Abstract
Liver cancer stem cells (CSCs) have been identified and shown to have self-renewal and differentiation properties; however, the biology of these hepatic CSCs remains largely unknown. Here, we analyzed transcriptome gene expression profiles of liver CSCs and non-CSCs from hepatocellular carcinoma (HCC) cells lines and found that the transcription factor (TF) ZIC2 is highly expressed in liver CSCs. ZIC2 was required for the self-renewal maintenance of liver CSCs, as ZIC2 depletion reduced sphere formation and xenograft tumor growth in mice. We determined that ZIC2 acts upstream of the TF OCT4 and that ZIC2 recruits the nuclear remodeling factor (NURF) complex to the OCT4 promoter, thereby initiating OCT4 activation. In HCC patients, expression levels of the NURF complex were consistent with clinical severity and prognosis. Moreover, ZIC2 and OCT4 levels positively correlated to the clinicopathological stages of HCC patients. Altogether, our results indicate that levels of ZIC2, OCT4, and the NURF complex can be detected and used for diagnosis and prognosis prediction of HCC patients. Moreover, these factors may be potential therapeutic targets for eradicating liver CSCs.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide, ranking the third leading cause of cancer-related deaths (1). The high rate of recurrence and heterogeneity are the 2 major features of HCC (2). The cancer stem cell model (CSC model) proposes that only a rare subset of cancer cells within tumor bulk display the capacity to self-renew, differentiate, and generate a new tumor (3). These CSCs are responsible for sustaining tumor growth and are resistant to conventional treatments (4), accounting for a hierarchical organization of heterogeneous cancer cells and a high rate of cancerous recurrence. Recently, several surface markers, including CD13, CD133, CD24, EpCAM, CD44, and CD90, can be used to enrich liver CSCs (5, 6). However, the hepatic CSC biology remains largely unknown.
Liver CSCs, like other CSCs, behave as stem cells in that they can self-renew and differentiate into heterogeneous tumor cells. Master transcription factor–mediated (TF-mediated) transcriptional regulation is one of the key regulatory mechanisms to determine cell-fate changes (7, 8). OCT4, NANOG, and SOX2 have been defined as the core pluripotent TFs in embryonic stem cells (ESCs). Four TFs (OCT4, SOX2, KLF4, and c-MYC) have been defined as being able to convert differentiated fibroblasts into induced pluripotent stem cells (iPSCs) (9, 10). These findings indicate that core TFs play a central role in establishing and maintaining the pluripotency of stem cells. OCT4, a member of the POU family, exerts a fundamental role in the maintenance of naive pluripotency as a master TF in mammalian development. OCT4 deficiency results in preimplantation lethality of mouse embryos (11). OCT4 is also sufficient to initiate reprogramming of mouse and human somatic cells without other reprogramming factors (12, 13), suggesting that OCT4 plays a crucial role in somatic cell reprogramming. Recent studies have reported that OCT4 is highly expressed in urothelial cancer and ovarian cancer tissues (14, 15). These observations suggest that OCT4 may play an important role in tumorigenesis.
The zinc finger TF ZIC2 belongs to a gene family originally identified by their homology to the drosophila odd-paired genes (16). In both mice and humans, heterozygous deletions or other mutations of ZIC2 cause severe brain malformation, indicating its critical role in the development of the CNS (17). ZIC2 has been reported to be highly expressed in some tumors and implicated in tumorigenesis (18, 19). However, how ZIC2 regulates liver cancer and liver CSCs remains elusive.
Chromatin remodeling is a prerequisite for gene transcription (20), which relies on ATP-dependent chromatin-remodeling complexes. The nuclear remodeling factor complex (NURF complex), a major member of the ISWI subfamily, specifically targets the chromatin via association with sequence-specific TFs and modified histones (21, 22). The NURF complex contains 3 components in mammalian cells, including BPTF, SNF2L, and RBBP4 (also known as RBAP46/48) (23). The NURF complex plays a critical role in the regulation of embryonic development, differentiation, and stemness maintenance (24, 25). The NURF complex can also modulate the canonical WNT pathway, probably through regulating the chromatin structures of targeting genes to make TFs more accessible (26). Recently, we demonstrated that the NURF complex participates in the regulation of self-renewal of hematopoietic stem cells (HSCs) in a c-MYC–dependent manner (27). In this study, we show that ZIC2 is highly expressed in liver CSCs and is required for the self-renewal of liver CSCs. ZIC2 recruits the NURF complex to trigger OCT4 activation, which sustains the stemness of liver CSCs.
Results
ZIC2 is highly expressed in liver CSCs.
CD13 and CD133 are widely used as liver CSC markers (6, 28). The CD13+CD133+ cell subset exhibited slow growth ability compared with their counterpart CD13–CD133– cells. We previously demonstrated that the CD13+CD133+ cell subset was successfully enriched in HCC cell lines and HCC primary samples (29). We isolated CD13+CD133+ cell populations from Huh7 and Hep3B cells, since liver CSCs and their sphere-forming abilities were much stronger than those of CD13–CD133– cell subsets (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI81979DS1). We previously performed transcriptome microarray analysis of CD13+CD133+ cells (hereafter called liver CSCs) and CD13–CD133– cells (hereafter called non-CSCs) sorted from HCC cell lines (GSE66529) (29).We analyzed differential noncoding RNA transcripts in liver CSCs compared with non-CSCs and showed that lncTCF7 is highly expressed in liver CSCs, playing a critical role in the maintenance of liver CSC self-renewal (29).
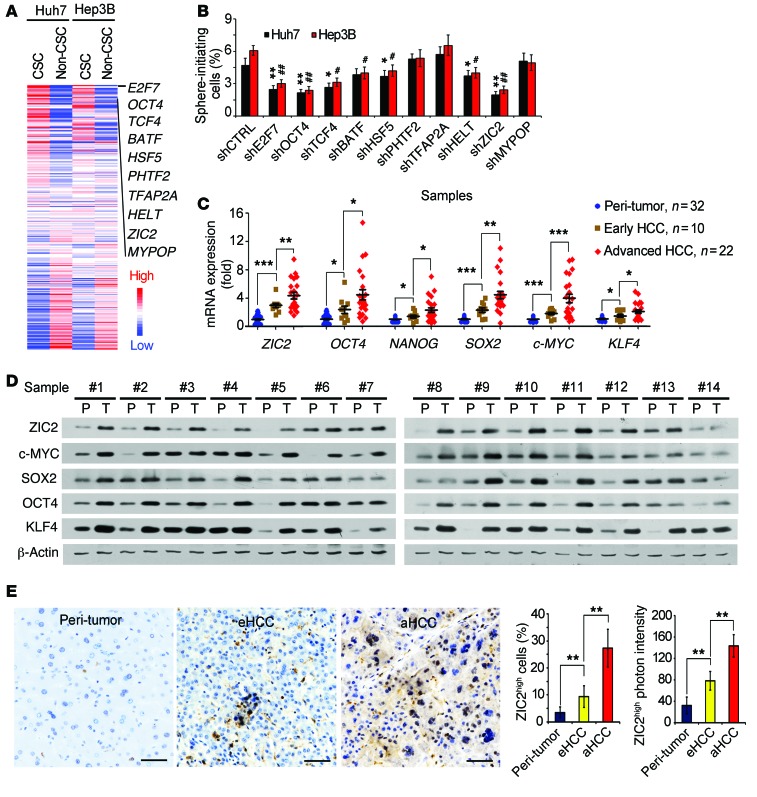
Here, we wanted to identify which TFs were involved in the maintenance of liver CSC self-renewal. We analyzed TFs based on gene expression profiles of liver CSCs and non-CSCs derived from Huh7 and Hep3B cells. We chose top-10 TFs that were highly expressed in liver CSCs compared with non-CSCs (Figure 1A). We then knocked down these selected TFs and performed in vitro sphere-formation assays. Among these 10 TFs, ZIC2 depletion displayed the strongest inhibitory effect on oncosphere formation (Figure 1B). We next detected ZIC2 expression in HCC patient samples and noticed that ZIC2 was highly expressed in HCC samples, including early and advanced stages (Figure 1C). In parallel, major pluripotency factors, including OCT4, NANOG, SOX2, c-MYC, and KLF4, were also elevated in HCC samples. These observations were validated by immunoblotting (Figure 1D). Moreover, elevated ZIC2 expression was further confirmed by IHC staining (Figure 1E). High expression of ZIC2 in HCC tumors was also confirmed by Wurmbach’s cohort (GSE6764) (ref. 30 and Supplemental Figure 1C). Additionally, we also noticed that ZIC2 was highly expressed in nondifferentiated HCC cell lines versus in differentiated HCC cell lines (GSE36133) (ref. 31 and Supplemental Figure 1D). Finally, high expression of ZIC2 was in agreement with poor prognosis of HCC patients (GSE10186) (ref. 32 and Supplemental Figure 1E). Altogether, ZIC2 is highly expressed in HCC tumor tissues.
Figure 1. ZIC2 is highly expressed in HCC tumor tissues.
(A) Heatmap of gene expression levels of 283 TFs. Top 10 highly expressed TFs were listed. (B) Top 10 TFs were depleted in Huh7 and Hep3B cells. Their sphere formation was tested via in vitro assays. Significance was calculated vs. shCtrl. (C and D) ZIC2 expression levels were verified in HCC patient samples by quantitative RT-PCR (C) and immunoblotting (D). P, peri-tumor; T, tumor. (E) HCC samples were stained for IHC assay. Representative images were shown in the left panel; the ratios of ZIC2 highly expressed cells and ZIC2 photon intensity were calculated using Image-Pro Plus 6, and statistical results were shown in the right panel. eHCC, early HCC; aHCC, advanced HCC. Scale bars: 50 μm. Data are shown as means ± SD. Two-tailed Student’s t test was used for statistical analysis. *P < 0.05; **P < 0.01; ***P < 0.001; #P < 0.05; and ##P < 0.01. Data are representative of at least 3 independent experiments.
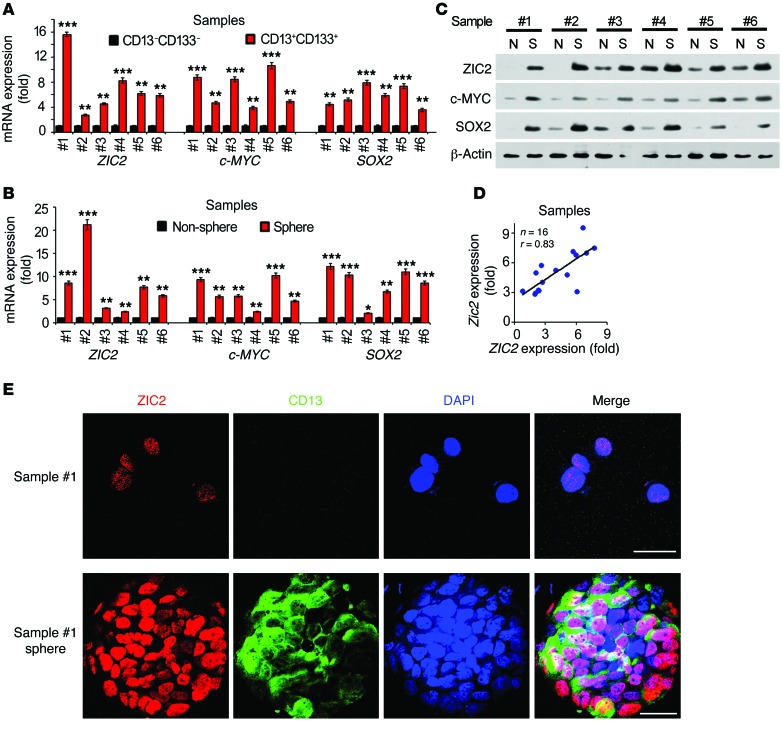
We next tested expression levels of ZIC2 in liver CSCs enriched from both HCC cell lines and patient samples. We found that ZIC2 was highly expressed in liver CSCs compared with non-CSCs derived from HCC samples (Figure 2A and Supplemental Figure 2A). In addition, we performed sphere-formation experiments using HCC cell lines and HCC primary sample cells, and detected expression levels of ZIC2. We found that ZIC2 expression was dramatically increased in the oncosphere cells generated by both patient samples and HCC cell lines (Figure 2, B and C, and Supplemental Figure 2B). Moreover, ZIC2 expression was positively correlated with CD13 expression (Figure 2D). Finally, ZIC2 was localized as puncta staining in the nucleus of tumor cells, and similar staining pattern appeared in oncosphere cells (Figure 2E). Taken together, ZIC2 is highly expressed in the nucleus of HCC tumor tissues and liver CSCs.
Figure 2. ZIC2 is elevated in liver CSCs.
(A) ZIC2 is highly expressed in CD13+CD133+ primary cells. (B and C) ZIC2 is more highly expressed in oncospheres derived from HCC sample cells than nonsphere tumor cells by quantitative RT-PCR (B) and immunoblotting (C). (D) ZIC2 and CD13 expression was detected by quantitative RT-PCR, followed by correlation analysis. (E) HCC primary cells and spheres were permeabilized and stained with anti-ZIC2 and anti-CD13 antibodies, then counterstained with DAPI followed by confocal microscopy. Scale bars: 20 μm. Data are shown as means ± SD. Two-tailed Student’s t test was used for statistical analysis. *P < 0.05; **P < 0.01; and ***P < 0.001. Data are representative of at least 3 independent experiments.
ZIC2 is required for self-renewal of liver CSCs.
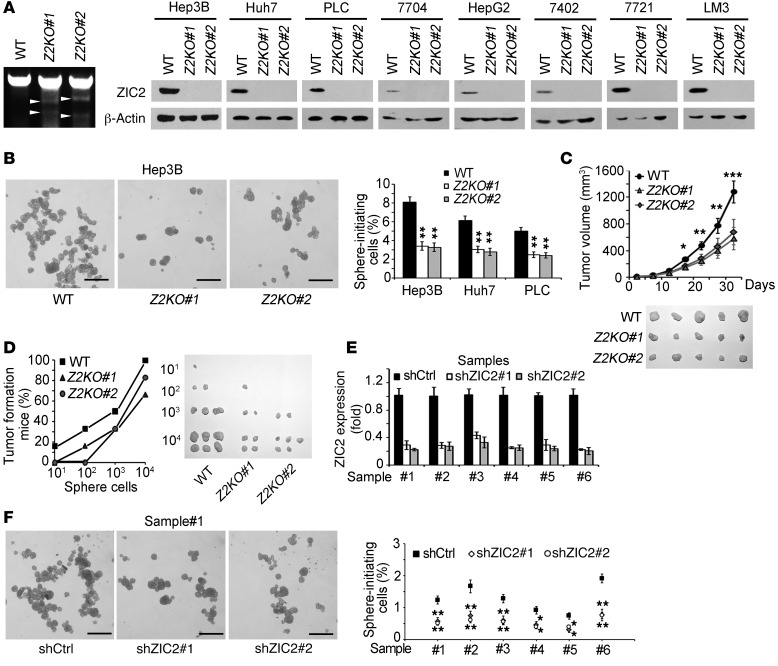
To further determine the pathological role of ZIC2 in liver CSCs, we knocked out ZIC2 in 8 HCC cell lines using a CRISPR/Cas9 system (Figure 3A). Two ZIC2 KO cell strains were established for each cell line. ZIC2 deletion dramatically declined sphere formation in Huh7, Hep3B, and PLC cells, which had sphere-formation capacity (Figure 3B and Supplemental Figure 3A). Additionally, ZIC2-deficient Hep3B cells remarkably decreased xenograft tumor growth (Figure 3C). We then performed sphere formation and digested oncospheres formed by ZIC2-deficient or control cells into single-cell suspension, then s.c. implanted 1 × 104, 1 × 103, 1 × 102, and 10 cells into BALB/c nude mice. Tumor-initiating capacity was evaluated after 3 months of tumor implantation. ZIC2 deficiency dramatically reduced tumor-initiating capacity (Figure 3D) and declined CSC ratios by a limiting dilution xenograft analysis (Supplemental Table 1A). Similar observations were achieved in other cell lines with ZIC2 deletion. We also silenced ZIC2 in 6 HCC primary cells using a lentivirus-infection system and established ZIC2 knockdown cells (Figure 3E). Two pairs of shRNA sequences obtained similar knockdown efficiency for each sample cell. ZIC2 knockdown significantly decreased sphere formation and xenograft tumor growth (Figure 3F and Supplemental Figure 3, B and C). Similar observations were achieved in all the 6 HCC samples.
Figure 3. ZIC2 deficiency abrogates self-renewal of liver CSCs and tumor propagation.
(A) ZIC2-deficient cells were established using a CRISPR/Cas9 system. T7 endonuclease I cleavage confirmed the efficiency of sgRNA (white arrowheads), and ZIC2 KO efficiency was detected by Western blot. ZIC2 was deleted in 8 cell lines. (B) ZIC2 deficiency impairs self-renewal of HCC cells. ZIC2 KO cells and control cells were cultured for sphere formation. (C) ZIC2-deficient and control Hep3B cells (1 × 106) were injected into BALB/c nude mice. Tumor sizes were measured every 5 days. (D) ZIC2 deficiency reduces tumor-initiating capacity. The indicated Hep3B cells were implanted into BALB/c nude mice for tumor initiation. Percentages of tumor-formation mice were calculated (left panel), and established tumors were shown (right panel). (E) ZIC2 was silenced in HCC primary tumor cells. (F) ZIC2-silenced cells (shZIC2) and scramble control cells (shCtrl) were cultured for sphere formation. Six HCC samples obtained similar observations. Z2KO, ZIC2 KO cell. Scale bars: 500 μm (B and F). Data are shown as means ± SD. Two-tailed Student’s t test was used for statistical analysis. *P < 0.05; **P < 0.01; and ***P < 0.001. Data are representative of at least 3 independent experiments.
We next overexpressed ZIC2 in HCC primary cells and cell lines using lentivirus infection (Supplemental Figure 3D and data not shown). We observed that ZIC2 overexpression in HCC primary cells remarkably increased sphere formation and xenograft growth (Supplemental Figure 3, E and F). We tested 6 HCC samples with similar results. Similar results were obtained by ZIC2 overexpression in cell lines such as Hep3B, Huh7, and PLC cells (Supplemental Figure 3G and data not shown). Altogether, ZIC2 is required for the maintenance of liver CSC self-renewal and tumor propagation.
ZIC2 initiates activation of OCT4 signaling.
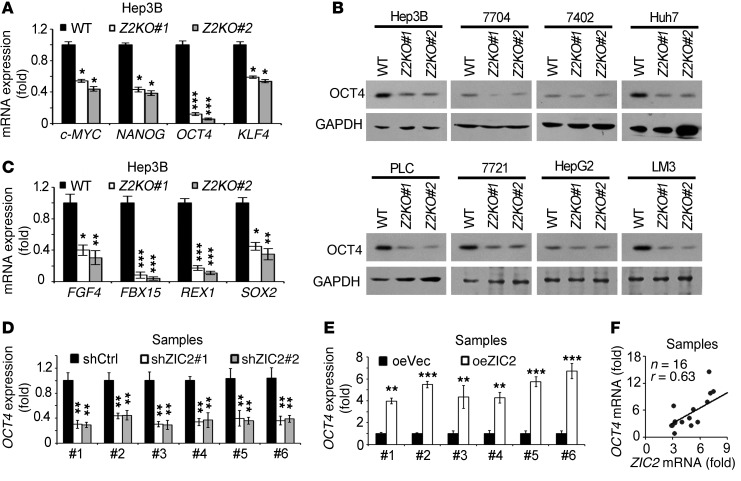
To further determine the underlying mechanism of ZIC2 in the regulation of liver CSCs, we analyzed key TFs related to stemness in ZIC2 KO Hep3B cells. We noticed that OCT4 mRNA was almost undetectable in ZIC2-deficient cells (Figure 4A). Moreover, OCT4 protein levels were remarkably reduced in ZIC2-deficient cells using Western blot (Figure 4B). In addition, ZIC2 deficiency dramatically declined the downstream target genes of OCT4 (Figure 4C). Similar results were obtained in ZIC2-silenced HCC primary cells (Figure 4D).
Figure 4. ZIC2 initiates OCT4 activation.
(A) ZIC2 deficiency reduces OCT4 expression. The indicated core stemness factors were analyzed in ZIC2 KO or control Hep3B cells. (B) OCT4 expression was detected by immunoblotting in ZIC2-deleted cells. (C) The expression levels of OCT4 downstream genes were detected in ZIC2 KO and control cells. (D) ZIC2 knockdown in HCC primary cells reduces OCT4 expression. (E) ZIC2 overexpression increases OCT4 expression. ZIC2 was overexpressed in HCC primary cells followed by OCT4 detection. (F) Correlation analysis of OCT4 and ZIC2 were analyzed based on their expression levels in HCC samples by quantitative RT-PCR. Z2KO, ZIC2 KO cell; oeVec, overexpressed vector; oeZIC2, overexpressed ZIC2. Data are shown as means ± SD. Two-tailed Student’s t test was used for statistical analysis. *P < 0.05; **P < 0.01; and ***P < 0.001. Data are representative of at least 3 independent experiments.
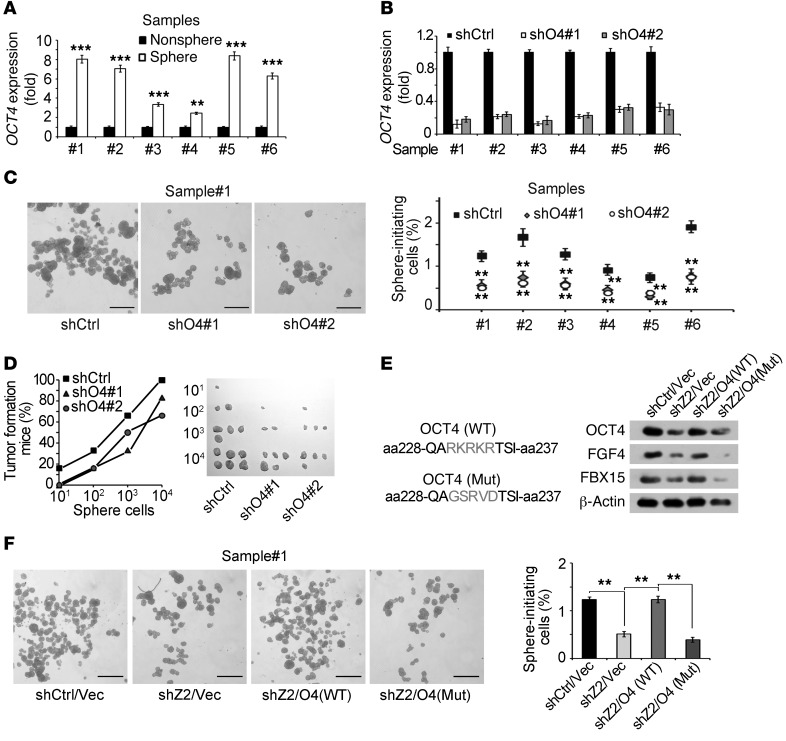
ZIC2 overexpression in HCC primary cells and cell lines remarkably increased OCT4 expression (Figure 4E and data not shown). Additionally, ZIC2 expression levels were positively correlated with the OCT4 expression levels in HCC tumor tissues (Figure 4F). Expression levels of OCT4 and its target genes were dramatically elevated in liver cancer cells and liver CSCs formed by HCC primary cells and Hep3B cells (Figure 1C, Figure 5A, and Supplemental Figure 4, A–C). Finally, OCT4 expression levels were consistent with the CD13 expression levels (Supplemental Figure 4D). These data suggest that OCT4 is activated in liver CSCs, which was in agreement with the transcriptome microarray data (Figure 1A).
Figure 5. OCT4 activation is required for the maintenance of liver CSCs.
(A) OCT4 is highly expressed in spheres generated by HCC primary cells. (B and C) OCT4 was knocked down in HCC primary cells (B) and OCT4-silenced primary cells were cultured for sphere-formation assays (C). (D) The indicated cells were digested into single cells and then injected into BALB/c nude mice with different dilutions. (E and F) ZIC2-depleted primary cells were established and rescued with functional OCT4 and dominant-negative mutant OCT4 (E), then self-renewal was measured in sphere-formation medium (F). Z2, ZIC2; O4, OCT4; Vec, vector; Mut, mutation. Scale bars: 500 μm (C and F). Data are shown as means ± SD. Two-tailed Student’s t test was used for statistical analysis. **P < 0.01 and ***P < 0.001. Data are representative of at least 3 independent experiments.
To further confirm the role of OCT4 in the maintenance of liver CSC self-renewal, we knocked down OCT4 in HCC primary cells by 2 pairs of OCT4 shRNAs (Figure 5B). OCT4 knockdown dramatically reduced the capacity of sphere formation (Figure 5C) and it attenuated xenograft tumor initiating capacity and liver CSC ratios (Figure 5D and Supplemental Table 1B). We silenced OCT4 in 6 HCC primary sample cells with similar observations. In parallel, similar observations were achieved in Hep3B cells (Supplemental Figure 4E). Taken together, OCT4 signaling is essential for the maintenance of liver CSC stemness.
We next overexpressed ZIC2 in OCT4-silenced HCC primary cells. We found that ZIC2 overexpression in OCT4-silenced HCC cells did not promote sphere formation or xenograft tumor growth (Supplemental Figure 5, A and B). Furthermore, we knocked down ZIC2 in OCT4-silenced HCC cells and observed that depletion of ZIC2 in OCT4-silenced HCC cells did not affect either sphere formation or xenograft tumor growth (data not shown). Additionally, ZIC2 knockdown in OCT4-overexpressing HCC cells did not impair OCT4-mediated elevated sphere formation or xenograft tumor growth (Supplemental Figure 5, C and D). Moreover, we rescued WT OCT4 or a dominant-negative mutant OCT4 (33) expression in ZIC2-depleted primary cells (Figure 5E). We noticed that WT OCT4 was able to rescue the self-renewal of ZIC2-silenced cells (Figure 5F), whereas the dominant-negative mutant OCT4 had no such activity. We tested 6 HCC samples with similar results (Supplemental Figure 5E). We concluded that ZIC2 drives self-renewal of liver CSCs upstream of OCT4.
ZIC2 binds to the OCT4 promoter for its activation.
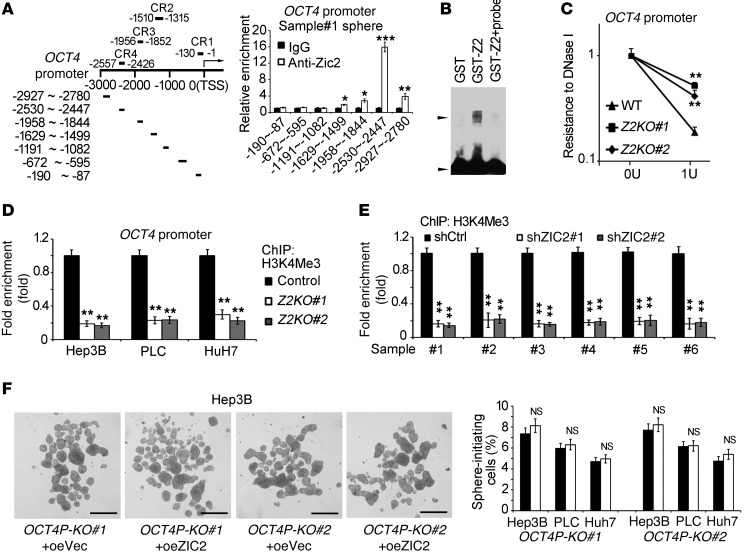
We next performed a ChIP assay in primary oncospheres using anti-ZIC2 antibody. We analyzed a 3-kb locus upstream from the transcription start site (TSS) of the OCT4 gene promoter. We observed that a –2,530~–2,447 bp region of the OCT4 promoter upstream from the TSS was occupied by ZIC2 protein (Figure 6A). We tested 3 HCC primary specimens with similar results. Similar results were achieved in Hep3B cells (Supplemental Figure 6A). To confirm that OCT4 was activated by ZIC2, we performed luciferase reporter assays. We identified a –2,550~–2,430 bp fragment of the OCT4 promoter (CR4 region) bound to the ZIC2 protein (Supplemental Figure 6, B and C). Finally, the ZIC2 binding of the OCT4 promoter was confirmed by a gel EMSA (Figure 6B).
Figure 6. ZIC2 binds to the OCT4 promoter.
(A) Spheres derived from HCC primary cell were lysed and performed a ChIP assay. Detected fragments of the OCT4 promoter were illustrated in the left panel. Enrichment of different regions of the OCT4 promoter was detected by quantitative RT-PCR (right panel). (B) ZIC2 binds to the promoter of OCT4. rGST-ZIC2 and the [r-32P]dATP-labeled OCT4 promoter fragment were incubated followed by an EMSA assay. Probe, non–[r-32P]dATP-labeled OCT4 promoter fragment. Upper black arrowhead denotes the binding of ZIC2 and probes, and lower black arrowhead denotes free probes. (C) ZIC2 deficiency increases resistance to DNase I digestion at the OCT4 locus. (D) ZIC2-deficient sphere cells were applied to a ChIP assay using anti-H3K4Me3 antibody. (E) ZIC2-depleted primary cells were applied to a ChIP assay using anti-H3K4Me3 antibody. (F) Sphere formation was analyzed in the ZIC2 binding sequence of OCT4 promoter–deficient cells. OCT4P-KO, ZIC2 binding region of OCT4 promoter KO. CR, conserved region; oeVec, overexpressed vector; oeZIC2, overexpressed ZIC2. Scale bars: 500 μm. Data are shown as means ± SD. Two-tailed Student’s t test was used for statistical analysis. *P < 0.05; **P < 0.01; and ***P < 0.001. Data are representative of at least 3 independent experiments.
We next wanted to determine the physiological function of the ZIC2 binding to the OCT4 promoter. We examined chromatin accessibility of the OCT4 locus by a standard DNase I digestion assay. We found that ZIC2 KO remarkably attenuated chromatin accessibility of the OCT4 locus (Figure 6C). Moreover, ZIC2 deficiency significantly reduced the H3K4 tri-methylation (H3K4Me3) level on the OCT4 locus (Figure 6D), a major hallmark of transcription activation. Six HCC primary samples showed similar observations (Figure 6E). By contrast, ZIC2 overexpression dramatically enhanced both the chromatin accessibility of the OCT4 locus and the H3K4Me3 level of the OCT4 locus (Supplemental Figure 6, D–F). These results suggest that ZIC2 binds to the promoter of OCT4 that initiates its activation.
We next explored whether ZIC2 sustains the self-renewal of liver CSCs via binding to OCT4 promoter. We deleted the ZIC2 binding sequence of OCT4 promoter in Hep3B cells using a CRISPER/Cas9 lentivirus system, which was confirmed by DNA sequencing (Supplemental Figure 6G). We found that ZIC2 overexpression did not enhance OCT4 expression in the OCT4 promoter binding sequence–deleted Hep3B cells (Supplemental Figure 6H). Consequently, these cells failed to promote sphere formation (Figure 6F and Supplemental Figure 6I) and xenograft tumor propagation (Supplemental Figure 6J). These data indicate that ZIC2 binding to the OCT4 promoter is necessary for ZIC2-initiated self-renewal of liver CSCs.
ZIC2 interacts with the NURF complex.
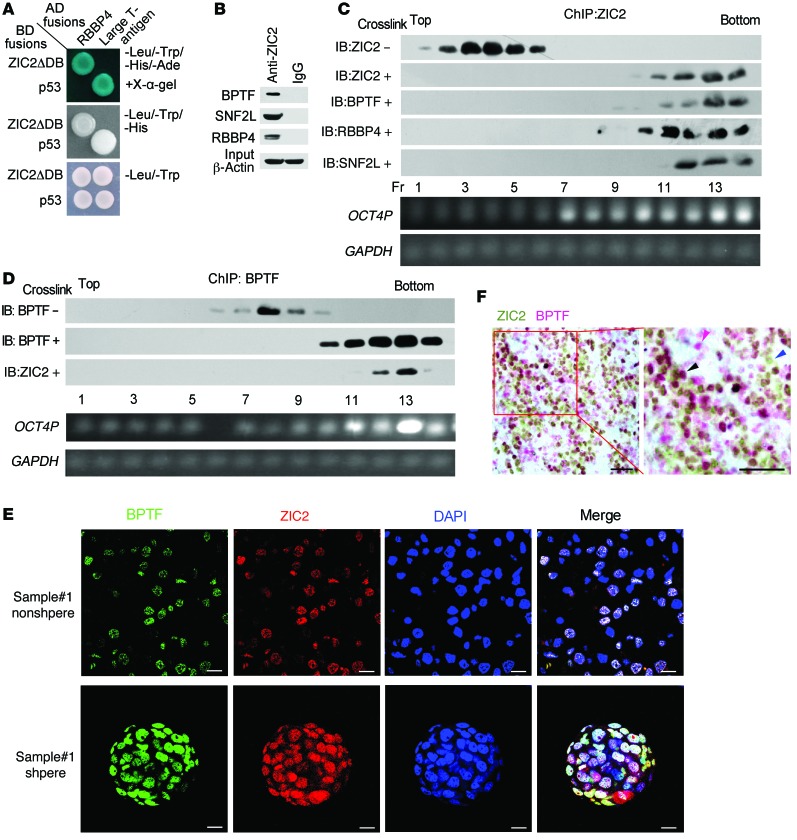
To further elucidate the molecular mechanism of ZIC2 in regulating OCT4 expression, we screened interacting candidates using ZIC2 as bait with a yeast 2 hybrid system. We deleted the DNA-binding domain of ZIC2 (aa 255-415) to subclone into pGBKT7 plasmid (BD-ZIC2ΔDB). We identified that RBBP4 gene was an interactor for ZIC2 and further confirmed by yeast 2 hybrid assays (Figure 7A). RBBP4 is a component of the NURF complex. Their direct interaction was verified by a pulldown assay (Supplemental Figure 7A). With a coimmunoprecipitation (Co-IP) assay, anti-ZIC2 antibody could precipitate RBBP4 in primary sphere cell lysates, as well as the other 2 components BPTF and SNF2L (Figure 7B). In addition, we next performed ChIP immunoblots with elution gradients. With crosslinking treatment, ZIC2 coeluted with the components of the NURF complex (Figure 7C). In parallel, BPTF was also associated with ZIC2 in a chromatin-binding fashion (Figure 7D). These data indicate that the interaction of ZIC2 with the NURF complex is chromatin embedded.
Figure 7. ZIC2 interacts with the NURF complex.
(A) ZIC2 associates with RBBP4. Yeast strain AH109 was cotransfected with Gal4 DNA-binding domain (BD) fused the DNA binding region–deleted (aa 255-415) ZIC2 gene (BD-ZIC2ΔDB), and Gal4-AD–fused RBBP4. Selected clones were detected for β-galactosidase activity. p53 and large T antigen were used as a positive control. (B) Anti-ZIC2 antibody precipitated the NURF complex from primary sphere lysates by Co-IP assay. (C and D) HCC sample sphere cells were treated with 1% formaldehyde for crosslinking. Then anti-ZIC2 (C) or anti-BPTF antibody (D) was incubated with treated lysates for ChIP assays, followed by size fractionation with sucrose gradient ultracentrifugation. Eluate gradients were used for Western blot and PCR assays. (E) ZIC2 colocalizes with BPTF. HCC primary nonsphere cells and spheres were stained for BPTF and ZIC2, counterstained with DAPI. SNF2L and RBBP4 were also merged with ZIC2 (data not shown). Scale bar: 10 μm. (F) ZIC2 and BPTF were costained by IHC staining. Blue arrowhead denotes ZIC2, red arrowhead indicates BPTF, and black arrowhead indicates merge of ZIC2 and BPTF. Scale bars: 50 μm. Data are representative of at least 3 independent experiments.
Colocalization of ZIC2 with the NURF complex in the nucleus was visualized in HCC primary cells and their oncosphere cells by confocal microscopy (Figure 7E). Similar observations were obtained in Hep3B cells and their oncospheres (Supplemental Figure 7B). IHC staining showed merged staining of ZIC2 with the NURF complex (Figure 7F and data not shown).
We next tested expression levels of the NURF complex in liver CSCs. We noticed that the components of the NURF complex were highly expressed in liver CSCs derived from HCC samples and oncosphere cells generated from HCC samples (Supplemental Figure 8, A and B). Additionally, the expression levels of the 3 components of the NURF complex were positively correlated with the CD13 expression level in HCC samples (Supplemental Figure 8C). Finally, the expression levels of the 3 components of the NURF complex were significantly elevated in CD13hi samples compared with CD13lo samples provided by the Wang’s cohort dataset (GSE14520) (ref. 34 and Supplemental Figure 8D). Overall, these data indicate that ZIC2 interacts with the NURF complex in the nucleus of HCC cells.
The NURF complex is required for self-renewal of liver CSCs.
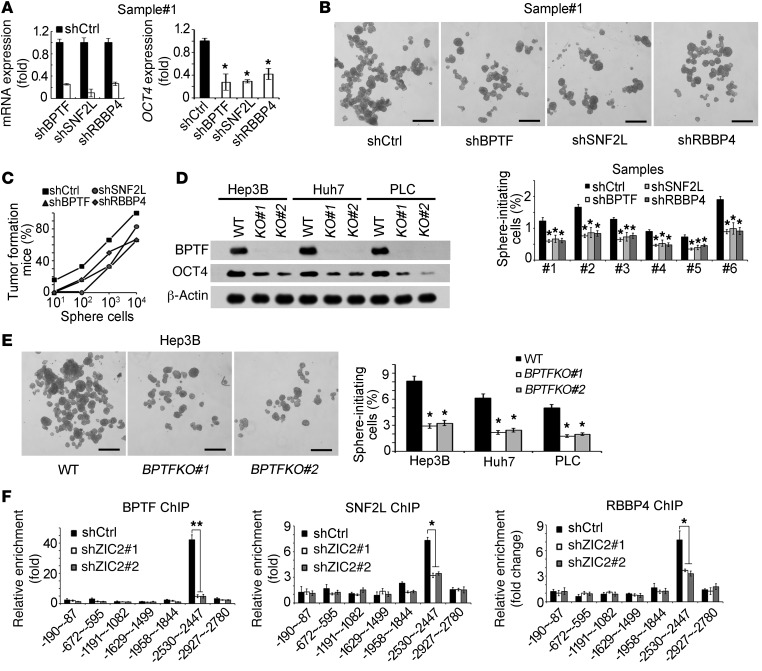
To explore the role of the NURF complex in the regulation of liver CSCs, we silenced each component of the NURF complex in HCC primary cells (Figure 8A, left panel). Knockdown of the NURF components remarkably suppressed OCT4 expression in HCC primary cells (Figure 8A, right panel). Similar results were obtained using Hep3B cells (Supplemental Figure 9A). Furthermore, knockdown of the NURF complex significantly reduced chromatin accessibility of the OCT4 locus (Supplemental Figure 9B). The NURF complex bound to the OCT4 promoter by ChIP assays (Supplemental Figure 9C). Coincidently, the NURF complex occupied the CR4 region of the OCT4 promoter, the same region as ZIC2. Importantly, the CR4 region of the OCT4 promoter displayed much lower levels of H3K4Me3 in NURF complex–silenced primary cells (Supplemental Figure 9D). These data suggest that the NURF complex enhances OCT4 expression.
Figure 8. The NURF complex is required for self-renewal of liver CSCs.
(A) The NURF complex enhances OCT4 expression. The NURF components were knocked down followed by detection of OCT4 expression. (B) Sphere formation was performed with NURF complex silenced primary cells. Six HCC samples obtained similar results. (C) Oncospheres generated by BPTF-, SNF2L-, and RBBP4-silenced primary cells were digested for gradient injection. Percentage of tumor-formation mice was calculated. (D) BPTF-deficient cells were established by a CRISPR/CAS9 system. BPTF deficiency was verified by Western blot. (E) BPTF KO cells were cultured for sphere-formation assays. Similar results were achieved in the tested 3 cell lines. (F) ChIP assays were performed using the indicated anti-NURF antibodies, and then OCT4 promoter enrichment regions were examined by quantitative RT-PCR. Scale bars: 500 μm (B and E). Data are shown as means ± SD. Two-tailed Student’s t test was used for statistical analysis. *P < 0.05 and **P < 0.01. Data are representative of at least 3 independent experiments.
Of note, knockdown of the NURF components in HCC primary cells and cell lines dramatically reduced sphere formation and xenograft growth (Figure 8B and Supplemental Figure 9, E and F). Additionally, depletion of the NURF components in HCC primary cells markedly reduced tumor initiating capacity (Figure 8C) and CSC ratios by a limiting-dilution xenograft analysis (Supplemental Table 1C). We tested 6 HCC samples and achieved similar results. To further verify the relatively physiological role of the NURF complex, we knocked out BPTF, a core component of the NURF complex, in HCC cell lines using a CRISPR/Cas9 system (Figure 8D). We found that BPTF deficiency remarkably impaired OCT4 expression and sphere formation (Figure 8, D and E, and Supplemental Figure 9G), which was in agreement with the NURF complex–silencing results.
We next rescued WT or mutant OCT4 expression in NURF-depleted HCC primary cells (Supplemental Figure 10A). We found that WT OCT4 could predominantly restore the self-renewal of NURF-depleted cells, whereas mutant OCT4 had no such effect (Supplemental Figure 10, B and C), suggesting that the NURF complex promotes liver CSC self-renewal via OCT4 signaling. Importantly, depletion of ZIC2 abolished its binding capacity to the NURF complex, leading to disassociation from the OCT4 promoter (Figure 8F). However, depletion of the NURF complex did not affect the binding capacity of ZIC2 to the OCT4 promoter (Supplemental Figure 10D), suggesting that the NURF complex directly regulated OCT4 expression. In sum, NURF complex–mediated OCT4 activation is required for the self-renewal maintenance of liver CSCs.
NURF complex levels are consistent with HCC severity and prognosis.
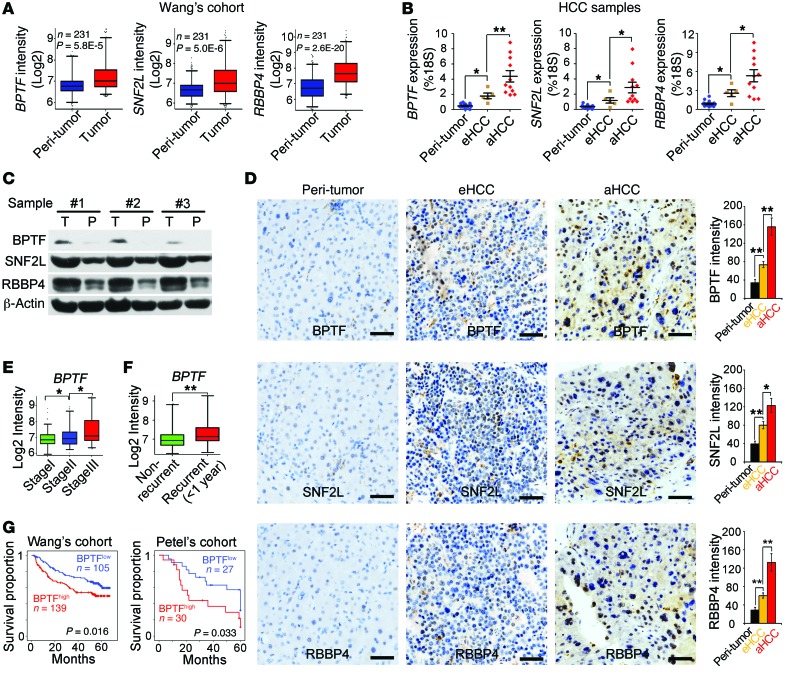
As shown above, the NURF complex was highly expressed in liver CSCs and involved in the regulation of liver CSCs. We further wanted to explore whether the NURF complex was implicated in HCC progression. Firstly, we analyzed NURF expression levels using Wang’s cohort (GSE14520). The 3 NURF complex components were significantly highly expressed in HCC tumor tissues (Figure 9A). We tested the NURF complex expression in HCC samples through quantitative RT-PCR and immunoblotting. The NURF complex was highly expressed in HCC patients (Figure 9, B and C). Additionally, the expression levels of the NURF complex were well correlated with clinicopathological stages of HCC patients (Figure 9B). This result was further validated by IHC staining (Figure 9D).
Figure 9. NURF complex levels are consistent with HCC severity and prognosis.
(A) The NURF complex is highly expressed in HCC datasets provided by Wang’s cohort (GSE14520). (B and C) High expression of the NURF complex was verified in HCC samples by quantitative RT-PCR (B) and Western blot (C). eHCC, early HCC; aHCC, advanced HCC. (D) Expression of BPTF, SNF2L, and RBBP4 was observed by IHC staining (left panel). Photon intensity was calculated using Image-Pro Plus 6 (right panel). Scale bars: 50 μm. (E and F) High BPTF expression is consistent with late clinicopathological stages (E) and early recurrence (F) of HCC patients provided by Wang’s cohort (GSE14520). (G) Expression levels of the NURF complex are correlated with prognosis prediction of HCC patients. HCC samples were divided into 2 groups according to BPTF expression levels followed by Kaplan-Meier survival analysis. For A, E, and F, data are shown as box-and-whisker plots. Whiskers below and above boxes extend to the 5th and 95th percentiles, respectively. Horizontal lines within boxes represent median levels of gene intensity. Boxes represent interquartile range (IQR); upper and lower edges correspond to the 75th and 25th percentiles, respectively. For B and D, data are shown as means ± SD. Two-tailed Student’s t test was used for statistical analysis. *P < 0.05 and **P < 0.01. Experiments were repeated at least 3 times.
Base on analyzing Wang’s cohort (GSE14520), we observed that the BPTF expression levels were consistent with clinicopathological stages of HCC patients (Figure 9E). Additionally, higher BPTF expression levels represented higher frequency of 1-year recurrence of HCC patients (Figure 9F). Finally, we analyzed Wang’s cohort (GSE14520) (34) and Petel’s cohort (E-TABM-36) (35) with survival information. We found that patients with higher expression levels of BPTF displayed worse prognosis (Figure 9G). In sum, expression levels of the NURF complex in tumor tissues are consistent with clinical severity and prognosis of HCC patients.
Discussion
Twenty years ago, Dick and colleagues firstly isolated the CD34+/CD38– leukemic subpopulation cells as leukemic CSCs (36). Subsequently, CSCs have been identified in many other solid tumors, including breast, lung, brain, liver, colon, prostate, and bladder cancers (3, 6, 37). CSCs harbor similar characteristics associated with normal tissue stem cells, including self-renewal, differentiation, and the ability to form a new tumor. CSCs may be responsible for cancer relapse and metastasis due to their invasive and drug-resistant capacities (38). Several surface markers have been identified to enrich liver CSCs, whose heterogeneous markers may represent different cellular origins (39). However, it remains largely unknown about hepatic CSC biology. In this study, we analyzed differential TFs from our transcriptome microarray datasets derived from liver CSCs (29). We found the TF ZIC2 is highly expressed in liver CSCs. ZIC2 is required for the self-renewal maintenance of liver CSCs upstream of OCT4 activation signaling. ZIC2 recruits the NURF complex to the OCT4 promoter that initiates OCT4 activation. Expression levels of the NURF complex are consistent with clinical severity and prognosis of HCC patients.
Elucidation of signaling pathways that modulate the maintenance and survival of liver CSCs is important for the understanding of hepatic CSC biology and the development of novel therapies against HCC. Several signaling pathways, such as WNT, TGF-β, AKT, and STAT3 pathways, have been defined to be implicated in the regulation of liver CSCs (39). Not surprisingly, some liver CSC subsets and normal tissue stem cells may share core regulatory genes and common signaling pathways. In this study, we found that OCT4 is highly expressed in liver CSCs and is essential for the maintenance of liver CSCs. The TF OCT4, encoded by the Pou5f1 gene, is a member of the POU family. OCT4 is required for the stemness properties of ESCs (11), suggesting it is an essential factor in early development of mammalian embryogenesis. Importantly, OCT4 harbors oncogenic potential (40, 41). Moreover, it has been reported that OCT4 is involved in the maintenance of lung CSCs, as well as the regulation of tumor progression in bladder cancer (42, 43). Here, we showed that OCT4 is required for the self-renewal maintenance of liver CSCs, suggesting that OCT4 plays a critical role in the regulation of liver CSCs and tumor propagation.
OCT4 acts as a gatekeeper in the beginning of mammalian development; it regulates the transcription of many genes, such as FOXO15, ZNF42, REX1, FGF4, SOX2, NANOG, and so on. However, how OCT4 expression is regulated remains largely unknown. Here, we observed that ZIC2 can bind to the fourth region (CR4) of OCT4 promoter, which triggers OCT4 expression in liver CSCs. The ZIC TF family members (ZIC1-5) contain highly conserved C2H2-class zinc finger motifs and are involved in early embryonic development. ZIC2 mutations cause human holoprosencephaly (44). A recent report showed that ZIC2 is highly expressed in cervical cancer and associated with activation of Hedgehog signaling (18). Here, we found that ZIC2 is constitutively expressed in liver CSCs and participates in the regulation of liver CSC maintenance. Actually, ZIC2 is also highly expressed in ESCs, iPSCs, and 2/3 partial hepatectomy–regenerated (PH-regenerated) liver cells (data not shown). These data suggest that ZIC2 plays a crucial role in the maintenance and survival of both normal stem cells and CSCs.
Due to the analogy to tissue-specific stem cells, which confer maintaining adult tissues, CSCs contribute to long-term growth and are responsible for the maintenance and growth of tumors (45). The clonal evolution model proposes that tumor cells progressively accumulate mutations, some of which harbor increased fitness and survival advantage that generate CSCs (46). Transcriptional regulation is one of the key regulatory mechanisms determining cell fate, which includes cis-regulatory elements and transacting factors. Among all transacting factors, chromatin-remodeling regulation plays a pivotal role in gene transcription (20, 47), which depends on ATP-dependent chromatin remodeling complexes. Based on a common SWI2/SNF2-related catalytic ATPase subunit, the chromatin remodeling complexes are classified into 4 major subfamilies, which are SWI/SNF, ISWI, CHD, and INO80 (48). The NURF complex is a core member of the ISWI subfamily. The NURF complex consists of 4 subunits in drosophila (NURF301, NUFF55, NURF38, and ISWI) and 3 components in mammalian cells (BPTF, RBBP4, and SNF2L) (49). The NURF complex can regulate the canonical Wnt signaling pathway via modulating the chromatin structures of targeting genes to make transcriptional regulators more accessible (26). We previously showed that the NURF complex can be recruited on the c-MYC promoter for its activation, which regulates the self-renewal of HSCs (27). Here, we showed that ZIC2 can recruit the NURF complex to the OCT4 promoter for its initiation, which modulates the maintenance and survival of liver CSCs. These findings indicate that the NURF complex is implicated in the tumorigenesis of HCC.
Based on our online-cohort dataset analysis and experimental results, the NURF complex is highly expressed in HCC tumors and liver CSCs. The NURF complex knockdown or deletion impairs the self-renewal maintenance of liver CSCs and tumor initiation. Importantly, the expression levels of the NURF complex are positively correlated with clinical severity and prognosis of HCC patients. Moreover, ZIC2 and OCT4 are also required for the maintenance and survival of liver CSCs, which are positively related to the clinicopathological stages of HCC patients. In summary, ZIC2, OCT4, and the NURF complex can be detected for the diagnosis and prognosis prediction of HCC patients, which might be used as targets for eradicating liver CSCs for future therapy.
Methods
Gene expression analyses.
Cohort datasets were downloaded from NCBI or EBI. R language and Bioconductor was used for background correction, normalization, calculation of gene expression, and annotation (50). Genes and expression lists generated by R3.1.0 were used for further analysis.
HCC cell lines.
Human HCC cell line Hep3B, PLC, LM3, 7721, HepG2, and Huh7 were provided by Zeguang Han (Shanghai Jiaotong University School of Medicine, Shanghai, China); 7704 and 7402 were provided by Pengyuan Yang (Institute of Biophysics, Chinese Academy of Sciences, Beijing, China). HCC cells were grown in DMEM medium supplemented with 10% FBS (Invitrogen), 100 μg/ml penicillin, and 100 U/ml streptomycin.
Antibodies and regents.
Anti–β-actin (catalog A1978), anti-Flag (catalog F1804) Abs were purchased from Sigma-Aldrich. Anti-ZIC2 (catalog ARP35821_P050) antibody was obtained from Aviva Systems Biology. Anti-OCT4 (catalog 2750) antibody was purchased from Cell Signaling Technology. Anti-SNF2L (catalog 61465) antibody was purchased from Active Motif. Anti-BPTF (catalog bs-11641R) antibody was obtained from Bioss Inc. Anti-RBBP4 (catalog 20364-1-AP) antibody was purchased from Proteintech Group, Inc. FITC-conjugated CD13 antibody (catalog 11-0138) was obtained from eBioscience. Phycoerythrin-conjugated (PE-conjugated) CD133 (catalog 130098826) was purchased from Miltenyi Biotec. Alexa488-conjugated donkey anti-mouse IgG (catalog R37114), Alexa594-conjugated donkey anti-mouse IgG (catalog R37115), and Alexa594-conjugated donkey anti-rabbit IgG (catalog R37119) Abs were obtained from Invitrogen. Alkaline phosphatase–conjugated (AP-conjugated) secondary antibody (catalog BA1011) was obtained from Wuhan Boster Biological Technology, Ltd. HRP-conjugated secondary antibody (catalog sc-2004, sc-2005) was obtained from Santa Cruz Biotechnology, Inc. bFGF (catalog GF446-50UG) was obtained from Millipore. EGF (catalog E5036-200UG), PEG5000 (catalog 175233-46-2), and DAPI (catalog 28718-90-3) were purchased from Sigma-Aldrich. N2 supplement (catalog 17502-048) and B27 (catalog 17504-044) were obtained from Invitrogen. Dual-detection luciferase detection kit (catalog E1910) was purchased from Promega.
Quantitative RT-PCR.
mRNA was extracted according to standard protocols provided by Invitrogen. The total mRNA served as a template, and reverse-transcription PCR was followed by standard protocols from Promega Company. cDNA was examined for gene expression by quantitative RT-PCR. For ChIP assay, extracted DNA fragments and input genomic DNA served as a template. The primer sequences we used are shown in the Supplemental Table 2.
Western blot.
Western blot was performed as described (27). For details, specimens or spheres derived from primary cells were homogenized with RIPA buffer (150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP40, 1 mM EDTA, and 50 mM Tris [pH 8.0]) supplemented with cocktail protease inhibitor (Roche Diagnostics). After boiling for 15 minutes, supernatants were loaded for SDS-PAGE. Nitrocellulose membranes were incubated with primary antibodies for immunoblotting, then incubated and visualized by HRP-conjugated secondary antibodies.
CRISPR/Cas9 KO system.
ZIC2- and BPTF-deficient Hep3B cells were established using a CRISPR/Cas9 system according to the standard protocol provided by Zhang’s lab (51). Briefly, single guide RNA (sgRNA) was generated by online CRISPR Design Tool (http://tools.genome-engineering.org) and cloned into pSpCas9 (BB)-2A-GFP vector. After confirming the cutting efficiency of sgRNA, pSpCas9 vectors were transfected into Hep3B cells. Sorted GFP+ Hep3B cells were seeded into 96-well plates for monoclonalization. Four weeks later, the cell clones derived from single cells were detected for gene expression. The sgRNA sequences of ZIC2 were 5′-CCATCACCACTCCGCCGCGG-3′ and 5′-TTCACGGTCCTGCATCTCGG-3′. The sgRNA sequences of BPTF were 5′-CCCCCCGATGGGTCCGGACG-3′ and 5′-CCCGCCTCGGCTCCGACATG-3′.
For deletion of the ZIC2 binding sequence of OCT4 promoter in Hep3B cells, we used LentiCRISPRv2 (catalog 52961), pVSVg (catalog 8454), and psPAX2 (catalog 12260) plasmids purchased from AddGene. We produced lentivirus in 293T cells and infected Hep3B cells with lentivirus, followed by puromycin selection and monoclonalization. sgRNA sequences of the OCT4 promoter were: OCT4 promoter KO #1: 5′-GTGCCGTGATGGTTCTGTCC-3′ (F) and 5′-GGAGGAACATGCTTCGGAAC-3′ (R); OCT4 promoter KO #2: 5′-CCTCTGAGAG GCCGTCTTCT-3′ (F) and 5′-TCGGCCTTTAACTGCCCAAA-3′ (R). Selected cell clones with deletion of the ZIC2 binding sequence of OCT4 promoter were confirmed by DNA sequencing.
IHC assay.
Formalin-fixed tumor tissue sections were deparaffinized in xylene (10 minutes, twice), rehydrated in graded alcohols (100%, 100%, 95%, 85%, and 70% alcohols), and finally submerged in distilled water. After treated in 3% Hydrogen Peroxide (H2O2) for 15 minutes, the slides were processed for antigen retrieval in Tris-EDTA buffer (10 mM, pH 8.0), 121°C for 5 minutes, and then cooled down slowly. After blocking with 10% goat serum for 30 minutes, the sections were incubated in primary antibodies overnight. After washing 3 times with PBS, the sections were incubated in HRP or AP-conjugated secondary antibodies, and subsequent detection was performed using the standard substrate detection of HRP or AP. Then, the sections were stained with hematoxylin and dehydration in graded alcohols and xylene. For double staining, there was no counterstaining with hematoxylin.
Co-IP assay.
For Co-IP, sphere cells (5 × 106) were harvested, washed 3 times with PBS, and then treated with RIPA buffer for 30 minutes at 4°C. Precipitation was removed from cell lysate by centrifugation, and the supernatants was precleared for 1 hour by protein A/G beads (Santa Cruz Biotechnology Inc.). Then, the precleared protein A/G beads were removed and primary antibodies were added for incubation overnight. New protein A/G beads were added for IP. Finally the precipitate was collected and detected as described (27).
Lentivirus production and cell infection.
ZIC2 plasmid was provided by Eloísa Herrera (Instituto de Neurociencias de Alicante, Campus San Juan, Avd. Ramón y Cajal s/n, Alicante 03550, Spain). We constructed Pbplv-GFP-Flag-ZIC2 using ZIC2 plasmid as a template. For virus production, we transfected 293T cells with Pbplv-GFP or Pbplv-GFP-Flag-ZIC2 plasmid, along with package plasmids. Hep3B and HCC primary cells were infected by the virus supernatants. After sorting for GFP+ cells, we established ZIC2 overexpression cells. ZIC2-, OCT4-, and NURF complex–silenced Hep3B or HCC primary cells were established using pSicoR-GFP shRNA vectors by a similar strategy. shRNA sequences are listed in Supplemental Table 3.
Sphere formation assay.
Hep3B cells (1,000) or HCC primary cells (5,000) were seeded in Ultra Low Attachment 6-well plates (Corning Inc.) and cultured in Dulbecco’s modified Eagle’s medium/F12 supplemented with B27, N2 (Invitrogen), 20 ng/ml EGF, and 20 ng/ml Basic Fibroblast Growth Factor (Millipore). Cells were incubated in a CO2 incubator 2 weeks later; spheres were counted under stereomicroscope (Olympus). We collected medium containing nonsphere cells and spheres into an eppendorf tube and let stand for 5 minutes for sphere/nonsphere separation. The pellets were spheres, and supernatants were nonsphere cells. The spheres were fixed for immunofluorescence staining or digested for Co-IP and Western blot assays. Supernatants were then removed into a new eppendorf tube with transferpettor and collected by centrifugation at 4,000 rpm for 5 minutes. Pellets were nonsphere cells and used directly for subsequent experiments. We cultured these nonsphere cells under the same nonadherent conditions as sphere cells.
Flow cytometry.
Cells were labeled with FITC-conjugated CD13 and PE-conjugated CD133 antibodies followed by CSC isolation. For FACS analysis, cells were incubated either with fluorescence-conjugated antibodies or with primary antibodies and further fluorescence-conjugated secondary antibodies.
Yeast 2-hybrid.
Yeast 2-hybrid screening was performed using Matchmaker Gold Yeast 2-Hybrid system (Takara Bio Inc.) followed by the guidelines of the manufacturer’s instruction (52). Briefly, the DNA binding region–deleted (aa 255-415) ZIC2 gene was cloned into pGBKT7 plasmid (BD-ZIC2ΔDB). BD-ZIC2ΔDB and plasmids containing Gal4-activating domain (AD) fused human liver library (Takara Bio Inc.) were cotransformed into the AH109 yeast strain. Then, the transformed yeasts were grown on plates with SD medium lacking adenine, histidine, tryptophan, and leucine. Selected clones were isolated and identified by DNA sequencing. Recovery of the plasmids and β-gal assay were performed as described (52).
Immunofluorescence staining.
Cells were fixed by 4% paraformaldehyde (PFA) for 20 minutes and penetrated by 0.5% tritonX-100 for 30 minutes. After blocking with 10% FBS, primary antibodies were added and incubated overnight at 4°C. After washing 3 times with PBS, fluorescence-conjugated secondary antibodies were added for observation by confocal microscopy. For HCC tissue samples, specimens were treated with 3% hydrogen peroxide and then Tris-EDTA buffer for antigen retrieval before blocking with FBS.
ChIP.
We performed ChIP assays according to the standard protocol (Uptate Biotechnology, Inc.). Briefly, primary or Hep3B spheres were digested with Trypsin/EDTA and incubated in 1% formaldehyde for 10 minutes at 37°C. They were then cracked by warm SDS lysis buffer for 10 minutes on ice, followed by ultrasonic to shear DNA to lengths between 200 and 1,000 bp. Chromatin was precleared with protein A/G beads, followed by incubation with the indicated antibodies for ChIP assay as described (27).
ChIP-Immunobloting assay.
Oncosphere cells were treated with (+crosslink) or without (–crosslink) 1% formaldehyde for 10 min, cracked with SDS lysis buffer, and treated by ultrasonication. After IP with the indicated antibodies and beads, the eluate (500 μl) was layered onto 35 ml 10%–40% (W/V) gradient sucrose. Beckman SW28 rotor was used for ultracentrifugation at 24,000 rpm for 30 hours. Then, fractions were carefully collected into 2-ml tubes, and the elution gradients were concentrated to 100 μl using centrifugal concentration tubes. Finally, the samples were analyzed using Western blotting or PCR assays.
DNase I sensibility assay.
Cell nuclei were isolated and lysed for DNase I digestion as described (53). After digestion for 5 minutes at 37°C, total DNA was extracted and quantitative RT-PCR was performed with the indicated promoter-specific primers.
Gel EMSA assay.
Recombinant GST-ZIC2 or GST protein was expressed in E. coli BL21 (λDE3) and purified with GST beads. The DNA fragment (–2550~–2430 bp) of OCT4 promoter was produced by Sangon company (Shanghai, China), and labeled with [r-32P]dATP according to standard protocol. Probes and recombinant proteins were incubated in binding buffer, and mobility shift assay was performed using gel electrophoresis.
Luciferase reporter assay.
Luciferase reporter assay was performed according to the standard protocol of Dual-Luciferase Reporter Assay system (Promega). Briefly, indicated fragments of OCT4 promoter were cloned into the pGL3 luciferase reporter vector and transfected into ZIC2 overexpression and control Hep3B cells. For each sample, 1 ng pRL-TK was transfected for loading control. Thirty-six hours later, cells were collected and lysed by lysis buffer followed by detection of luciferase activity.
In vivo xenograft experiments.
Six-week-old female BALB/c normal mice and nude mice were obtained from the Animal Center of the Chinese Academy of Medical Sciences (Beijing, China). For tumor-initiating assays, 6 BALB/c nude mice were used every sample. Tumor-initiating cell frequencies were calculated using extreme limiting dilution analysis (54). Tumor cells (1 × 106) were injected into 6-week-old BALB/c nude mice. Every 5 days, tumor volume was calculated as described (55). For tumor-formation assay, spheres were digested into single cells by Trypsin/EDTA, and indicated gradient cells were injected into BALB/c nude mice. Tumor formation was evaluated at the third month. For xenograft tumor propagation, 5 BALB/c nude mice were used for one sample.
Statistics.
Tumor-initiating cell frequencies were calculated using extreme limiting dilution analysis (54). Statistical significance was evaluated using a 2-tailed Student’s t test. P values less than 0.05 were considered significant.
Study approval.
Human liver cancer specimens were obtained from the Department of Hepatobiliary Surgery, PLA General Hospital (Beijing, China) with informed consent, according to the Institutional Review Board approval. All experiments involving mice were approved by the institutional committee of Institute of Biophysics, Chinese Academy of Sciences.
Supplementary Material
Acknowledgments
We thank Junying Jia (Institute of Biophysics, CAS) and Jing Li (Cnkingbio Co. Ltd, Beijing , China) for technical support. This work was supported by the National Natural Science Foundation of China (81330047, 91419308, 81272270, 81402459, and 81101531); the State Projects of Essential Drug Research and Development (2012ZX09103301-041); 973 Program of the Ministry of Science and Technology of China (2015CB553705, 2010CB911902); and the Strategic Priority Research Programs of the Chinese Academy of Sciences (XDA01010407).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(10):3795–3808. doi:10.1172/JCI81979.
References
- 1.Han ZG. Functional genomic studies: insights into the pathogenesis of liver cancer. Annu Rev Genomics Hum Genet. 2012;13:171–205. doi: 10.1146/annurev-genom-090711-163752. [DOI] [PubMed] [Google Scholar]
- 2.Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330):314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 3.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10(6):717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8(10):806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 5.Yang ZF, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13(2):153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Haraguchi N, et al. CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest. 2010;120(9):3326–3339. doi: 10.1172/JCI42550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475(7356):390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 8.Ladewig J, Koch P, Brüstle O. Leveling Waddington: the emergence of direct programming and the loss of cell fate hierarchies. Nat Rev Mol Cell Biol. 2013;14(4):225–236. doi: 10.1038/nrm3543. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor OCT4. Cell. 1998;95(3):379–391. doi: 10.1016/S0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 12.Kim JB, et al. OCT4-induced pluripotency in adult neural stem cells. Cell. 2009;136(3):411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Kim JB, et al. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461(7264):649–653. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- 14.Wezel F, Pearson J, Kirkwood LA, Southgate J. Differential expression of OCT4 variants and pseudogenes in normal urothelium and urothelial cancer. Am J Pathol. 2013;183(4):1128–1136. doi: 10.1016/j.ajpath.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, et al. Follicle-stimulating hormone inhibits apoptosis in ovarian cancer cells by regulating the OCT4 stem cell signaling pathway. Int J Oncol. 2013;43(4):1194–1204. doi: 10.3892/ijo.2013.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benedyk MJ, Mullen JR, DiNardo S. odd-paired: a zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes Dev. 1994;8(1):105–117. doi: 10.1101/gad.8.1.105. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Frigola C, Carreres MI, Vegar C, Mason C, Herrera E. ZIC2 promotes axonal divergence at the optic chiasm midline by EphB1-dependent and -independent mechanisms. Development. 2008;135(10):1833–1841. doi: 10.1242/dev.020693. [DOI] [PubMed] [Google Scholar]
- 18.Chan DW, et al. ZIC2 synergistically enhances Hedgehog signalling through nuclear retention of Gli1 in cervical cancer cells. J Pathol. 2011;225(4):525–534. doi: 10.1002/path.2901. [DOI] [PubMed] [Google Scholar]
- 19.Marchini S, et al. The zinc finger gene ZIC2 has features of an oncogene and its overexpression correlates strongly with the clinical course of epithelial ovarian cancer. Clin Cancer Res. 2012;18(16):4313–4324. doi: 10.1158/1078-0432.CCR-12-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154(3):490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442(7098):86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 22.Ruthenburg AJ, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145(5):692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97(7):833–842. doi: 10.1016/S0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 24.Landry J, et al. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4(10): doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cherry CM, Matunis EL. Epigenetic regulation of stem cell maintenance in the Drosophila testis via the nucleosome-remodeling factor NURF. Cell Stem Cell. 2010;6(6):557–567. doi: 10.1016/j.stem.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song H, Spichiger-Haeusermann C, Basler K. The ISWI-containing NURF complex regulates the output of the canonical Wingless pathway. EMBO Rep. 2009;10(10):1140–1146. doi: 10.1038/embor.2009.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia P, et al. WASH is required for the differentiation commitment of hematopoietic stem cells in a c-Myc-dependent manner. J Exp Med. 2014;211(10):2119–2134. doi: 10.1084/jem.20140169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma S, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132(7):2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, et al. The long noncoding RNA lncTCF7 primes self-renewal of liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Wurmbach E, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 31.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoshida Y, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69(18):7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan G, Qin B, Liu N, Scholer HR, Pei D. Identification of a nuclear localization signal in OCT4 and generation of a dominant negative mutant by its ablation. J Biol Chem. 2004;279(35):37013–37020. doi: 10.1074/jbc.M405117200. [DOI] [PubMed] [Google Scholar]
- 34.Roessler S, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142(4):957–966 e912. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyault S, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45(1):42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 36.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 37.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 38.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15(9):1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 39.Ji J, Wang XW. Clinical implications of cancer stem cell biology in hepatocellular carcinoma. Semin Oncol. 2012;39(4):461–472. doi: 10.1053/j.seminoncol.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gidekel S, Pizov G, Bergman Y, Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4(5):361–370. doi: 10.1016/S1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 41.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121(3):465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 42.Chang CC, Shieh GS, Wu P, Lin CC, Shiau AL, Wu CL. Oct-3/4 expression reflects tumor progression and regulates motility of bladder cancer cells. Cancer Res. 2008;68(15):6281–6291. doi: 10.1158/0008-5472.CAN-08-0094. [DOI] [PubMed] [Google Scholar]
- 43.Chen YC, et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One. 2008;3(7): doi: 10.1371/journal.pone.0002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown SA, et al. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat Genet. 1998;20(2):180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]
- 45.Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13(10):727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- 46.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krasteva V, Buscarlet M, Diaz-Tellez A, Bernard MA, Crabtree GR, Lessard JA. The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood. 2012;120(24):4720–4732. doi: 10.1182/blood-2012-04-427047. [DOI] [PubMed] [Google Scholar]
- 48.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 49.Alkhatib SG, Landry JW. The nucleosome remodeling factor. FEBS Lett. 2011;585(20):3197–3207. doi: 10.1016/j.febslet.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: the R Foundation for Statistical Computing; 2011. [Google Scholar]
- 51.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia P, et al. RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res. 2014;24(8):943–958. doi: 10.1038/cr.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye B, et al. Pcid2 inactivates developmental genes in human and mouse embryonic stem cells to sustain their pluripotency by modulation of EID1 stability. Stem Cells. 2014;32(3):623–635. doi: 10.1002/stem.1580. [DOI] [PubMed] [Google Scholar]
- 54.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1–2):70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 55.Zhu P, et al. C8orf4 negatively regulates self-renewal of liver cancer stem cells via suppression of NOTCH2 signalling. Nat Commun. 2015;6:7122. doi: 10.1038/ncomms8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.