocyte differentiation. Journal of cellular physiology. 2006;207:735–745. doi: 10.1002/jcp.20615. [DOI] [PubMed] [Google Scholar]
- 41.Neumann E, Khawaja K, Muller-Ladner U. G protein-coupled receptors in rheumatology. Nature reviews. Rheumatology. 2014;10:429–436. doi: 10.1038/nrrheum.2014.62. [DOI] [PubMed] [Google Scholar]
- 42.Abramson SB, Attur M, Amin AR, Clancy R. Nitric oxide and inflammatory mediators in the perpetuation of osteoarthritis. Current rheumatology reports. 2001;3:535–541. doi: 10.1007/s11926-001-0069-3. [DOI] [PubMed] [Google Scholar]
- 43.Pelletier J, Jovanovic D, Fernandes JC, Manning P, Connor JR, Currie MG, Martel-Pelletier J. Reduction in the structural changes of experimental osteoarthritis by a nitric oxide inhibitor. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 1999;7:416–418. doi: 10.1053/joca.1998.0229. [DOI] [PubMed] [Google Scholar]
- 44.Pelletier JP, Lascau-Coman V, Jovanovic D, Fernandes JC, Manning P, Connor JR, Currie MG, Martel-Pelletier J. Selective inhibition of inducible nitric oxide synthase in experimental osteoarthritis is associated with reduction in tissue levels of catabolic factors. The Journal of rheumatology. 1999;26:2002–2014. [PubMed] [Google Scholar]
- 45.Studer R, Jaffurs D, Stefanovic-Racic M, Robbins PD, Evans CH. Nitric oxide in osteoarthritis. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 1999;7:377–379. doi: 10.1053/joca.1998.0216. [DOI] [PubMed] [Google Scholar]
- 46.Ustunel I, Ozenci AM, Sahin Z, Ozbey O, Acar N, Tanriover G, Celik-Ozenci C, Demir R. The immunohistochemical localization of notch receptors and ligands in human articular cartilage, chondroprogenitor culture and ultrastructural characteristics of these progenitor cells. Acta histochemica. 2008;110:397–407. doi: 10.1016/j.acthis.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Grogan SP, Miyaki S, Asahara H, D'Lima DD, Lotz MK. Mesenchymal progenitor cell markers in human articular cartilage: normal distribution and changes in osteoarthritis. Arthritis research & therapy. 2009;11:R85. doi: 10.1186/ar2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nature medicine. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blaise R, Mahjoub M, Salvat C, Barbe U, Brou C, Corvol MT, Savouret JF, Rannou F, Berenbaum F, Bausero P. Involvement of the Notch pathway in the regulation of matrix metalloproteinase 13 and the dedifferentiation of articular chondrocytes in murine cartilage. Arthritis and rheumatism. 2009;60:428–439. doi: 10.1002/art.24250. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe N, Tezuka Y, Matsuno K, Miyatani S, Morimura N, Yasuda M, Fujimaki R, Kuroda K, Hiraki Y, Hozumi N, Tezuka K. Suppression of differentiation and proliferation of early chondrogenic cells by Notch. J Bone Miner Metab. 2003;21:344–352. doi: 10.1007/s00774-003-0428-4. [DOI] [PubMed] [Google Scholar]
- 51.Chen S, Tao J, Bae Y, Jiang MM, Bertin T, Chen Y, Yang T, Lee B. Notch gain of function inhibits chondrocyte differentiation via Rbpj-dependent suppression of Sox9. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2012 doi: 10.1002/jbmr.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haller R, Schwanbeck R, Martini S, Bernoth K, Kramer J, Just U, Rohwedel J. Notch1 signaling regulates chondrogenic lineage determination through Sox9 activation. Cell death and differentiation. 2012;19:461–469. doi: 10.1038/cdd.2011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyazawa K, Mori A, Yamamoto K, Okudaira H. Transcriptional roles of CCAAT/enhancer binding protein-beta, nuclear factor-kappaB, and C-promoter binding factor 1 in interleukin (IL)-1beta-induced IL-6 synthesis by human rheumatoid fibroblast-like synoviocytes. The Journal of biological chemistry. 1998;273:7620–7627. doi: 10.1074/jbc.273.13.7620. [DOI] [PubMed] [Google Scholar]
- 54.Wongchana W, Palaga T. Direct regulation of interleukin-6 expression by Notch signaling in macrophages. Cellular & molecular immunology. 2012;9:155–162. doi: 10.1038/cmi.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zanotti S, Canalis E. Interleukin 6 mediates selected effects of Notch in chondrocytes. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2013;21:1766–1773. doi: 10.1016/j.joca.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nature cell biology. 2004;6:547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- 57.Wei F, Moore DC, Wei L, Li Y, Zhang G, Wei X, Lee JK, Chen Q. Attenuation of osteoarthritis via blockade of the SDF-1/CXCR4 signaling pathway. Arthritis research & therapy. 2012;14:R177. doi: 10.1186/ar3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henry SP, Jang CW, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis. 2009;47:805–814. doi: 10.1002/dvg.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128:5099–5108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- 61.Stanger BZ, Datar R, Murtaugh LC, Melton DA. Direct regulation of intestinal fate by Notch. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic acids research. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hilton MJ, Tu X, Long F. Tamoxifen-inducible gene deletion reveals a distinct cell type associated with trabecular bone, and direct regulation of PTHrP expression and chondrocyte morphology by Ihh in growth region cartilage. Developmental biology. 2007;308:93–105. doi: 10.1016/j.ydbio.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.