Abstract
Adoptively transferred tumor-infiltrating T lymphocytes (TILs) that mediate complete regression of metastatic melanoma have been shown to recognize mutated epitopes expressed by autologous tumors. Here, in an attempt to develop a strategy for facilitating the isolation, expansion, and study of mutated antigen–specific T cells, we performed whole-exome sequencing on matched tumor and normal DNA isolated from 8 patients with metastatic melanoma. Candidate mutated epitopes were identified using a peptide-MHC–binding algorithm, and these epitopes were synthesized and used to generate panels of MHC tetramers that were evaluated for binding to tumor digests and cultured TILs used for the treatment of patients. This strategy resulted in the identification of 9 mutated epitopes from 5 of the 8 patients tested. Cells reactive with 8 of the 9 epitopes could be isolated from autologous peripheral blood, where they were detected at frequencies that were estimated to range between 0.4% and 0.002%. To the best of our knowledge, this represents the first demonstration of the successful isolation of mutation-reactive T cells from patients’ peripheral blood prior to immune therapy, potentially providing the basis for designing personalized immunotherapies to treat patients with advanced cancer.
Introduction
Cancer results from the accumulation of mutations, some of which lead to uncontrolled cell growth and tumor metastasis and are termed drivers, along with other mutations, termed passengers, that may not contribute to the tumorigenic phenotype (1). Recent advances in next-generation sequencing enabled the rapid assessment of the mutational landscape of human cancers and revealed that melanomas can harbor between fewer than 100 and more than 2,000 nonsynonymous mutations (2–5). Driver as well as passenger mutations could potentially lead to the expression of mutated proteins recognized by the immune system as foreign (6–11). When encountering peptides presented by MHC molecules on the surface of target cells, T cells are able to recognize single amino acid differences between mutated and nonmutated epitopes, providing an opportunity for the development of novel adoptive immunotherapies (12).
The administration of autologous tumor-infiltrating T lymphocytes (TILs) can lead to complete, durable tumor regressions in patients with metastatic melanoma (13, 14). Understanding the nature and the specificity of these tumor-reactive lymphocytes is crucial for the successful dissemination of adoptive immunotherapy. Studies carried out since the early 1990s, primarily using cDNA library screening methods, demonstrated that tumor-reactive T cells recognized epitopes derived from a variety of molecules (15). The majority of tumor antigens identified using these approaches could be grouped into several general categories: differentiation antigens, whose expression is limited to a single tissue; cancer germline antigens that are not expressed in normal adult tissues with the exception of testis; and antigens derived from tumor-specific somatically mutated genes. In this regard, we recently evaluated the ability of TILs to recognize candidate mutated epitopes identified using whole-exome sequencing of autologous tumors in conjunction with HLA class I–binding algorithms. Using this approach, we identified 7 mutated peptide epitopes recognized by 3 therapeutic bulk TIL cultures that mediated objective tumor regressions (6). We also demonstrated that mutation-reactive T cells present in TILs could be identified by screening autologous antigen-presenting cells that were transfected with tandem minigene libraries expressing cancer mutations identified from patients’ tumors (9). Importantly, we recently demonstrated that mutation-specific T cells can also be found in solid tumors other than melanoma (16). Tumor-infiltrating CD4+ T cells derived from a patient with cholangiocarcinoma were found to recognize a mutated epitope derived from ERBB2IP, and administration of a bulk lymphocyte population containing a high percentage of T cells reactive with this epitope mediated an objective clinical response without toxicity (16).
Strategies that target mutated tumor-specific antigens may have advantages over those that target nonmutated self-antigens. Many tissue-specific differentiation antigens have been identified; however, targeting these molecules with potent effector T cells has been associated with autoimmunity, leading in some cases to dose-limiting toxicities (17, 18). In addition, T cells recognizing mutated (“foreign”) antigens are not subject to central tolerance and thus may potentially express higher-affinity T cell receptors (TCRs) than do those directed against self-antigens (7, 8).
Here, we sought to develop an approach that would facilitate the isolation, expansion, and study of T cells specific for mutated cancer antigens. Nonsynonymous somatic mutations were initially identified by carrying out whole-exome and RNA-sequencing (RNA-seq) analyses of tumors from metastatic melanoma patients. Large panels of MHC tetramers containing candidate mutated peptides that were predicted using MHC-binding algorithms to bind with high affinity to patients’ HLA molecules were then generated using a previously described UV-mediated peptide exchange strategy (19). Screening of fresh human tumor digests and bulk TIL cultures for their ability to bind mutated tetramers resulted in the identification of mutation-reactive T cells in bulk populations from 5 of the 8 patients evaluated using this approach. Furthermore, this report demonstrates that T cells specific for neoantigens, which represented as little as 0.002% of the peripheral T cell population prior to therapy, can be sorted and expanded in vitro by a factor of 1,000-fold or more. Cells isolated from peripheral blood or in vitro–cultured TILs using this approach can provide a readily accessible source of functional TCRs that can potentially be used to genetically engineer autologous T cells and provide the means to develop personalized adoptive immunotherapies for cancer.
Results
Use of whole-exome sequencing approach to identify and isolate T cells specific for mutated antigens from fresh tumor digests.
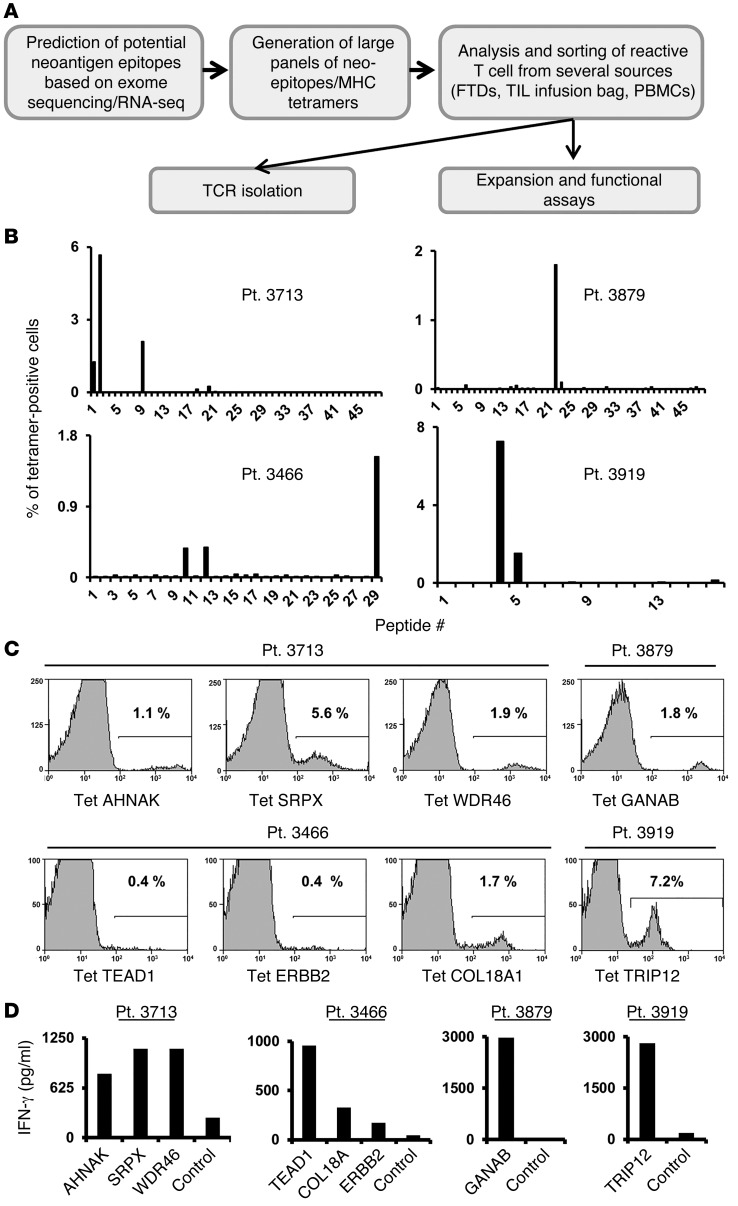
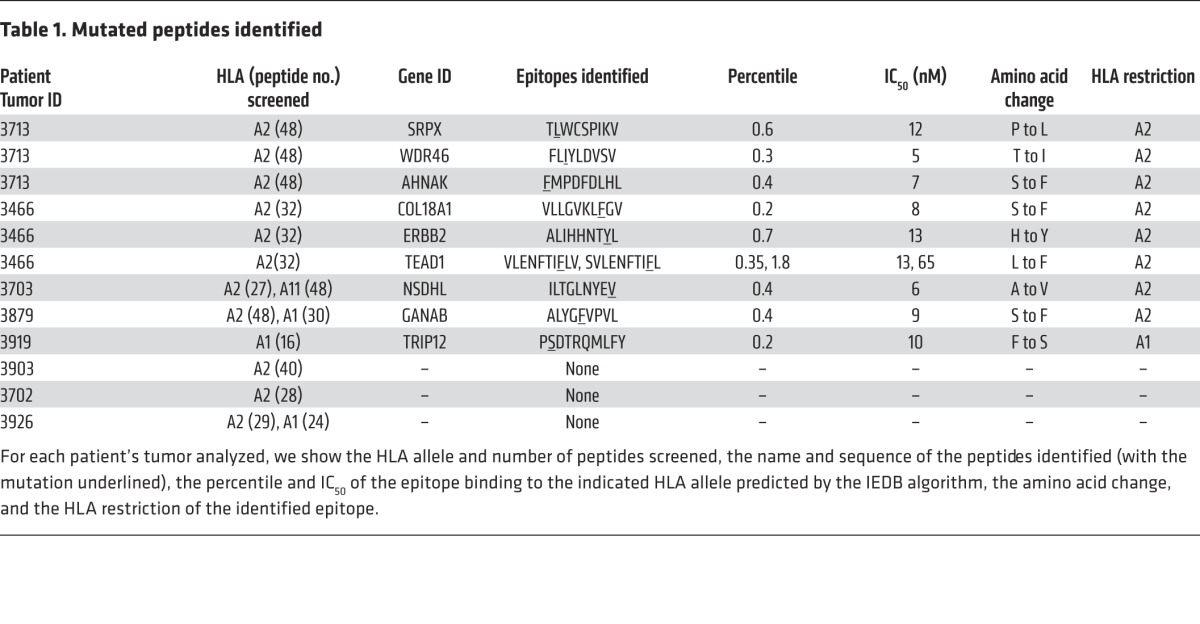
A schematic representation of the experimental approach followed in this study is presented in Figure 1A. Initially, DNA isolated from tumors that were resected from 8 patients with metastatic melanoma was subjected to whole-exome sequencing to identify nonsynonymous mutations. In addition, RNA was isolated to carry out RNA-seq analysis from the same tumor samples in an attempt to limit candidate peptides to those derived from expressed gene products. Liberal filters were used to generate nonsynonymous somatic exome variant calls to minimize the chance of false-negatives, as described in the Methods section. The number of exome variants initially identified from each of the tumors varied from a low of approximately 300 to a high of over 10,000, as shown in Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI82416DS1. After filtering expressed transcripts, mutated protein products were used to predict potential epitopes restricted to patients’ HLA class I alleles using a peptide-binding algorithm (20). Between 16 and 48 of the mutated peptides that were predicted to bind with high affinity to HLAA*01:01 or A*02:01 were then synthesized (Supplemental Table 2) and used to generate panels of fluorescent MHC tetramers using a previously described UV-mediated peptide exchange procedure(19).
Figure 1. Analysis of FTDs.
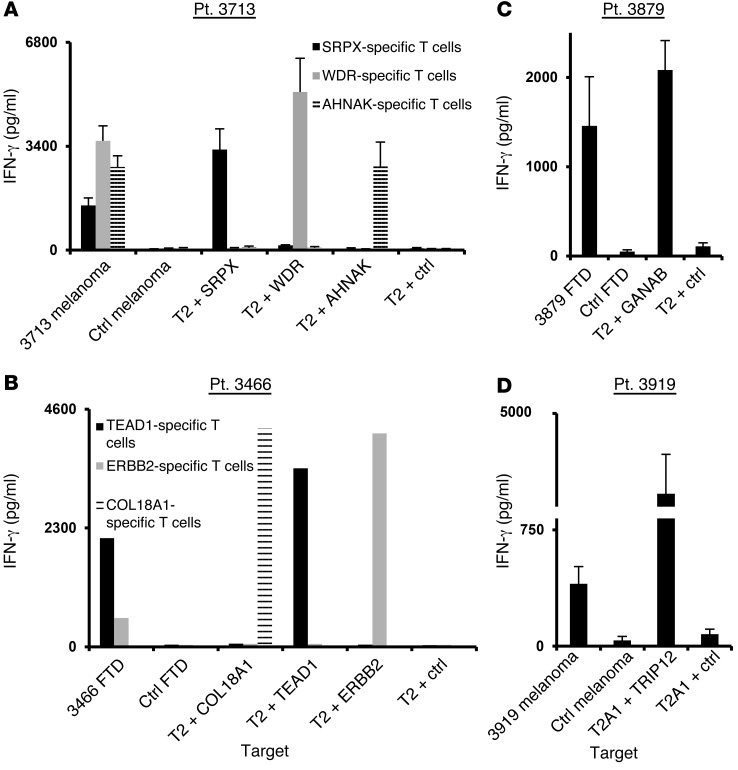
(A) Schematic representation of the experimental strategy. (B) Approximately 3 × 104 to 5 × 104 cells from FTDs were incubated for 3 to 4 days in lymphocyte medium and then stained with panels of mutated epitope MHC tetramers. The cells were analyzed by flow cytometry (gated on the lymphocyte population). For each patient, the frequency of tetramer-positive cells is indicated (y axis) for each peptide screened (x axis). Patients 3713, 3466, and 3879 were screened for binding to HLA-A*02:01 tetramers and patient 3919 for binding to HLA-A1 tetramers. (C) Tetramer (Tet) staining for selected mutated epitopes identified for each patient. The percentage of positive cells is indicated. (D) Cultured FTD cells were incubated with T2 cells pulsed with the predicted mutated epitopes or incubated with T2 cells pulsed with irrelevant peptide (Ctrl, see Methods) for 16 hours, and IFN-γ secretion was measured by ELISA. Pt., patient.
Analysis of short-term in vitro cultures derived from fresh tumor digests (FTDs) obtained from 4 of the 7 patients with available fresh tumor samples contained T cells that bound to 1 or more of the candidate tetramers containing mutated candidate epitopes (Figure 1B). The FTD cultures from patients 3713 and 3466 each bound to 3 HLA-A2 tetramers, and the FTD cultures from patients 3879 and 3919 each bound to a single HLA-A2 and A1 tetramer, respectively. The percentages of tetramer-positive cells detected in these cell populations ranged from 0.4%, observed with the TEAD1 and ERBB2 epitopes recognized by FTD from patient 3466, and 7.2% for the HLA-A1–restricted TRIP12 epitope recognized by FTD from patient 3919 (Figure 1C and Table 1). Further evaluation demonstrated that the FTD cultures recognized T2 cells pulsed with the cognate mutated epitopes (Figure 1D). In some cases, identification of the optimal T cell epitopes required screening of additional peptides that were not present in the initial screening assays. The mutated TEAD1 peptide with the highest-ranking score, VLENFTIFLV, ranked in the top 0.35% of a random set of natural peptides and had a predicted IC50 of 13 nM. While tetramer staining and reactivity were observed with a TEAD1 peptide, VLENFTIFLV, that was predicted to possess a relatively high affinity for HLA-A2 (Supplemental Figure 1), staining was significantly improved when using a 9-mer peptide that was shifted by 1 amino acid toward the amino terminus of the TEAD1 protein SVLENFTIFL (Supplemental Figure 1). This peptide, however, only ranked in the top 1.8% of peptides and was predicted to possess a binding affinity of 65 nM.
Table 1. Mutated peptides identified.
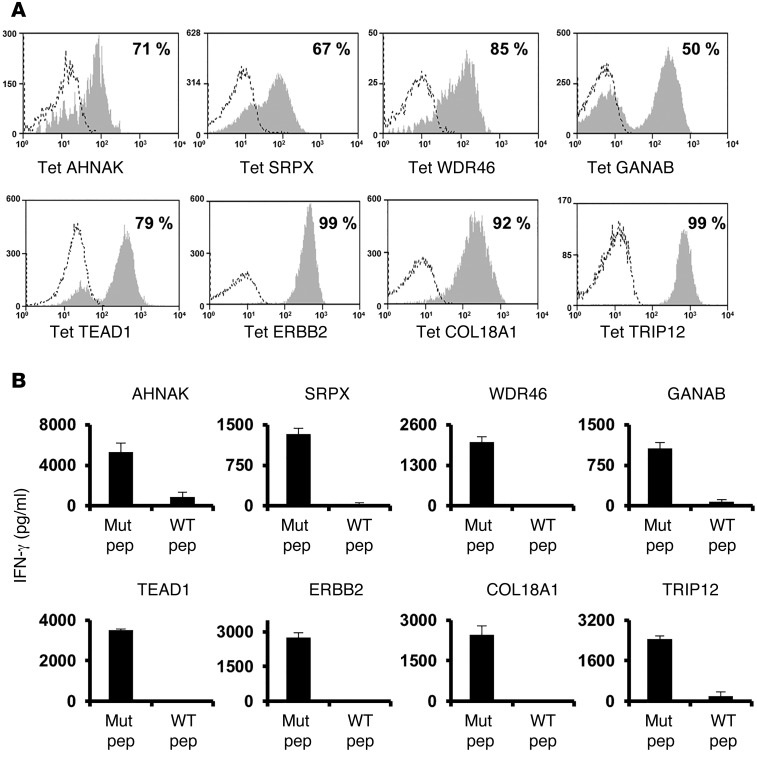
Approximately 103 to 8 × 104 tetramer-positive T cells were then electronically sorted and stimulated using anti-CD3 Ab in the presence of irradiated feeder cells, which led to an approximately 1,000-fold expansion 7–10 days after stimulation. Tetramer staining revealed that between 50% and 99% of the T cells that were sorted and expanded using this approach bound to the original tetramer used to isolate the cells (Figure 2A). Each of the expanded T cell cultures recognized the appropriate cognate-mutated epitope, but not the WT version, in cocultures with pulsed T2 cells (Figure 2B), indicating that this screening and isolation strategy represents a robust method for expanding antigen-reactive T cells from FTDs.
Figure 2. Sorting and expansion of mutated antigen–specific T cells.
(A) Tetramer-positive T cells were sorted from FTDs and expanded in vitro by stimulation with OKT3. Ten days later, T cells were assessed for tetramer binding. (B) T cells that were sorted and expanded in vitro were cocultured with T2 cells pulsed with the cognate mutated epitope or cocultured with T2 cells pulsed with the corresponding WT peptide. Sixteen hours after the start of the coculture, IFN-γ secretion was measured by ELISA. Results are presented as the mean ± SEM (n = 3). P < 0.05 by Student’s t test for the difference in IFN-γ secretion between mutated (Mut pep) and WT peptides (WT pep).
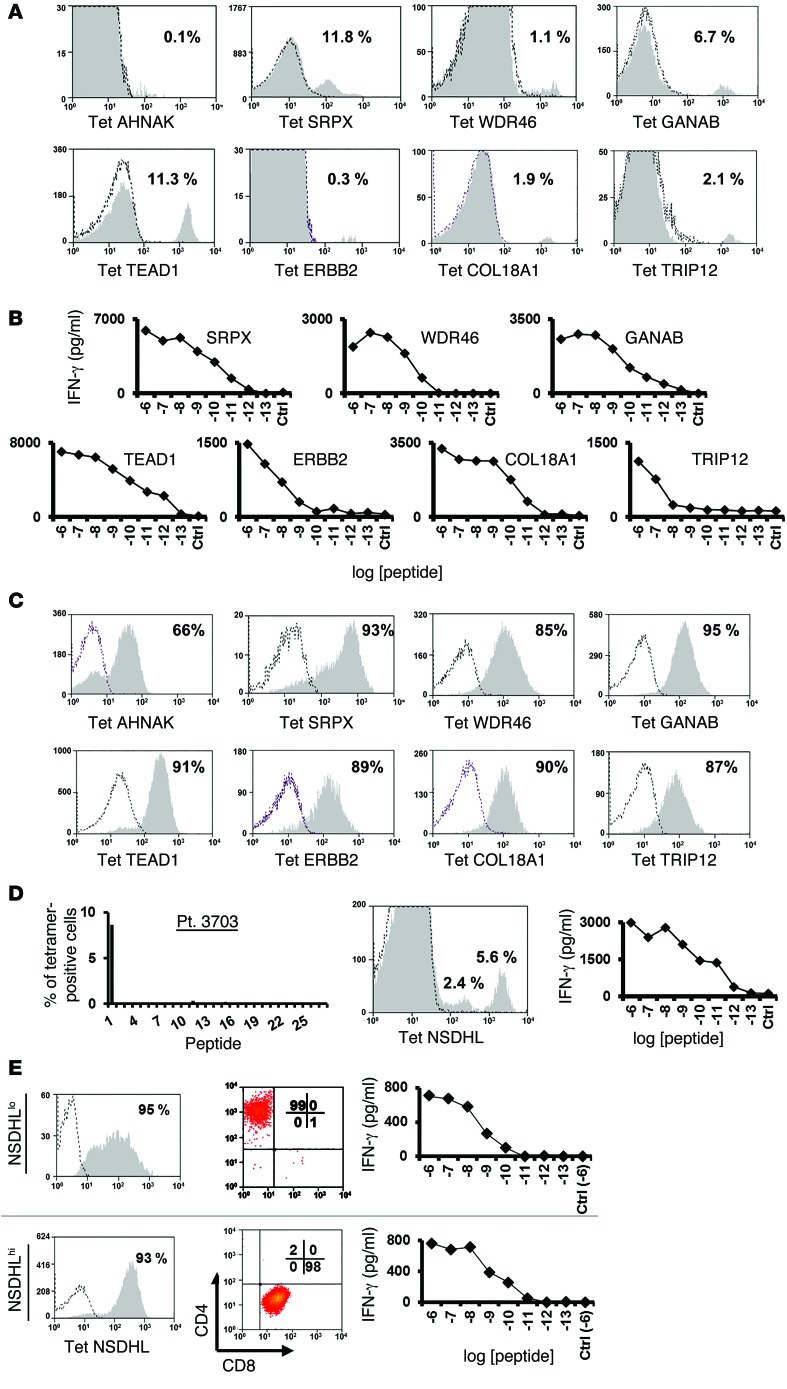
Isolation of tetramer-reactive T cells from treatment TIL cultures.
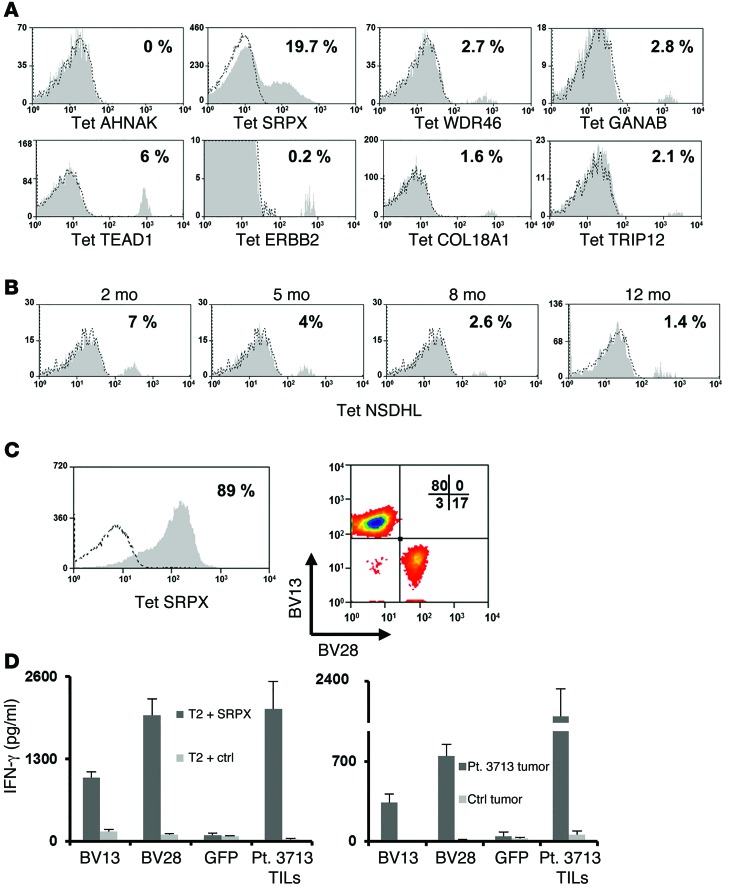
The proportion of T cells specific for the mutated epitopes in bulk TIL cultures used to treat the patients described above were then evaluated for their ability to recognize the mutated epitopes identified using FTD cultures. All of the specificities identified in the FTDs (Figure 1) were also detected in the expanded TIL cultures by tetramer staining (Figure 3A). With the exception of AHNAK, TILs secreted IFN-γ in response to all of the epitopes identified using FTD cultures (Figure 3B). The relatively low frequency of AHNAK-specific cells in the TIL culture, which was estimated to be approximately 0.1%, might explain the lack of reactivity in this assay; nevertheless, we could perform a peptide titration assay using AHNAK-specific T cells that were sorted and expanded from infusion TILs (Supplemental Figure 2). The relative frequencies of these cell populations often differed dramatically between the FTDs and the infusion TILs. For example, the frequency of AHNAK-specific cells decreased from 1.1% to 0.1 % following in vitro expansion, whereas TEAD1-specific T cells increased from 0.4% to 11% after expansion. As above, mutated antigen–specific T cells could be sorted to purity levels ranging from 66% to 95% (Figure 3C) and recognized the appropriate peptides (data not shown).
Figure 3. Treatment TILs recognize mutated antigens.
(A) Bulk-treatment TIL cultures were stained with the indicated tetramer and analyzed by flow cytometry. (B) TILs were cocultured with T2 cells pulsed with the indicated mutated epitope at different concentrations (ranging from 10–6 to 10–13 M) or cocultured with T2 cells pulsed with irrelevant peptide (Ctrl, at 10–6 M). Sixteen hours after the start of the coculture, IFN-γ secretion was measured by ELISA. (C) Tetramer-positive cell populations were sorted from infusion TILs and expanded using OKT3. Ten days after the start of the expansion, T cells were assessed for tetramer binding. (D) Bulk-treatment TILs for patient 3703 were analyzed for HLA-A*02:01/mutated epitope tetramer binding. The frequency of tetramer-positive cells (y axis) for each peptide screened (x axis) is indicated (left panel). A positive mutated epitope, NSDHL, was identified (middle panel), and a representative staining plot of 2 tetramer-positive cell populations is shown (right panel). (E) NSDHLhi and NSDHLlo cell populations were sorted and expanded, and 10 days after the start of the expansion, T cells were assessed for NSDHL-tetramer binding (left panels). Additionally, these sorted cell populations were assessed for CD4 and CD8 expression (middle panels). The NSDHLhi cell population, which consisted of 98% CD8+ T cells, and the NSDHLlo cell population, which consisted of 99% CD4+ T cells, were analyzed for peptide reactivity in coculture with T2 cells pulsed with titrated doses of the mutated NSDHL epitope. Sixteen hours after the start of the coculture, IFN-γ secretion was measured in the culture supernatant by ELISA (right panels). Results are representative of 2 independent experiments.
One of the limitations of these studies is a lack of FTDs due to the low viability or relatively small size of some resected tumor samples; nevertheless, for some patients such as number 3703, for whom no FTD was available, expanded TILs were available for testing. Screening of a panel of MHC tetramers containing 27 HLA-A*02:01– and 48 HLA-A*11:01–restricted mutated candidate epitopes identified by sequencing DNA from 3703 uncultured fresh tumor revealed that TILs from patient 3703 recognized a single HLA-A2–restricted mutated NSDHL epitope(Figure 3D). Two cell populations that differed in their intensity of binding to the mutated NSDHL tetramer were electronically sorted and expanded in vitro. Analysis of these cells following expansion indicated that T cells that bound low levels of NSDHL (NSDHLlo) predominantly expressed CD4, whereas the NSDHL cell population that bound high levels of NSDHL tetramer (NHDHhi) predominantly expressed CD8 (Figure 3E). Their functional avidities were then evaluated by examining their response to T2 cells that were pulsed with between 10–6 M and 10–13 M concentrations of the NSDHL peptide. The results indicated that the NSDHLhi and NSDHLlo cell populations possessed similar avidities, recognizing targets pulsed with a minimum of 10–10 M peptide (Figure 3E).
Mutated antigen–specific T cells can be isolated from peripheral blood lymphocytes.
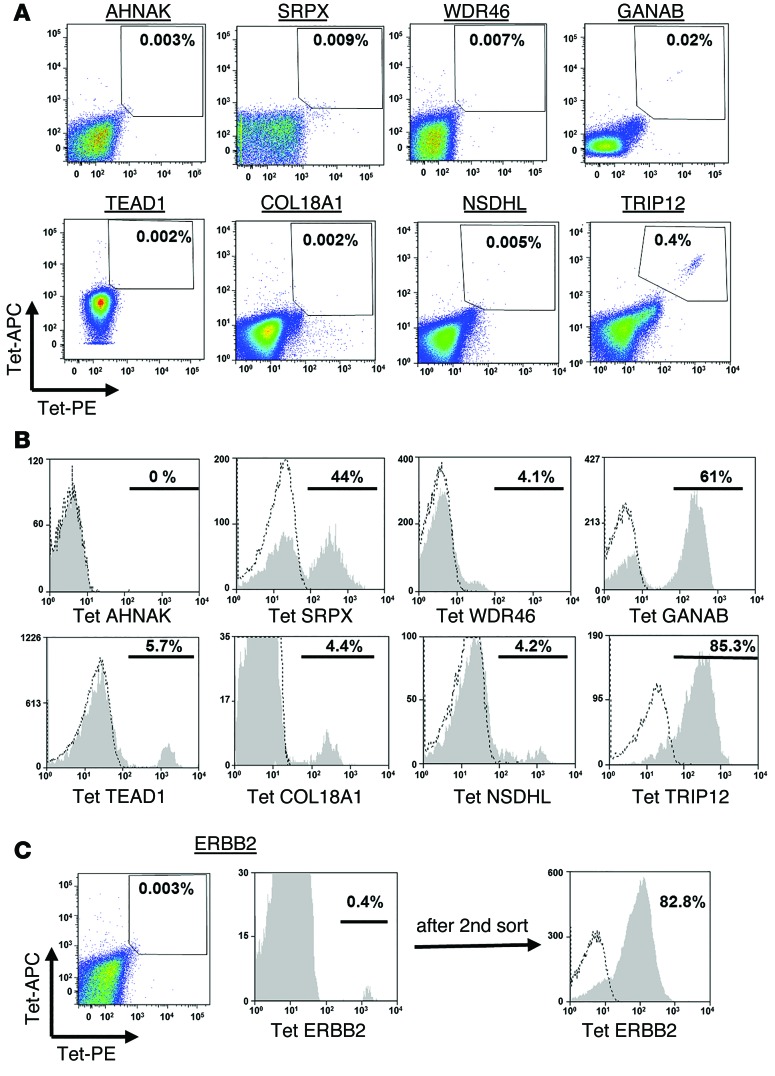
In an attempt to deal with the limited availability of fresh tumor samples from some patients, efforts were made to isolate T cells reactive with the mutated epitopes from the patients’ peripheral blood mononuclear cells (PBMCs). As the frequency of such cells was expected to be extremely low, peripheral lymphocytes obtained prior to TIL treatment were incubated with MHC tetramers that had been labeled with the allophycocyanin (APC) and phycoerythrin (PE) fluorophores, allowing cell separations to be carried out using a 2D gating strategy. The levels of tetramer-specific T cells detected in peripheral blood were generally indistinguishable from background staining, with the exception of the TRIP12 HLA-A2 tetramer, which bound to 0.4% of the PBMCs from patient 3919 (Figure 4A). Following 10 days of in vitro expansion of electronically sorted cells stained with the 2 fluorophores, between 0.4% and 46 % of the cells bound to the appropriate tetramer, with the exception of cells reactive with the mutated AHNAK epitope (Figure 4, B and C). In addition, T cells reactive with the ERBB2-mutated epitopes only constituted 0.4% of the T cells obtained after a single expansion in vitro; however, following a second round of enrichment performed with a cell sorter, 82% of the expanded population of T cells recognized the mutated ERBB2 epitope (Figure 4C). These data indicate that T cells specific for mutated tumor antigens can be isolated and expanded from the peripheral blood of most melanoma patients.
Figure 4. Sorting and expansion of mutated antigen–specific T cells from peripheral blood.
(A) Cells (1 × 107 to 4 × 107) from pretreatment pheresis samples were thawed and incubated overnight in lymphocyte medium, followed by staining simultaneously with the indicated PE- and APC-labeled tetramers. The frequency of double-positive cells is indicated. (B) The tetramer-positive cell population was sorted and expanded in vitro, and 10 days after expansion, these cells were assessed for tetramer binding. The percentage of positive cells (gated on the CD8+ cell population) is indicated. (C) Similarly, mutated ErbB2–specific cells were sorted from peripheral blood cells. As the relative frequency of these cells was low after the first expansion (left histogram), a second sorting step was performed, and these cells were expanded and then analyzed for ErbB2-tetramer binding (right histogram).
Evaluation of T cell reactivity against mutated epitopes.
Additional studies were then carried out to determine whether T cells that had been sorted using MHC tetramers could efficiently recognize both target pulsed with the mutated epitopes and cognate tumor cells. To that end, mutation-reactive T cells were cocultured with either peptide-pulsed T2 or autologous tumor cells, if available. All the T cell populations secreted high levels of IFN-γ in response to the cognate epitope presented by T2 cells (Figure 5, A–D), but failed to respond to the corresponding WT peptides (data not shown). Moreover, all of the T cells tested recognized autologous tumor cells, but not HLA-matched control melanoma cells, with the exception of COL18A1-reactive T cells, which failed to secrete significant levels of IFN-γ when cocultured with autologous fresh tumor cells from patient 3466. This may be due to a defect in processing this particular epitope, as T cells reactive with the mutated TEAD1 and ERBB2 epitopes recognized fresh tumor cells from patient 3466, indicating that these antigens were endogenously processed and presented by the tumor cell line (Figure 5B).
Figure 5. Tumor reactivity of T cells specific for mutated antigens.
T cells that were isolated using mutated tetramer complexes were analyzed for peptide and tumor reactivity. T cells from patients 3713 (A), 3466 (B), 3879 (C), and 3919 (D) were cocultured with either peptide-pulsed T2/T2A1 cells (as indicated) or tumor cells. Sixteen hours after the start of the coculture, IFN-γ secretion was measured by ELISA in the culture supernatant. Results for patients 3713, 3879, and 3919 are presented as the mean ± SEM (n = 3). P > 0.05 by Student’s t test for the difference in IFN-γ secretion compared with the negative control.
Persistence of mutated antigen–specific T cells and TCR isolation.
Studies were then carried out to evaluate the persistence of T cells reactive with mutated epitopes following adoptive TIL transfer. Specific T cells recognizing all of the mutated epitopes identified using the tetramer-binding approach, with the exception of the AHNAK peptide, could be detected in patients’ peripheral blood 2–12 months after treatment (Figure 6, A and B). Interestingly, the frequencies of the respective T cell populations in the post-treatment samples were similar to those in the original TIL cultures (Figure 3), and T cells reactive with some epitopes, such as the NSDHL peptide, could be detected 1 year after treatment.
Figure 6. Persistence of and TCR isolation from T cells specific for mutated antigens.
(A) Cells obtained from pheresis in patients 2–3 months following adoptive TIL transfer were stained with the indicated MHC tetramer and analyzed for binding. The percentage of tetramer-positive cells among the gated lymphocyte population is indicated. (B) Similarly, pheresis samples from patient 3703 obtained 2, 5, 8, and 12 months following treatment were analyzed for NSDHL tetramer binding. The percentage of positive T cells among the gated lymphocytes is indicated. (C) Sorted T cells specific for mutated SRPX (left panel) were assessed for TCR BV13 and BV28 expression (right panel). (D) The α and β chains for the BV13 (CDR3b: CASSFFGSNQPQHF) and BV28 (CDR3b: CASGSSGQGEYGELFF) TCR were cloned from expanded T cells sorted from patient 3713 infusion bag cells stained with SRPX tetramer. OKT3-stimulated PBMCs were transfected with RNAs encoding the appropriate α and β pairs or with GFP. Transfected T cells were cocultured with either mutated SRPX–pulsed T2 cells (left panel) or tumor cells from patient 3713 (right panel), along with the appropriate negative controls (right panel: T2 pulsed with MART126-35 -2L epitope and melanoma cells from patient 3466). Bulk-treatment TILs from patient 3713 were used as a positive control for effector cells. Sixteen hours after the start of the coculture, IFN-γ secretion was measured in the culture supernatant. Results are presented as the mean ± SEM (n = 3) for 2 individual donors. P < 0.05 by Student’s t test for the difference in IFN-γ secretion compared with the negative control.
Another potential immunotherapeutic application of this tetramer-based isolation approach would be to clone the T cell receptor α (TRA) and β (TRB) chains expressed by mutated antigen–specific T cell populations (12). In an attempt to validate this approach, T cells reactive with the HLA-A2–restricted mutated SRPX epitope were isolated from the TIL population that had been administered to patient 3713. Staining with anti-TRB Abs indicated that approximately 80% of the SRPX-reactive T cells expressed BV13, whereas nearly 20% of the T cells expressed BV28. (Figure 6C). The TRA and TRB chains were then cloned from these 2 populations of tetramer-reactive T cells, and OKT3-stimulated PBMCs were transfected with in vitro–transcribed RNA encoding the appropriate TRA and TRB chains. Cells transfected with TCRs isolated from either population specifically secreted IFN-γ in response to T2 cells pulsed with the SRPX peptide and the autologous 3713 tumor cell line but not when cocultured with irrelevant control targets (Figure 6D). These findings indicate that it may be feasible to develop treatments based on the transfer of TCRs isolated from T cells sorted using tetramers bearing mutated epitopes to autologous peripheral T cells.
Discussion
Objective clinical response rates of up to 70% have been observed in melanoma patients treated with expanded populations of TILs that were selected on the basis of reactivity with autologous tumor cells (13), and recent studies have indicated that melanoma TILs frequently recognize somatically mutated gene products (6, 9). Ongoing studies are being conducted in additional patients to evaluate the prevalence of neoantigen-reactive T cells in TILs derived from other melanoma patients, which may represent an alternative to the use of autologous tumor cell targets in screening assays for the selection of cultures with tumor reactivity. The adoptive transfer of TILs containing high numbers of CD4+ T cells that recognize a mutated HLA class II–restricted neoantigen led to a partial regression of multiple lung metastases in a patient with cholangiocarcinoma (16), and current efforts are focused on replicating these results in other patients.
One of the factors potentially limiting the effectiveness of this approach, however, is the frequency of neoantigen-reactive T cells in bulk populations of TILs. Attempts to address this issue have focused on purifying tumor-reactive T cells from bulk TILs and PBMCs using HLA class I tetramers generated with candidate neoepitopes identified by whole-exome sequencing. The present study demonstrates that it is possible to design large panels of MHC tetramers presenting mutated epitopes and use them to identify and sort tumor-reactive T cells from tumor samples and from patients’ peripheral blood, where they are represented at levels estimated to be as low as 0.002% of the total lymphocyte population. While T cells specific for mutated epitopes could be identified and isolated from 5 of 8 patients in our study (Table 1), it may be possible to enhance the efficiency of this process. The strategy presented herein relies on several factors such as the precise determination of tumor genomic data, the fidelity of epitope prediction algorithms, and the availability of T cells. Robust algorithms capable of accurately predicting peptide immunogenicity (21) and proteasomal processing (22) are needed to determine the optimal threshold for the inclusion of epitopes in the initial screening phase. Identification of the optimal TEAD1 epitope, which was not present in the initial screening assays carried out with TILs from patient 3466, illustrates one of the problems encountered using current peptide prediction algorithms. Further improvements could take advantage of advanced tetramer screening methods such as combinatorial tetramer staining and mass cytometric analysis to facilitate characterization of the mutated epitopes (23–25). Additional conditional MHC ligands (26) for less frequent HLA alleles are also needed to expand the use of this approach to a wider patient population. These studies may be facilitated by identification of factors that influence the generation of neoantigen-reactive T cells. Although the expression of tumor antigens may bear some relationship to the induction of T cell responses, many additional factors such as the influence of processing mechanisms and the overall avidity of the TCR for particular peptide-MHC complexes can also influence the recognition of these epitopes.
Analysis of phenotypic and functional characteristics of tetramer-positive tumor-reactive T cells detected in fresh tumor and peripheral blood prior to therapy will also provide important clues to the factors that influence their ability to mediate tumor regression, although these studies are hampered by the relatively low frequency and unknown purity of many of the T cell populations isolated using these methods. Recent advances in single-cell whole-transcriptome analysis, coupled with functional TCR analysis, will hopefully lead to a better understanding of the factors that influence the in vivo activity of tumor-reactive T cells.
Comparisons of the frequencies of mutated epitope–specific T cells in the FTDs and in expanded TIL cultures provide evidence that individual clonotypes may undergo significant expansion or contraction following in vitro culture. For example, T cells reactive with the AHNAK epitope were almost lost following the in vitro expansion protocol and did not appear to persist in the patient after transfer. These findings indicate that some specificities present at early stages of TIL cultures may contract over time and would warrant the use of FTDs or briefly expanded cultures in the epitope/tetramer screening phases. It may also be possible to identify additional T cell specificities by screening peripheral lymphocytes using pools of tetramers containing multiple candidate epitopes. In addition, use of an initial CD3+ T cell enrichment step may enhance the efficiency of a subsequent sort carried out with a peptide-MHC tetramer.
Interestingly, phenotypic analysis demonstrated that approximately 30% of the T cells reactive with an HLA-A2–restricted NSDHL epitope isolated from a patient’s TILs expressed CD4, a phenomenon that was not observed with the other populations of T cells examined in this study (data not shown). Two HLA class I–restricted CD4+ T cells that recognize nonmutated tumor antigens were previously identified (27, 28); however, unlike the results presented in this report, corresponding populations of CD8+ T cells were not identified in the same patients studied to identify tumor-reactive CD4+ T cells. Multiple neoantigens have been identified as targets of CD4+ T cells (29); however, all of the previous targets were recognized in the context of HLA class II gene products. While they generally appear to possess reduced cytolytic activity relative to CD8+ T cells, tumor-reactive CD4+ T cells have been shown to play a significant role in mediating tumor regression in both animal models and patients (16, 30–33). The physiological role and significance of such nonclassical MHC class I–restricted T cells and their involvement in patients’ antitumor response is unclear, but recognition of class I–expressing tumor cells in vivo could lead to the secretion of cytokines that modulate the function of tumor-promoting cells in the tumor microenvironment and support cytotoxic T lymphocyte (CTL) activity (30, 34).
Our preliminary data also demonstrate that multiple clonotypes specific for the same mutated epitope can be isolated from individual patients. A more detailed analysis of the phenotypes of these clonotypes and the biophysical properties of the TCR might shed light on the extent, depth, and requirements for efficient antitumor immune responses. From a practical standpoint, the ability to quickly isolate mutated epitope–specific TCRs, as shown herein, is of importance if one wants to harness the power of T cell redirection using TCR gene transfer (12). The clinical implementation of this approach will require a more flexible platform for TCR expression such as a transposon-based system (35), which is not dependent on labor-intensive testing of viral supernatants. The majority of candidate epitopes were restricted to a single patient tumor sample; however, peptides encompassing the common driver mutations NRAS Q61R and RAC1 P29S were identified as candidate HLA-A1 epitopes in tumors from patients 3879 and 3919. In addition, NRAS Q61R and RAC1 P29S mutations were both observed in a tumor sample from patient 3702, and an HLA-A2–binding peptide encompassing the RAC1 P29S mutation was identified as a candidate epitope for patients 3702 and 3879. Previous studies demonstrated that the HLA-A1–restricted NRAS peptide evaluated in the current study was recognized by tumor-reactive T cells (36), and although T cells reactive with either of these epitopes were not identified in these patients, these cells could potentially be present at low levels in tumor or peripheral blood of other patients expressing the appropriate HLA alleles. Additionally, recent studies demonstrate that immune checkpoint inhibitors can generate and/or amplify an immune response against mutated antigens (37–40). As a result of their systemic nature, these treatments can lead to severe autoimmune toxicities (41, 42). The occurrence of severe autoimmune toxicities following treatment with an approach that targets neoantigens using MHC tetramer–sorted lymphocytes or cells engineered to express mutated epitope–specific TCRs is unlikely, however, as expression of these antigens is strictly limited to tumor cells. Combining checkpoint inhibitors with the adoptive transfer of enriched populations of neoantigen-reactive T cells may also represent an effective treatment option for patients who progress following treatment with individual therapies.
In conclusion, our findings demonstrate that it is feasible to use MHC tetramers to isolate T cells specific for mutated epitopes in patients with metastatic melanoma and lay the groundwork for designing novel and personalized immunotherapeutic strategies for the treatment of advanced cancers.
Methods
Subjects and cell lines.
Lymphocytes were cultured in complete lymphocyte medium (50% AIM V, 50% RPMI 1640; Invitrogen, Life Technologies) supplemented with 5% human serum and 300 IU IL-2. Melanoma tumor cells were generated at the Surgery Branch of the NCI and cultured in RPMI 1640 supplemented with 5% FCS. T2 (CRL-1992; ATCC) is a lymphoblastoid cell line deficient in transporter associated with antigen processing (TAP) function, facilitating the binding of cell-surface HLA-A*02:01 molecules with exogenously pulsed peptides. T2A1 is an HLA-A1 transfectant of T2 that can also present HLA-A1–restricted epitopes. All cells were maintained in an incubator at 37°C and 5% CO2. Bulk TIL cultures were expanded from FTDs as previously described (43, 44).
Exome-sequencing and RNA-seq analyses.
Whole-exome sequencing was carried out at Personal Genome Diagnostics Inc. (Baltimore, Maryland, USA) and the Genome Technology Access Center (Washington University, St. Louis, Missouri, USA), as previously described (6, 9). Transcriptome analysis was performed on RNA isolated from fresh tumor cells or tissue culture cell lines. Briefly, total RNA was isolated from fresh tumor fragments using an RNAeasy kit (91355; QIAGEN) and RNA-seq libraries were prepared using SureSelect RNA Library Preparation kits (95051; Agilent Technologies) according to the manufacturers’ instructions. Samples were run on a MiSeq (92122; Illumina), and approximately 4 × 107 paired-end reads were obtained per sample. Variants were called if there were more than 2 variant reads, a minimum of 10% variant allele frequency (VAF), and less than 1% VAF in the normal DNA. In addition, common SNPs with a VAF of greater than 1% were eliminated from the analysis. Expression was evaluated by determining the fragment per kilobase per million reads (FPKM) values from the RNA-seq analysis for transcripts encoding individual epitopes, and the candidates that were screened were limited to those encoded by transcripts with a minimum FPKM of 1. A summary of the number of nonsynonymous somatic variant exome calls is shown in Supplemental Table 1. Whole genomic data were deposited in the NCBI’s Sequence Read Archive (SRA) database (accession no. SRP062169).
Epitope prediction and generation of MHC tetramers.
For tumor from patient 3713, peptides encoded by variant transcripts with an FPKM value of greater than 3 were evaluated, whereas for tumors from patients 3466, 3703, 3879, 3919, 3903, 3702, and 3926, peptides encoded by variants with a minimum FPKM of 1 were evaluated. Candidate 9 and 10 amino acid peptides containing mutated residues that were predicted to bind with high affinity to patients’ MHC class I molecules were identified using the Immune Epitope Database (IEDB) prediction algorithm (http://tools.immuneepitope.org/mhci/) (20) and were synthesized by Peptide 2.0 or Atlantic Peptides as crude peptides. The mutated and corresponding WT peptides that were recognized by T cells were then synthesized and HPLC purified to greater than 95% by Peptide 2.0 or Atlantic peptides.
Biotinylated HLA monomers (for HLA-A*02:01, A*01:01, and A*11:01) loaded with UV-cleavable epitopes (26) were produced by the NIH Tetramer Core Facility at Emory University (Atlanta, Georgia, USA). The sequences of the peptides used in this study are detailed in Supplemental Table 2 and were synthesized by Peptide 2.0. Large panels of MHC tetramers were generated using the protocol detailed by Rodenko et al. (19). Briefly, HLA loaded with UV-sensitive peptide monomers were subjected to UV light in the presence of 50 μM individual candidate synthetic peptides for 1 hour on ice. Then, the monomers were tetramerized in the presence of fluorescent (PE or APC) streptavidin (Life Technologies) and kept at 4°C for further use.
FACS analysis and Abs.
Prior to tetramer analysis, T cells were incubated in PBS containing 5% FCS and dasatinib (50 nM) for 30 minutes at 37°C to enhance tetramer binding (45). Between 2 × 104 and 5 × 104 cells were stained with 1 ng/μl MHC tetramer in PBS containing 5% FCS for 45 minutes at room temperature. Following 2 washes in PBS, immunofluorescence was analyzed as the relative log fluorescence of live cells gated on the on populations of cells with relatively low levels of forward and side scatter. Immunofluorescence of the gated lymphocyte population was measured using a FACSCanto (BD) or CyAn-ADP (Beckman Coulter) flow cytometer. Fluorophore-labeled anti-human CD8 (clone SK1), CD4 (clone L200), BV13 (clone AF23), and BV28 (clone CH92) Abs were purchased from BD-Immunotech.
Sorting and rapid expansion protocol.
Following a 60-minute incubation with fluorophore-conjugated Abs or tetramers, T cells were isolated using a FACSAria cell sorter (BD) and collected in sterile PBS containing 50% FCS. T cells were expanded to large numbers using a rapid-expansion protocol (REP) with IL-2, anti-CD3 Ab, and irradiated feeder cells (46).
Analysis of T cell responses.
T cells were tested for reactivity in cytokine release assays using commercially available ELISA kits for IFN-γ (Thermo Scientific) at an effector-to-target ratio of 1:1. Briefly, between 2 × 104 to 105 T cells were incubated for 16 hours at 37°C with a similar number of tumor cells or peptide-pulsed T2 cells. Unless indicated otherwise, T2 cells were pulsed with 1 μM specific peptide or control (pp65, NLVPMVATV) peptide for 2 hours at 37°C, and then washed 2–3 times prior to the start of the coculture.
TCR isolation and expression.
RNA was isolated from approximately 106 tetramer-sorted cells. A 5′-RACE (rapid amplification of cDNA ends) procedure was performed using the SMARTer RACE kit (Clontech) and primers for the constant regions. The PCR products were cloned into a TOPO cloning vector (Invitrogen, Life Technologies). Twenty to forty clones for each chain were sequenced. TRA and TRB chains identified using the IMGT/V-Quest website (http://www.imgt.org/IMGT_vquest/vquest) (47) and subsequently cloned in a plasmid for mRNA production (48–50). Briefly, in vitro–transcribed mRNA encoding the identified TRA and TRB chains was generated using the AmpliCap-Max T7 High Yield Message Maker Kit (CELLSCRIPT) and purified using an RNA Clean-Up and Concentration Kit (Norgen Biotek Corp.). PBMCs were separated by centrifugation on a Ficoll-Hypaque cushion and stimulated with 50 ng/ml OKT3 and 300 IU/ml IL-2. Electroporation was performed at 400 V and 500 μs using an ECM 830 Square Wave Electroporation System (BTX Instrument Division, Harvard Apparatus Inc.). Transfections were performed using 1–2 μg in vitro–transcribed mRNA encoding the appropriate TRA and TRB chains per 106 PBMCs.
Statistics.
Data samples were compared using a 2-tailed Student’s t test where indicated, and a P value of less than 0.05 was considered statistically significant.
Study approval.
All of the patients were enrolled in a randomized trial comparing adoptive TIL transfer plus nonmyeloablative chemotherapy with or without 1,200 cGy total body irradiation (11C0123), with the exception of patient 3466, who was enrolled in a trial evaluating the response to unselected TILs (07C0176). Patients were eligible for these trials if they were 18 years or older; had measurable metastatic melanoma and an Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1; a life expectancy of more than 3 months; and no evidence of active systemic infections, coagulation disorders, or other active major medical or cardiovascular or immunodeficiency diseases. Patients had progressive metastatic melanoma upon their entrance into the study, and at least 4 weeks had elapsed after any previous treatment. All patients had a metastatic lesion of greater than 2 cm in diameter that could be resected for growth of TILs and had cells that grew in culture to sufficient quantities with in vitro antitumor activity as described in Methods. All materials from patients were obtained during the course of clinical trials approved by the IRB of the NCI, and all patients provided written informed consent prior to their participation in this study.
Supplementary Material
Acknowledgments
We thank Arnold Mixon and Shawn Farid for their excellent technical assistance during the cell-sorting procedures; the Adelson Medical Research Foundation (AMRF) for their generous support; and the NIH Tetramer Core Facility (contract HHSN272201300006C) for the provision of MHC tetramers.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(10):3981–3991. doi:10.1172/JCI82416.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walia V, Mu EW, Lin JC, Samuels Y. Delving into somatic variation in sporadic melanoma. Pigment Cell Melanoma Res. 2012;25(2):155–170. doi: 10.1111/j.1755-148X.2012.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robbins PF, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19(6):747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Rooij N, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31(32):e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heemskerk B, Kvistborg P, Schumacher TN. The cancer antigenome. EMBO J. 2013;32(2):194–203. doi: 10.1038/emboj.2012.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu YC, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Cancer Res. 2014;20(13):3401–3410. doi: 10.1158/1078-0432.CCR-14-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu YC, et al. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J Immunol. 2013;190(12):6034–6042. doi: 10.4049/jimmunol.1202830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Buuren MM, Calis JJ, Schumacher TN. High sensitivity of cancer exome-based CD8 T cell neo-antigen identification. Oncoimmunology. 2014;3: doi: 10.4161/onci.28836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blankenstein T, Leisegang M, Uckert W, Schreiber H. Targeting cancer-specific mutations by T cell receptor gene therapy. Curr Opin Immunol. 2015;33:112–119. doi: 10.1016/j.coi.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg SA, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Cancer Res. 2011;17(13):4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg SA. Decade in review-cancer immunotherapy: Entering the mainstream of cancer treatment. Nat Rev Clin Oncol. 2014;11(11):630–632. doi: 10.1038/nrclinonc.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Cancer. 2014;14(2):135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 16.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344(6184):641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson LA, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114(3):535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkhurst MR, et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol Ther. 2011;19(3):620–626. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodenko B, et al. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat Protoc. 2006;1(3):1120–1132. doi: 10.1038/nprot.2006.121. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y, et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40(Web Server issue):W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calis JJ, et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol. 2013;9(10): doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desai DV, Kulkarni-Kale U. T-cell epitope prediction methods: an overview. Methods Mol Biol. 2014;1184:333–364. doi: 10.1007/978-1-4939-1115-8_19. [DOI] [PubMed] [Google Scholar]
- 23.Newell EW. Higher throughput methods of identifying T cell epitopes for studying outcomes of altered antigen processing and presentation. Front Immunol. 2013;4: doi: 10.3389/fimmu.2013.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newell EW, Sigal N, Nair N, Kidd BA, Greenberg HB, Davis MM. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol. 2013;31(7):623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515(7528):572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 26.Toebes M, et al. Design and use of conditional MHC class I ligands. Nat Med. 2006;12(2):246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura MI, et al. MHC class I-restricted recognition of a melanoma antigen by a human CD4+ tumor infiltrating lymphocyte. Cancer Res. 1999;59(24):6230–6238. [PubMed] [Google Scholar]
- 28.Somasundaram R, et al. CD4(+), HLA class I-restricted, cytolytic T-lymphocyte clone against primary malignant melanoma cells. Cancer. 2000;85(1):253–259. doi: 10.1002/(sici)1097-0215(20000115)85:2<253::aid-ijc17>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 29.Linnemann C, et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med. 2015;21(1):81–85. doi: 10.1038/nm.3773. [DOI] [PubMed] [Google Scholar]
- 30.Cohen CJ, et al. Recognition of fresh human tumor by human peripheral blood lymphocytes transduced with a bicistronic retroviral vector encoding a murine anti-p53 TCR. J Immunol. 2005;175(9):5799–5808. doi: 10.4049/jimmunol.175.9.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muranski P, Restifo NP. Adoptive immunotherapy of cancer using CD4(+) T cells. Curr Opin Immunol. 2009;21(2):200–208. doi: 10.1016/j.coi.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunder NN, et al. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med. 2008;358(25):2698–2703. doi: 10.1056/NEJMoa0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreiter S, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520(7549):692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuball J, et al. Cooperation of human tumor-reactive CD4+ and CD8+ T cells after redirection of their specificity by a high-affinity p53A2. Immunity. 2005;22(1):117–129. doi: 10.1016/j.immuni.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Peng PD, et al. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther. 2009;16(8):1042–1049. doi: 10.1038/gt.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linard B, et al. A ras-mutated peptide targeted by CTL infiltrating a human melanoma lesion. J Immunol. 2002;168(9):4802–4808. doi: 10.4049/jimmunol.168.9.4802. [DOI] [PubMed] [Google Scholar]
- 37.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kvistborg P, et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci Transl Med. 2014;6(254): doi: 10.1126/scitranslmed.3008918. [DOI] [PubMed] [Google Scholar]
- 39.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rizvi NA, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong YC, Flynn JC. Opportunistic autoimmune disorders potentiated by immune-checkpoint inhibitors anti-CTLA-4 and anti-PD-1. Front Immunol. 2014;5: doi: 10.3389/fimmu.2014.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33(17):1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Topalian SL, Muul LM, Solomon D, Rosenberg SA. Expansion of human tumor infiltrating lymphocytes for use in immunotherapy trials. J Immunol Methods. 1987;102(1):127–141. doi: 10.1016/S0022-1759(87)80018-2. [DOI] [PubMed] [Google Scholar]
- 44.Dudley ME, et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J Clin Oncol. 2005;23(10):2346–2357. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lissina A, et al. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J Immunol Methods. 2009;340(1):11–24. doi: 10.1016/j.jim.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riddell SR, Watanabe KS, Goodrich JM, Li CR, Agha ME, Greenberg PD. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257(5067):238–241. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- 47.Giudicelli V, Brochet X, Lefranc MP. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc. 2011;2011(6):695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y, et al. High-efficiency transfection of primary human and mouse T lymphocytes using RNA electroporation. Mol Ther. 2006;13(1):151–159. doi: 10.1016/j.ymthe.2005.07.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bialer G, Horovitz-Fried M, Ya’acobi S, Morgan RA, Cohen CJ. Selected murine residues endow human TCR with enhanced tumor recognition. J Immunol. 2010;184(11):6232–6241. doi: 10.4049/jimmunol.0902047. [DOI] [PubMed] [Google Scholar]
- 50.Haga-Friedman A, Horovitz-Fried M, Cohen CJ. Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J Immunol. 2012;188(11):5538–5546. doi: 10.4049/jimmunol.1103020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.