Abstract
Organohalide contaminants such as triclosan and triclocarban have been well documented in municipal wastewater treatment plants (WWTPs), but the degradation of these contaminants is not well understood. One possible removal mechanism is organohalide respiration by which bacteria reduce the halogenated compound. The purpose of this study was to determine the abundance of organohalide-respiring bacteria in eight WWTP anaerobic digesters. The obligate organohalide respiring Dehalococcoides mccartyi was the most abundant and averaged 3.3 × 107 copies of 16S rRNA genes per gram, while the Dehalobacter was much lower at 2.6 × 104 copies of 16S rRNA genes per gram. The genus Sulfurospirillum spp. was also detected at 1.0 × 107 copies of 16S rRNA genes per gram. No other known or putatively organohalide-respiring strains in the Dehalococcoidaceae family were found to be present nor were the genera Desulfitobacterium or Desulfomonile.
Keywords: triclosan, triclocarban, reductive dehalogenation, Dehalococcoides mccartyi, Dehalobacter
Introduction
A wide variety of halogenated compounds (organohalides or organohalogens) are found in wastewater coming from a variety of sources such as cleaning agents, disinfectants, industrial runoff, and pesticides.1 Compounds such as polychlorinated biphenyls, chlorophenols, triclocarban, triclosan, and diclofenac have been quantified through municipal wastewater treatment plants (WWTPs).2,3 Because of the carcinogenic, toxic, and/or estrogenic nature of many of these compounds,4–6 as well as the potential for the spread of antibiotic resistance by some such as triclosan,7 the presence of these compounds in wastewater raises concerns for the land application of WWTP biosolids and downstream water quality.
In WWTPs, the removal of organohalides from the water varies; however, there is typically a reduction of the concentration throughout the system.8–11 For example, triclosan was found in one WWTP to have an influent concentration of 0.93–2.11 μg L−1, and the effluent concentration was found to be 0.07–0.15 μg L−1.2 A measurement of adsorbed organic halogen compounds showed a significant reduction as it passed through another WWTP.9 Though much of the halogenated compounds are being removed from the water during treatment, these compounds are predominately sorbed onto the solids and moved into solids treatment processes such as anaerobic digesters.11 Some degradation of organohalides is found likely to occur from mass balances.11 One study, for example, showed that 93% of triclosan is removed from the influent, with 41% sorbed to the sludge and 52% transformed or elsewise lost in the process.2
Though the concentrations of organohalides in WWTPs are becoming well documented,12 the degradation of these compounds in WWTPs is not well understood. One possible removal mechanism is through organohalide respiration, where the organohalide is dehalogenated, and thus typically transformed to less toxic or nontoxic compounds.13 A wide range of organohalide-respiring bacteria has been isolated and many of their genomes have been sequenced.14 In a WWTP, these processes may occur in anaerobic digesters, where reducing conditions would be favorable to organohalide-respiring physiology. Indeed, the organohalide-respiring strain Dehalococcoides mccartyi st. 195 as well as others were isolated from enrichment cultures stemming from WWTP digesters.15–17 Some of the contaminants in WWTPs such as polychlorinated biphenyls and chlorophenols are well known to be degraded by organohalide respirers18,19 and have been found to be dechlorinated in anaerobic digesters.9,20 Dechlorination products of the two chlorinated compounds with the highest concentrations in WWTPs, triclocarban, and triclosan, have also recently been identified in sludge treatment processes,21 and organohalide respiration of these compounds has been suggested by some studies,22 but not yet conclusively proven.
The abundance and diversity of organohalide-respiring bacteria in WWTP anaerobic digesters have not been directly studied. A meta-analysis23 of public database-deposited sequences from WWTP digesters and metagenomics of digesters24,25 have produced scant insight toward the abundance of organohalide-respiring bacteria as well. The purpose of this study was to determine the abundance of organohalide-respiring bacteria in WWTP anaerobic digesters.
Methodologies
Anaerobic digester sludge was collected from seven WWTPs in the central and northeastern Oklahoma region. One WWTP operated a two-stage anaerobic digestion process, and samples were collected from both the first- and the second-stage digester. Samples were collected in August 2014 in 1 L plastic bottles that were filled, tightly capped, and transported to the laboratory within two hours, where they were placed in a refrigerator.
DNA was extracted from between 0.25 and 0.5 g of sludge using the PowerSoil DNA extraction kit (MoBio Laboratories) according to manufacturer’s recommendations. Previously published quantitative PCR (qPCR) methodologies were used to quantify the known obligate organohalide-respiring genera of D. mccartyi,26 the Dehalobium chlorocoercia DF-1/o-17 group,27 Dehalogenimonas,28 and Dehalobacter.29 Also targeted were the facultative organohalide-respiring Desulfitobacteria,29 Geobacter,30 Desulfomonile tiedjei,31 Desulfovibrio,32 and Sulfurospirillum.33 The total bacteria 16S rRNA genes were also quantified.34 The putatively dechlorinating Gopher group35 and a less specific Dehalococcoides-like Chloroflexi group36 were also attempted for amplification. The primers for each qPCR protocol are listed in Table 1. Each qPCR mixture totaled 10 μL and contained 1 × iTaq SYBR Green Supermix with Rox (Bio-Rad Laboratories), 300 nM of each primer, and 0.5 μL of undiluted DNA extract or standard. Primers were purchased from Life Technologies. Analysis was on a CFX Connect Real Time System with CFX Manager software (Bio-Rad Laboratories). For all phylogenetic targets other than D. mccartyi, the thermocycling protocol was 95°C for three minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds. For D. mccartyi, the thermocycling protocol was 95°C for 3 minutes followed by 40 cycles of 95°C for 15 seconds and 56°C for 30 seconds. A melting curve analysis was performed after each complete run to ensure that primer-dimers were not amplified and that the amplification was specific. Standards for each qPCR were prepared from plasmid extracts containing the 16S rRNA gene of interest and quantified from the QuantiFluor dsDNA dye system (Promega Corporation). Each sample and standard was analyzed in triplicate qPCR runs. Each qPCR analysis was performed with negative controls (water blanks) and positive controls (DNA extract from mixed cultures with the targeted 16S rRNA gene).
Table 1.
Primers used to study organohalide-respiring organisms and Bacteria.
| PHYLOGENETIC TARGET | PRIMER | SEQUENCE | REFERENCE |
|---|---|---|---|
| Dehalococcoides mccartyi | 582F | 5′-CTGTTGGACTAGAGTACAGC-3′ | 26 |
| 728R | 5′-GTGACAACCTAGAAAACCGCCTT-3′ | ||
| “Dehalococcoides-like Chloroflexi” | 1150F | 5′-GGGCTACACACACGCTACAATGG-3′ | 36 |
| 1286R | 5′-GATATGCGGTTACTAGCAACTCCAAC-3′ | ||
| Dehalobium chlorocoercia DF1 | 866F | 5′-CGCTTTAAGTRTCCCGCC-3′ | 27 |
| 1265R | 5′-CCTATTGCTACCTGCTGTACC-3′ | ||
| Dehalogenimonas spp. | 634F | 5′-GGTCATCTGATACTGTTGGACTTGAGTATG-3′ | 28 |
| 799R | 5′-ACCCAGTGTTTAGGGCGTGGACTACCAGG-3′ | ||
| Dehalobacter spp. | 447F | 5′-GATTGACGGTACCTAACGAGG-3′ | 29 |
| 647R | 5′-TACAGTTTCCAATGCTTTACGG-3′ | ||
| Desulfitobacterium spp. | 406F | 5′-GTACGACGAAGGCCTTCGGGT-3′ | 29 |
| 619R | 5′-CCCAGGGTTGAGCCCTAGGT-3′ | ||
| Geobacter lovleyi | 564F | 5′-AAGCGTTGTTCGGAWTTAT-3′ | 30 |
| 840R | 5′-GGCACTGCAGGGGTCAATA-3′ | ||
| Desulfomonile spp. | 205F | 5′-GGGTCAAAGTCGGCCTCTCGACG-3′ | 31 |
| 628R | 5′-GCTTTCACATTCGACTTATCG-3′ | ||
| Desulfovibrio spp. | 691F | 5′-CCGTAGATATCTGGAGGAACATCAG-3′ | 32 |
| 826R | 5′-ACATCTAGCATCCATCGTTTACAGC-3′ | ||
| Sulfurospirillum spp. | 114F | 5′-GCTAACCTGCCCTTTAGTGG-3′ | 33 |
| 421R | 5′-GTTTACACACCGAAATGCGT-3′ | ||
| Gopher Group | 163F | 5′-TGACCYTGGCATCAGGGA-3′ | 35 |
| 441R | 5′-TATTTTACAACCCGAAGGCCTTCG-3′ | ||
| Bacteria | 341F | 5′-CCTACGGGAGGCAGCAG-3′ | 34 |
| 534R | 5′-ATTACCGCGGCTGCTGGC-3′ |
The primers, amplification efficiencies, standard curve R2 values, and standard ranges are listed in Table 2. Standard ranges represent standards between the lowest reproducible serially diluted standard and the highest standard in the linear range. Quantification limits are calculated to be the minimum concentration of 16S rRNA genes needed to be detected above the minimum standard. The D. chlorocoercia DF-1/o-17 group, Gopher group, and Desulfomonile were found not to be amplified from any digester samples under a range of melting temperatures (50°C–62°C), and standard curves were not produced for these methods. Furthermore, Desulfitobacterium were not detected above the quantification limits for any sample. For all genera that were above the quantification limit in the samples, clone libraries of the product were produced using the pGEM-T Easy Vector system (Pro-mega) from a minimum of two samples to verify specificity of the amplification. All sequences from clone libraries from the Dehalobacter, D. mccartyi, and Sulfurospirillum amplification were verified to be closely related to their targeted genera (>98% sequence identity) with a total of 12, 9, and 9 clones, respectively. Amplifications were found to be unspecific for Dehalogenimonas (9 total clones, all related to Bacteroidetes) and Dehalococcoides-like Chloroflexi (10 clones, 9 from various Firmicutes and 1 from the class Anaerolineae in Chloroflexi). The amplifications of Geobacter as well as Desulfovibrio yielded amplicons with a mixture of targeted and nontargeted genera with 6 out of 16 clones being specific to their targeted genus (>98%) for Geobacter and 2 out of 13 clones being specific for the Desulfovibrio amplification. The nontargeted Geobacter amplicons were from various genera in the Proteobacteria, while the nontargeted Desulfovibrio amplicons were Clostridia from the phylum Firmicutes and various genera of Anaerolineae from the Chloroflexi phylum.
Table 2.
Parameters of qPCR analysis.
| PHYLOGENETIC TARGET | EFFICIENCY, R2 | LINEAR RANGE (COPIES PER REACTION) | QUANTIFICATION LIMITS (COPIES PER g) |
|---|---|---|---|
| Dehalococcoides mccartyi | 104%, 0.99 | 1.9 × 101–1.9 × 106 | 7,600 |
| “Dehalococcoides-like Chloroflexi” | 105%, 0.99 | 1.9 × 102–1.9 × 108 | 76,000 |
| Dehalogenimonas spp. | 93%, 0.99 | 1.7 × 101–1.7 × 107 | 6,800 |
| Dehalobacter spp. | 110%, 0.99 | 1.2 × 101–1.2 × 105 | 4,800 |
| Desulfitobacterium spp. | 110%, 0.97 | 2.2 × 102–2.2 × 109 | 88,000 |
| Geobacter lovleyi | 76%, 0.98 | 5.8 × 101–5.8 × 106 | 23,200 |
| Desulfovibrio spp. | 99%, 0.98 | 1.3 × 102–1.3 × 109 | 5,200 |
| Sulfurospirillum spp. | 87%, 0.99 | 6.7 × 101–6.7 × 106 | 26,800 |
| Bacteria | 75%, 0.96 | 1.9 × 105–1.9 × 109 | 7.6 × 108 |
Terminal restriction fragment length polymorphism (TRFLP) was used in this study to identify nontargeted strains of the Dehalococcoidaceae family of Chloroflexi,37 which may contain nonisolated strains of organohalide-respiring bacteria. The PCR amplification and enzyme digestion were performed as described by Krzmarzick et al,36 except that the general Eubacteria primer 8F (5′-AGAGTTTGATCMTG GCTCAG-3′) was used in lieu of the primer Dhc553F. The fragment-size analysis of amplifications was performed at the DNA/Protein Core facility at Oklahoma State University with an ABI Model 3730 Analyzer with a MapMarker1000 size standard (BioVentures, Inc.). Clone libraries were performed on the PCR amplicons of the anaerobic digester samples using the pGEM-T Easy Vector system (Promega) and sequenced at the DNA/Protein core facility at Oklahoma State University with an ABI Model 3730 Analyzer. A total of 17 unique sequences were found, which corresponded to 15 OTUs (theoretical digestion sizes) representing all major groups (>1% of any sample’s total peak area) found in the TRFLP analysis in the samples. BLAST analysis of these sequences indicated that none of these sequences belonged to the Chloroflexi. Thus, any putative reductive dechlorinating Chloroflexi was of low abundance compared with the nontargeted groups that were amplified.
Results and Discussion
Dehalogenimonas spp., D. chlorocoercia DF-1, strain o-17, Desulfitobacterium spp., the Gopher group, D. tiedjei, and nonisolated and putatively organohalide-respiring strains in the Dehalococcoidaceae family were not found to be present in the qPCR and TRFLP methods used in this study in any of the sampled digesters. The amplification of nontargeted groups in the qPCR results for Dehalogenimonas and the Dehalococcoides-like Chloroflexi and the dominance of nontargeted groups in the TRFLP analysis preclude a conclusive determination that these organisms are not in digesters. These bacteria may still be present at some level but low in comparison to the nontargeted organisms that were amplified. For example, the species D. mccartyi was neither recovered in the TRFLP analysis nor in the clone libraries from the Dehalococcoides-like Chloroflexi qPCR analysis, though its 16S gene has 100% identities to the primers used in those methods. Yet the more specific qPCR method targeting this genus and clone libraries of this method verified its existence.
In a meta-analysis of sequences deposited in public databases, 12 sequences related to Dehalogenimonas spp. (out of >16,500 total sequences) were identified with anaerobic digester communities,23 indicating Dehalogenimonas spp. may be present in low levels of anaerobic digesters. The strains Dehalobium DF-1, o-17, Desulfitobacteria spp., Gopher group, and Desulfomonile may similarly be present in anaerobic digesters though not found in this study; indeed, D. tiedjei was isolated from anaerobic digester sludge.38
The Desulfovibrio and Geobacter genera were detected in the anaerobic digester samples. The quantification of Desulfovibrio ranged from 1.4 × 107 to 2.4 × 108 copies of 16S rRNA genes per gram, while the Geobacter ranged from 1.7 × 107 to 1.1 × 108 copies of 16S rRNA genes per gram (Table 3). These amplifications must be interpreted as maximal values as clone library analysis of their products indicated nontargeted genera were amplified as well. Furthermore, the sequences recovered from these amplifications that were closely related to their targeted genera were still <98% identical to the 16S rRNA genes of Geobacter lovleyii30 and Desulfovibrio dechloracetivorans39 strains, respectively, which are the facultative organohalide-respiring strains of their respective genera. Thus, the amount of these genera capable of organohalide respiration in anaerobic digesters may be minimal.
Table 3.
Number of 16S rRNA genes of Bacteria and organohalide respirers in WWTP anaerobic digesters (logarithmic units per gram of digester contents).
| SAMPLE | Bacteria | Dehalobacter | Dehalococcoides1 | Sulfurospirillum | Geobacter | Desulfovibrio |
|---|---|---|---|---|---|---|
| Digester 1 | 11.4 ± 0.042 | 4.45 ± 0.23 | 7.28 ± 0.02 | 6.92 ± 0.13 | <7.44 ± 0.19 | <7.56 ± 0.02 |
| Digester 2 | 11.1 ± 0.07 | 3.85 ± 0.10 | 7.14 ± 0.01 | 6.52 ± 0.05 | <7.23 ± 0.11 | <7.38 ± 0.01 |
| Digester 3 | 11.7 ± 0.03 | 4.04 ± 0.15 | 7.72 ± 0.02 | 7.33 ± 0.06 | <8.03 ± 0.09 | <7.39 ± 0.09 |
| Digester 4 | 11.3 ± 0.01 | 4.29 ± 0.08 | 7.55 ± 0.02 | 7.38 ± 0.09 | <7.83 ± 0.10 | <7.40 ± 0.05 |
| Digester 5 | 11.1 ± 0.12 | 4.24 ± 0.23 | 7.57 ± 0.09 | 6.97 ± 0.09 | <7.49 ± 0.32 | <7.34 ± 0.09 |
| Digester 6 | 11.2 ± 0.08 | 4.62 ± 0.06 | 7.52 ± 0.09 | 6.73 ± 0.03 | <7.49 ± 0.09 | <8.39 ± 0.01 |
| Digester 7 | 11.2 ± 0.24 | 4.77 ± 0.03 | 7.33 ± 0.05 | 6.55 ± 0.05 | <7.41 ± 0.53 | <7.15 ± 0.05 |
| Digester 8 | 11.6 ± 0.07 | BDL3 | 7.69 ± 0.07 | 6.80 ± 0.17 | <7.70 ± 0.11 | <7.45 ± 0.08 |
Notes:
D. mccartyi determined from 582F//728R primer pair in Table 1.
Mean ± standard deviation of qPCR analysis.
Below detection limit.
The quantification of D. mccartyi, Dehalobacter spp., Sulfurospirillum spp., and total bacteria are shown in Table 3 and Figure 1. The quantification of D. mccartyii, Dehalobacter spp., and Sulfurospirillum spp. are confirmed with clone library analysis of their amplification products. Total bacteria 16S rRNA genes in the anaerobic digesters ranged from 1.3 × 1011 to 5.4 × 1011 copies of 16S rRNA genes per gram with an average of 2.5 × 1011 16S rRNA genes per gram. The organohalide-respiring genera were orders of magnitude less. Dehalobacter was detected just over the quantification limit in seven digesters while being below the quantification limit in one digester. Dehalobacter ranged up to 5.9 × 104 copies of 16S rRNA genes per gram with an average of 2.6 × 104 copies of 16S rRNA genes per gram in the seven digesters with Dehalobacter. The obligate organohalide-respiring D. mccartyi was more abundant and ranged from 1.4 × 107 to 5.2 × 107 copies of 16S rRNA genes per gram with an average of 3.3 × 107 copies of 16S rRNA genes per gram. The genera containing the facultative organohalide-respiring bacterium Sulfurospirillum multivorans40 ranged from 3.3 × 106 to 2.4 × 107 copies of 16S rRNA genes per gram and averaged 1.0 × 107 copies of 16S rRNA genes per gram. Sulfurospirillum clones were more closely related to strains other than S. multivorans, which is the strain found to have reductive dehalogenase genes.40 The percent identity with S. multivorans was typically 96%–97%, while other Sulfurospirillum strains were related at up to 100% identity. Thus, these measurements may be true for the genus; they likely overestimate greatly the abundance of Sulfurospirillum capable of organohalide respiration.
Figure 1.
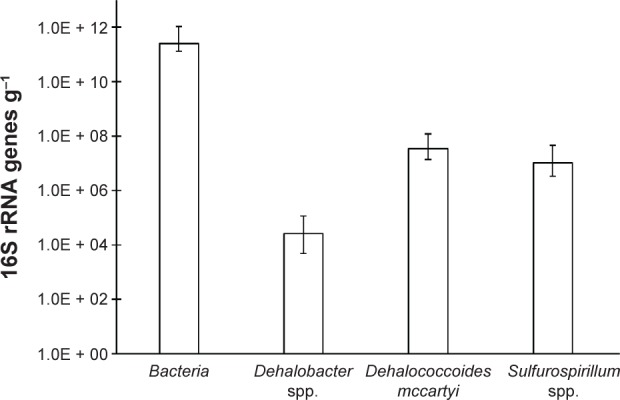
The averages and ranges of 16S rRNA genes per gram quantified with qPCR in eight anaerobic digester samples for bacteria, Dehalobacter spp., D. mccartyi, and Sulfurospirillum spp.
Despite concentrations of organohalides ranging in the mg/kg levels in anaerobic digester sludge,11 the microbial communities capable of organohalide respiration are relatively minimal, representing about 0.02% of the total bacterial community. Though low levels of microbial communities can have relevant implications for degradation, these levels of bacteria are rather low and may be a major contributing reason that the degradation of organohalide contaminants are limited. The differences of populations between digesters were less than one order of magnitude indicating that these populations may not vary significantly between WWTP anaerobic digesters operated in the United States. This small range in organohalide-respiring populations is in contrast to the several orders of magnitude range of organohalides found in anaerobic digesters.12 The concentrations of organohalides were not found in the digesters of this study, and thus, the relationship between organohalide abundance and organohalide-respiring populations is not known. Digesters 1 and 2 in Table 3 are, respectively, the primary and secondary digesters from a single WWTP. The secondary digester contained lower abundances than the primary digester for all organohalide-respiring bacteria and was generally on the low end of the ranges for all digesters studied.
The presence of obligate organohalide-respiring bacteria with broad dechlorination abilities14 such as Dehalococcoides and Dehalobacter is significant as it does indicate that dechlorination processes do occur in WWTPs, and this process is likely responsible for some of the decrease in organohalides as suggested by the discovery of triclosan and triclocarban dechlorination products.21 Though many conditions in digesters seem favorable for organohalide respirers, such as mesophilic temperatures41 and highly reducing conditions,42 other physiological conditions may be limiting their growth. The goal of reducing organohalides, however, may ultimately compete with (or complement) other goals in anaerobic digestion, such as increasing methane production.43 Still, further work is warranted to determine if operational conditions may be adjusted to favor organohalide respiration to reduce the concentrations of these chemicals in biosolids.
Footnotes
ACADEMIC EDITOR: Raul Rivas, Editor in Chief
PEER REVIEW: Three peer reviewers contributed to the peer review report. Reviewers’ reports totaled 511 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported by Oklahoma State University start-up funds to MJK. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Taught the study, which was a component of a graduate level course on molecular methods at Oklahoma State University, collected samples, and conceived and designed the experiments: MJK. Performed experiments, analyzed data, and jointly developed the structure and arguments of the manuscript: BJKS, MAB, BAF, TML, RKR, and MJK. Wrote the first draft of the manuscript: BJKS. Made critical revisions: MJK. All authors reviewed and approved the final manuscript.
REFERENCES
- 1.Schulz S, Hahn H. Generation of halogenated organic compounds in municipal wastewater. Water Sci Technol. 1998;37:303–309. [Google Scholar]
- 2.Samaras VG, Stasinakis AS, Mamais D, Thomaidis NS, Lekkas TD. Fate of selected pharmaceuticals and synthetic endocrine disrupting compounds during wastewater treatment and sludge anaerobic digestion. J Hazard Mater. 2013;244–245:259–267. doi: 10.1016/j.jhazmat.2012.11.039. [DOI] [PubMed] [Google Scholar]
- 3.Zhu S, Chen H. The fate and risk of selected pharmaceutical and personal care products in wastewater treatment plants and a pilot-scale multistage constructed wetland system. Environ Sci Pollut Res Int. 2014;21:1466–1479. doi: 10.1007/s11356-013-2025-y. [DOI] [PubMed] [Google Scholar]
- 4.Faroon OM, Keith S, Jones D, de Rosa C. Carcinogenic effects of polychlorinated biphenyls. Toxicol Ind Health. 2001;17:41–62. doi: 10.1191/0748233701th098oa. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson JL, Jacobson SW. Evidence for PCBs as neurodevelopmental toxicants to humans. Neurotoxicology. 1997;18:415–424. [PubMed] [Google Scholar]
- 6.Dann AB, Hontela A. Triclosan: environmental exposure, toxicity and mechanisms of action. J Appl Toxicol. 2011;3:285–311. doi: 10.1002/jat.1660. [DOI] [PubMed] [Google Scholar]
- 7.Schweizer HP. Triclosan: a widely used biocide and its link to antibiotics. FEMS Microbiol Lett. 2001;202:1–7. doi: 10.1111/j.1574-6968.2001.tb10772.x. [DOI] [PubMed] [Google Scholar]
- 8.Nelson ED, Do H, Lewis RS, Carr SA. Diurnal variability of pharmaceutical, personal care product, estrogen and alkylphenol concentrations in effluent from a tertiary wastewater treatment facility. Environ Sci Technol. 2011;45:1228–1234. doi: 10.1021/es102452f. [DOI] [PubMed] [Google Scholar]
- 9.Benabdallah El-Hadj T, Dosta J, Torres R, Mata-Álvarez J. PCB and AOX removal in mesophilic and thermophilic sewage sludge digestion. Biochem Eng J. 2007;36:281–287. [Google Scholar]
- 10.Bester K. Triclosan in a sewage treatment process—balances and monitoring data. Water Res. 2003;37:3891–3896. doi: 10.1016/S0043-1354(03)00335-X. [DOI] [PubMed] [Google Scholar]
- 11.Krzmarzick MJ, Novak PJ. Removal of chlorinated organic compounds during wastewater treatment: achievements and limits. Appl Microbiol Biotechnol. 2014;98:6233–6242. doi: 10.1007/s00253-014-5800-x. [DOI] [PubMed] [Google Scholar]
- 12.Heidler J, Halden RU. Fate of organohalogens in US wastewater treatment plants and estimated chemical releases to soils nationwide from biosolids recycling. J Environ Monit. 2009;11:2207–2215. doi: 10.1039/b914324f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Löffler FE, Sanford RA, Ritalahti KM. Reductively dechlorinating bacteria. Nature. 2005;204:581. doi: 10.1016/S0076-6879(05)97005-5. [DOI] [PubMed] [Google Scholar]
- 14.Richardson RE. Genomic insights into organohalide respiration. Curr Opin Biotechnol. 2013;24:489–505. doi: 10.1016/j.copbio.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Boyd SA, Shelton DR, Berry D, Tiedje JM. Anaerobic biodegradation of phenolic compounds in digested sludge. Appl Environ Microbiol. 1983;46:50–54. doi: 10.1128/aem.46.1.50-54.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suflita JM, Horowitz A, Shelton DR, Tiedje JM. Dehalogenation: a novel pathway for the anaerobic biodegradation of haloaromatic compounds. Science. 1982;218:1115–1117. doi: 10.1126/science.218.4577.1115. [DOI] [PubMed] [Google Scholar]
- 17.Freedman DL, Gossett JM. Biological reductive dechlorination of tetrachloroethylene and trichloroethylene under methanogenic conditions. Appl Environ Microbiol. 1989;55:2144–2151. doi: 10.1128/aem.55.9.2144-2151.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adrian L, Dudková V, Demnerová K, Bedard DL. “Dehalococcoides” sp. strain CBDB1 extensively dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl Environ Microbiol. 2009;75:4516–4524. doi: 10.1128/AEM.00102-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adrian L, Hansen SK, Fund JM, Görisch H, Zinder SH. Growth of Dehalococcoides strains with chlorophenols as electron acceptors. Environ Sci Technol. 2007;41:2318–2323. doi: 10.1021/es062076m. [DOI] [PubMed] [Google Scholar]
- 20.Mikesell MD, Boyd SA. Reductive dechlorination of the pesticides 2,4-D, 2,4,5 T, and pentachlorophenol in anaerobic sludges. J Environ Qual. 1985;14:337–340. [Google Scholar]
- 21.Pycke BFG, Roll IB, Brownawell BJ, et al. Transformation products and human metabolites of triclocarban and triclosan in sewage sludge across the United States. Environ Sci Technol. 2014;48:7881–7890. doi: 10.1021/es5006362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNamara PJ Krzmarzick MJ. Triclosan enriches for Dehalococcoides-like Chloroflexi in anaerobic soil at environmentally relevant concentrations. FEMS Microbiol Lett. 2013;344:48–52. doi: 10.1111/1574-6968.12153. [DOI] [PubMed] [Google Scholar]
- 23.Nelson MC, Morrison M, Yu Z. A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour Technol. 2011;102:3730–3739. doi: 10.1016/j.biortech.2010.11.119. [DOI] [PubMed] [Google Scholar]
- 24.Vanwonterghem I, Jensen PD, Ho DP, Batstone DJ, Tyson GW. Linking microbial community structure, interactions and function in anaerobic digesters using new molecular techniques. Curr Opin Biotechnol. 2014;27:55–64. doi: 10.1016/j.copbio.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Yu K, Xia Y, et al. Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl Microbiol Biotechnol. 2014;98:5709–5718. doi: 10.1007/s00253-014-5648-0. [DOI] [PubMed] [Google Scholar]
- 26.Duhamel M, Mo K, Edwards EA. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl Environ Microbiol. 2004;70:5538–5545. doi: 10.1128/AEM.70.9.5538-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watts JEM, Fagervold SK, May HD, Sowers KR. A PCR-based specific assay reveals a population of bacteria within the Chloroflexi associated with the reductive dehalogenation of polychlorinated biphenyls. Microbiology. 2005;151:2039–2046. doi: 10.1099/mic.0.27819-0. [DOI] [PubMed] [Google Scholar]
- 28.Yan J, Rash BA, Rainey FA, Moe WM. Detection and quantification of Dehalogenimonas and “Dehalococcoides” populations via PCR-based protocols targeting 16S rRNA genes. Appl Environ Microbiol. 2009;75:7560–7564. doi: 10.1128/AEM.01938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smits THM, Devenoges C, Szynalski K, Maillard J, Holliger C. Development of a real-time PCR method for quantification of the three genera Dehalobacter, Dehalococcoides, and Desulfitobacterium in microbial communities. J Microbiol Methods. 2004;57:369–378. doi: 10.1016/j.mimet.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Sung Y, Fletcher KE, Ritalahti KM, et al. Geobacter lovleyi sp. nov. strain SZ a novel metal-reducing and tetrachloroethene-dechlorinating bacterium. Appl Environ Microbiol. 2006;72:2775–2782. doi: 10.1128/AEM.72.4.2775-2782.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Fantroussi S, Mahillon J, Naveau H, Agathos SN. Introduction of anaerobic dechlorinating bacteria into soil slurry microcosms and nested-PCR monitoring. Appl Environ Microbiol. 1997;63:806–811. doi: 10.1128/aem.63.2.806-811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fite A, Macfarlane GT, Cummings JH, et al. Identification and quantitation of mucosal and faecel Desulfovibrios using real time polymerase chain reaction. Gut. 2004;53:523–529. doi: 10.1136/gut.2003.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duhamel M, Edwards EA. Microbial composition of chlorinated ethene-degrading cultures dominated by Dehalococcoides. FEMS Microbiol Ecol. 2006;58:538–549. doi: 10.1111/j.1574-6941.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- 34.Muyzer G, De Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krzmarzick MJ, Miller HR, Yan T, Novak PJ. Novel Firmicutes group implicated in the dechlorination of two chlorinated xanthones, analogues of natural organochlorines. Appl Environ Microbiol. 2014;80:1210–1218. doi: 10.1128/AEM.03472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krzmarzick MJ, McNamara PJ, Crary BB, Novak PJ. Abundance and diversity of organohalide-respiring bacteria in lake sediments across a geographical sulfur gradient. FEMS Microbiol Ecol. 2013;84:248–258. doi: 10.1111/1574-6941.12059. [DOI] [PubMed] [Google Scholar]
- 37.Löffler FE, Yan J, Ritalahti KM, et al. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov. within the phylum Chloroflexi. Int J Syst Evol Microbiol. 2013;63:625–635. doi: 10.1099/ijs.0.034926-0. [DOI] [PubMed] [Google Scholar]
- 38.Shelton D, Tiedje J. Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic acid. Appl Environ Microbiol. 1984;48:840–848. doi: 10.1128/aem.48.4.840-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun B, Cole JR, Sanford RA, Tiedje JM. Isolation and characterization of Desulfovibrio dechloracetivorans sp. nov., a marine dechlorinating bacterium growing by coupling the oxidation of acetate to the reductive dechlorination of 2-chlorophenol. Appl Environ Microbiol. 2000;66:2408–2413. doi: 10.1128/aem.66.6.2408-2413.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markus J, Schmitz RPH, Westermann M, Richter W, Diekert G. Growth substrate dependent localization of tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Arch Microbiol. 2006;186:99–106. doi: 10.1007/s00203-006-0125-5. [DOI] [PubMed] [Google Scholar]
- 41.Fletcher KE, Costanza J, Cruz-Garcia C, Ramaswamy NS, Pennell KD, Löffler FE. Effects of elevated temperature on Dehalococcoides dechlorination performance and DNA and RNA biomarker abundance. Environ Sci Technol. 2011;45:712–718. doi: 10.1021/es1023477. [DOI] [PubMed] [Google Scholar]
- 42.Amos BK, Ritalahti KM, Cruz-Garcia C, Padilla-Crespo E, Löffler FE. Oxygen effect on Dehalococcoides viability and biomarker quantification. Environ Sci Technol. 2008;42:5718–5726. doi: 10.1021/es703227g. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, Banaszak JE, Parameswaran P, Alder J, Krajmalnik-Brown R, Rittman BE. Focused-pulsed sludge pre-treatment increases the bacterial diversity and relative abundance of acetoclastic methanogens in a full-scale anaerobic digester. Water Res. 2009;43:4517–4526. doi: 10.1016/j.watres.2009.07.034. [DOI] [PubMed] [Google Scholar]


