Abstract
Objectives:
Osteoporosis is a metabolic bone disease of reduced bone mass density (BMD) and elevated risk of fracture due to an imbalance in bone formation and resorption. The risk and incidence of osteoporosis increase towards advanced age, particularly in postmenopausal women, and the risk is known to be affected by the variation in the expression of the associated regulatory genes. This work aimed to clarify the impact of variation in RUNX2 (runt domain transcription factor 2), which is an osteoblast-specific transcription factor that normally stimulates bone formation and osteoblast differentiation, regarding single-nucleotide polymorphism within RUNX2 promoter (P1) and risk of osteoporosis in postmenopausal Indonesian women.
Methods:
Using DNA sampling from blood, the variation at the single-nucleotide polymorphism (-330, G→T, rs59983488) at the RUNX2 P1 promoter was investigated using polymerase chain reaction–restriction fragment length polymorphism for 180 consenting postmenopausal Indonesian women. The subjects were examined for bone mass density and classification to normal and those with osteopenia or osteoporosis by T-scoring with dual-energy X-ray absorptiometry. Chi-square testing and logistic regression were mainly used for statistical assessment.
Results:
The results showed a general trend with increased risk of osteoporosis associated with the genotype TT (mutant type) and the corresponding T allele of the tested polymorphism of RUNX2 promoter P1. The trend was, however, not significant in multivariate testing adjusted for age and time after menopause.
Conclusion:
To confirm the potential risk with TT genotype would require testing of a much larger sample of subjects. As the tested single-nucleotide polymorphism only represents one of the relevant candidate locations of RUNX2, the results are taken nevertheless to suggest an impact by overall RUNX2 variation in the risk of osteoporosis in Indonesian postmenopausal women.
Keywords: Menopause, osteoporosis, RUNX2, polymorphism
Introduction
Permanent cessation of menstruation at menopause due to reduced or lost ovarian activity and provision of the oestrogen hormone will lead in the long run to faster rate of bone resorption than formation and increasing risk of osteoporosis.1–3 This metabolic bone disease is marked by reduction in bone mass density (BMD), deterioration of bone microarchitecture and increased bone fragility. The incidence of osteoporosis increases with age and time after menopause, and at a typical level of about 15%−40% remains a major hazard for ageing postmenopausal women practically everywhere in the world.4–6
While a combination of environmental and genetic factors will affect bone strength, genetic ones are implicated by twin and family studies to account for more than 50% of the variance in BMD.6 The variation implies corresponding shifts in the network of regulatory genes responsible for the action balance between the osteoblastic bone formation and osteoclastic bone resorption. One of the genes involved in bone formation is RUNX2 residing at the chromosomal location 6p21, spanning 124.7 kb and containing two promoters P1 and P2, and seven exons. The promoters result in two expressed isoforms of RUNX2 (runt domain transcription factor 2), of which RUNX2-I driven by P2 contains 514 amino acid residues, and RUNX2-II driven by P1 contains 528 residues. Both isoforms are found in human osteoblast cells and osteoblast precursors and are important for osteoblast differentiation and bone remodelling. Several other genes encoding bone matrix require RUNX2 expression, including those expressing alkaline phosphatase, osteopontin, bone sialoprotein and COL1A1.5–7 Mice deficient of the transcription factor RUNX2 do not show complete bone formation, and heterozygous RUNX2 knockout mice show abnormalities characteristic of cleidocranial dysplasia (CCD).8 Some mutations in RUNX2 can disrupt the DNA-binding activity of the transcription factor, while others can have more subtle impact on the RUNX2 biological activity that is generally complex and apparently not fully clarified as yet.
Population studies of polymorphisms of RUNX2 have also been carried out and some show association with BMD in postmenopausal women.9–11 Polymorphism in the promoter 1 (P1) of RUNX2 at -330 (G→T, rs59983488) single-nucleotide polymorphism (SNP) position has not been often analysed, but the study by Napierala et al.8 suggests an association with human CCD disease. The SNP of P1 (-330) upstream of the promoter region at the enhancer elements may affect transcription by stimulating the expression of RUNX2 through protein attached to the elements. Note that this SNP location is specific to humans and does not exist in mice.
To intervene the processes leading to osteoporosis, the most common approach is to inhibit some essential steps to osteoclast formation or activation.4,5 However, another or parallel approach could be to promote osteoblasts and osteoblastic bone formation, and in this respect, the roles of RUNX2 could have potential therapeutic value. On the other hand, recent work points towards the involvement of RUNX2 also in osteoclast activation and therefore to more complex, as yet unknown, detailed roles of RUNX2 in the balance of bone formation and resorption.12 Considering the large impact of osteoporosis hazard in the society, it is, hence, of considerable interest to clarify the regulatory role of RUNX2 and its genetic variation in more detail. This study aimed to investigate the association of a selected polymorphism of RUNX2 promoter (P1) with BMD and risk of osteoporosis in postmenopausal Indonesian women.
Methods
Study subjects, BMD assessment and ethical approval
In total, 180 postmenopausal Indonesian women were included after obtaining written informed consent signed by each participant. Confirmation of the menopausal status was based on subject interviews and correlating other data. No cases of surgical menopause or subjects on drugs affecting bone mass were included. Calcaneous BMD was assessed using dual-energy X-ray absorptiometry (DXA), and the results were classified in a conventional manner using T-score, which is the number of standard deviations (SD) below the mean level of young adults. At values above −1, T-score is categorised to the normal range, between −1 and −2.5 to osteopenia, and below −2.5 to osteoporosis. On this basis, 31 of the subjects were diagnosed with osteoporosis, 112 with osteopenia and 37 as normal (healthy). The study and applied methods were approved by the ethical committee of the Faculty of Dentistry, University of Indonesia.
Sampling and isolation of DNA
Venous blood samples of 2 mL were taken from each subject. For DNA isolation, blood samples were inserted into Falcon tubes containing red blood cell anticoagulant (1:3), and the tube was repeatedly inverted for 10 min. The tubes were then centrifuged for 15 min at 1500 rpm at room temperature, supernatant was discarded, and this was repeated 3 times. After that 2 mL of cell lysis solution (CLS) was added to the pellet and the solution in the up-down with the transfer pipette, and incubated in a water bath at 37°C for 60 min. Protein precipitation (PP) solution of 1.3 mL was added into the tube, vortexed and centrifuged at 40°C/3000 rpm for 5 min. The supernatant was poured into a new tube containing 2.3 mL of cold isopropanol, and the tube was inverted repeatedly until DNA in it appeared white. Then, 1.3 mL of cold 70% ethanol was added for DNA precipitation, removed and dried by inverting the tube. Washing was repeated two times. Furthermore, the tube was dried in tilted position. Then, 0.3 mL of Tris-EDTA buffer (TE) was added (adjusted by the amount of DNA) and incubated in a water bath at 37°C for 2 h. Finally, the solution was transferred into Eppendorf tubes with a micropipette (300 µL) and stored at −20°C.
Polymerase chain reaction amplification, restriction fragment length polymorphism and DNA sequencing
To assess the selected polymorphism in the RUNX2 promoter (P1), polymerase chain reaction (PCR) amplification was performed using a primer pair that includes the promoter P1. At position -330, forward primer 5′-AAA GCA AAG GAG GTT GAC CGG-3′ and reverse primer 5′-CCC TGC CCT TCT TTC TCT CTC-3′were used. PCR amplification was conducted in Elmer GeneAmp PCR System 9700. The 25 µL volume of each reagent consisted of 5 µL of genomic DNA and buffer solution containing 0.2 µM of each deoxyribonucleotide triphosphate (dNTP), 0.4 µM of each primer, 0.7U Taq polymerase (Promega), 1.5 mM MgCl2 and ddH2O. The PCR amplification included 35 cycles with an initial pre-denaturation at 94°C for 6 min, then the first cycle of denaturation at 94°C for 1 min, annealing at 62°C for 30 s and elongation at 72°C for 30 s. The final extension was done at 72°C for 5 min. The amplified fragments were separated by electrophoresis on 1.5% agarose gel (Promega) containing ethidium bromide 0.5 mg/mL in 1X Tris–acetate–EDTA (TAE) buffer solution (0.04 M Tris–acetate, 0.002 M ethylenediaminetetraacetic acid (EDTA), pH 8.0), applying 80V for 40 min. The length of the amplified target fragment is 225 bp.
Restriction fragment length polymorphism (RFLP) analysis was performed by inserting the restriction enzyme BsaJI (New England Biolabs (NEB), 1 µL, 10U/µL) into a tube containing the amplified DNA fragments, 2 µL RE10X buffer solution and 18 µL ddH2O. The mixture was incubated in a water bath at 60°C for 4 h, then inactivated by incubating at 80°C for 20 min. DNA fragments were subjected to electrophoresis on 3% agarose gel in 1X TAE at 90V for 60 min. In case of GG genotype (wild), cutting with the restriction enzyme resulted in two DNA bands of 205 bp and 20 bp in size. At the cutting site, mutant type will change from G to T, and GT (heterozygous) type is represented by three bands of 225 bp, 205 bp and 20 bp, and TT type is represented by one 225-bp band. DNA sequencing was used to confirm the position of nucleotide base changes by the cutting enzyme. DNA sequencing was performed by 1st BASE in Singapore to confirm polymorphic sites, using the PCR amplicons. The sequencing procedure involved PCR product purification, sequencing, precipitation, re-cycle sequencing and sequence analysis of the nucleotide bases.
Statistical analysis
To compare T-scores and grouping to normal subjects and those with osteopenia and osteoporosis according to genotypes and allotypes of the tested polymorphism of RUNX2, chi-square or Kruskal–Wallis tests were used for statistical analysis. Logistic regression was applied to assess the association of polymorphism and risk of osteoporosis. Statistical significance was assumed at p < 0.05.
Results
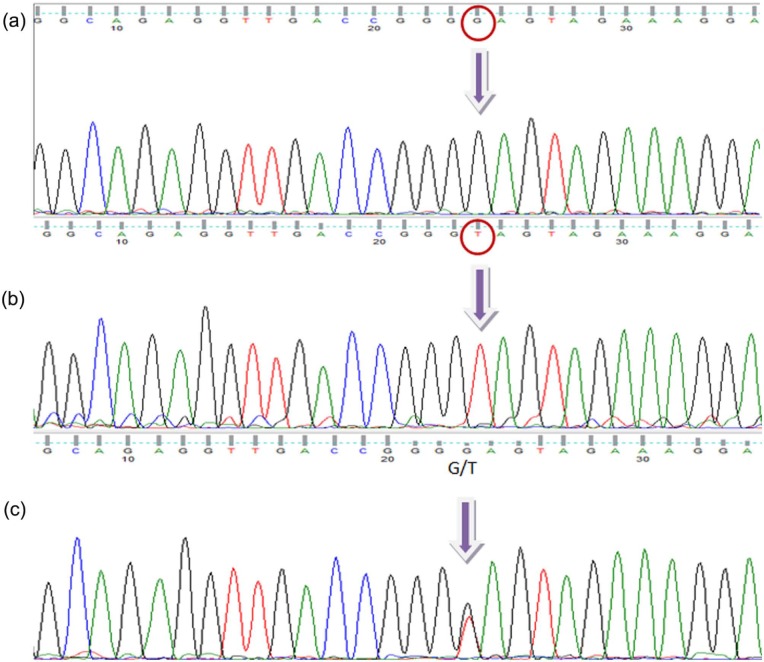
PCR-amplified DNA samples of the targeted RUNX2 P1 promoter region (-330) was shown to provide in electrophoresis the expected single band 225 bp in size (Figure 1(a)). After using the cutting enzyme BsaJI for polymorphism status, the resulting variants were GG genotype (wild type) with two bands, GT genotype (heterozygous) with three bands, and TT genotype (mutant) with a single band (Figure 1(b); note that in electrophoresis, the smallest fragments of 20 bp in size migrated out and are not shown). Example results of genotype sequencing for wild-type, heterozygous and mutant samples are shown in Figure 2. The mean T-scores and corresponding frequencies of normal subjects and those with osteopenia and osteoporosis according to genotype and allotype of the tested RUNX2 gene promoter polymorphism are shown in Table 1. The T-scores were similar for the GG and GT genotypes but significantly reduced for the TT genotype. From normal subjects to those with osteopenia and osteoporosis, there is a general trend of decreasing percentage in case of GG and GT genotypes, and a corresponding but stronger increase for the genotype TT. This was also reflected in the decrease in the percentages for allotype (allele) G and increase in the percentages for allotype (allele) T, as seen in Table 1.
Figure 1.
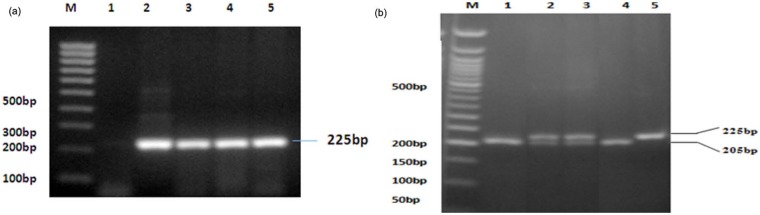
Results of PCR-RFLP: (a) PCR-amplified target fragment − M = 100-bp marker ladder; well 1 = negative control; 2 = positive control (GADPH primer); 3–5 = samples with 225-bp PCR product and (b) fragments after BsaJI enzyme cutting − M = 50-bp marker ladder, wells 1 and 4 = GG; 2–3 = GT; 5 = TT.
PCR-RFLP: polymerase chain reaction–restriction fragment length polymorphism; PCR: polymerase chain reaction; GADPH: glyceraldehyde 3-phosphate dehydrogenase.
Figure 2.
The results of sequencing: (a) = homozygous wild type (GG); (b) = homozygous mutant (TT); (c) = heterozygous (GT).
Table 1.
Frequency of genotypes and allotypes related to T-scores and grouping to normal subjects and those with osteopenia or osteoporosis; the p-values of 0.245 and 0.094 refer to comparison between normal and osteopenia/osteoporosis groups.
| Genotype |
Allele |
||||||
|---|---|---|---|---|---|---|---|
| GG | GT | TT | p | G | T | p | |
| Total N (%) | 110 (61.1) | 42 (23.3) | 28 (15.6) | 262 (72.8) | 98 (27.2) | <0.001a | |
| Age, mean (SD) | 53.2 (5.1) | 53.4 (4.3) | 54.3 (4.7) | – | – | – | |
| T-score, mean (SD) | −1.42 (0.99) | −1.38 (0.80) | −1.85 (0.74) | 0.007 | – | – | – |
| Normal N (%) | 25 (67.6) | 10 (27.0) | 2 (5.4) | 60 (81.1) | 14 (18.9) | ||
| Osteopenia N (%) | 68 (60.7) | 26 (23.2) | 18 (16.1) | 0.245 | 162 (72.3) | 62 (27.7) | 0.094 |
| Osteoporosis N (%) | 17 (54.8) | 6 (19.4) | 8 (25.8) | 40 (64.5) | 22 (35.5) | ||
SD: standard deviation; HWE: Hardy-Weinberg equilibrium.
Regarding HWE.
Logarithmic regression for comparison of the genotypes in the case of combined osteoporosis/osteopenia group gave a slightly elevated odds ratio (OR) of 2.39 (95% CI: 0.73–7.87) for the TT genotype in comparison with the wild type (GG), which is not statistically significant when the data are adjusted for age and time after menopause (p = 0.147).
Discussion
The genotype/allotype frequency distributions in this work deviated significantly from the Hardy–Weinberg equilibrium (p < 0.001). The reasons are not clear but disequilibrium appears not to be exceptional for RUNX2 polymorphisms.10 The mean T-scores of the GG and GT genotypes of the tested polymorphism were similar, but the fraction of subjects representing the GG genotype were about twice as common as those of GT genotype in all groups from normal (healthy) to osteoporotic (Table 1). In contrast, the least common TT genotype showed a significantly reduced level of mean T-score (p = 0.007). In case of GG and GT genotypes, there was a general trend of lower fraction of the subjects appearing in the osteoporosis group than in the group with normal range of BMD. In case of TT genotype, there was a corresponding, but more pronounced, increase of the fraction of subjects in the osteoporosis group. As expected, such a systematic trend was also reflected in the corresponding differences for allotype (allele) G and allotype (allele) T. The indicated risk of osteoporosis as OR appears to be elevated for genotype TT, but this is not statistically significant when adjusted for age and time after menopause. The sample was relatively small, comprising of 180 subjects in total, with 28 (15.6%) representing the TT genotype and only 8 of these with osteoporosis. However, as all observed trends are systematic from normal healthy subjects through osteopenia to osteoporosis, it is thought likely that in this case significance can be demonstrated by increasing the sample size. Indeed, several other studies have used significantly higher number of subjects, albeit partly because of lower frequencies of mutant genotypes.9–11 However, the results of this work also clearly indicate that multiple causes must be involved in the observed variation of BMD. This is not surprising as even for RUNX2, several different polymorphisms have been suggested to partially explain the observed variation in the genetic roots of osteoporosis (Table 2), and multiple other genes are known to influence the balance of osteoblastic bone formation and osteoclastic resorption.6–11
Table 2.
| Polymorphism | Promoter | Subjects (N) | Frequency | Reference |
|---|---|---|---|---|
| −336 G→A | P1 | Scottish (988 F) | AA 0.5%, GG 75.6% | Vaughan et al.11 |
| −1176 T→C | P1 | American, Canadian, European | Only with CCD | Napierala et al.8 |
| −1048 A→C | 100 with CCD | 1/180 (0.6%) | ||
| −334 C→A | (M and F) | Only with CCD | ||
| −330 G→T | 15/180 (8.3%) | |||
| −1025 T→C | P2 | Korean (729 F) | CC 1.2%, TT 83.7% | Lee et al.9 |
| −1492 A→T | P2 | TT 0%, AA 99.9% | ||
| −1025 T→C | P2 | Spanish (776 F) | CC 0.9%, TT 88.3% | Pineda et al.10 |
| −330 G→T | P1 | Indonesian (180 F) | TT 15.6%, GG 61.1% | This work |
M: male; F: female.
Many of the polymorphisms of RUNX2 studied in the previous investigations have represented relatively rare mutants that as such cannot alone reflect any major fraction of osteoporotic cases. In contrast, the polymorphism (-330 G→T) of the P1 region of RUNX2 is relatively common, with TT genotype indicated for 15.6% of the subjects in this work. As this mutant is common, it is, on the other hand, unlikely that it would confer very serious impact on the processes of bone formation. In agreement with this view, it has been reported that this polymorphism has no effect on the promoter activity in vitro.11
As osteoporosis in postmenopausal women is common (affecting 17.2% of the subjects on average in this work but more with advancing age) and at least by 50% explained by genetic factors, variation in individual genes shown by SNPs are unlikely to account for very large fraction of the total variation in BMD. As seen above, even for genes like RUNX2 that certainly can carry major influence on the bone homeostasis, the variation associated with individual SNPs is modest and at best can only explain a small part of the risk of osteoporosis. A more holistic view may be sought from genome-wide SNP analysis, suggesting that most of the genetic impact arises from chromosomes 4, 5, 7, 9, 11, 16 and 22, rather than from chromosome 6, where RUNX2 is residing.12
However, expression of individual regulatory genes can serve as a very valuable therapeutic target, as has been shown, for example, in experiments for receptor activator of nuclear factor kappa-B ligand (RANKL) inhibition against osteoporosis.5,6 In the case of RANKL, the emphasis is on reduction in osteoclastic bone resorption, but another potentially useful option is to promote the counteracting process of osteoblastic bone formation through activation of corresponding genes such as RUNX2.13–22 In addition, RUNX2 has been shown to be involved also in the regulation of the osteoclastic side of the balance, and this could lend further weight on future development of new agents or targets for this purpose. For this purpose, further work is needed to clarify the detailed regulatory mechanisms.
Conclusion
The results showed a general trend with increased risk of osteoporosis associated with the genotype TT (mutant type) and the corresponding T allele of the tested polymorphism (-330, G→T, rs59983488) of RUNX2 promoter P1 region in postmenopausal Indonesian women. The trend was however not significant in multivariate testing adjusted for age and time after menopause. In spite of the known essential role of RUNX2 in activating and mediating osteoblastic bone formation, the variation corresponding to the tested relatively common polymorphism can only explain a small fraction of the genetic risk of osteoporosis. RUNX2 activity may nevertheless provide targets or model sites to promote bone formation and combat osteoporosis.
Acknowledgments
The authors wish to acknowledge the skilful technical support by L. Yunaini.
Footnotes
Declaration of conflicting interests: The authors declare that there is no conflict of interest.
Funding: The authors acknowledge the financial support by the Directorate General of Higher Education, Ministry of Education and Culture of The Republic of Indonesia through Directorate Research and Public Services, University of Indonesia.
References
- 1. Nelson HD. Menopause. Lancet 2008; 371: 760–770. [DOI] [PubMed] [Google Scholar]
- 2. Raisz LG. Pathogenesis of osteoporosis: concept, conflict and prospects. J Clin Invest 2005; 115: 3318–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Duncan EL, Brown MA. Genetic studies in osteoporosis – the end of the beginning. Arthritis Res Ther 2008; 10: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet 2011; 377: 1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luhmann T, Germershaus O, Groll J, et al. Bone targeting for the treatment of osteoporosis. J Control Release 2012; 161: 198–213. [DOI] [PubMed] [Google Scholar]
- 6. Ralston SH, De Crombrugghe B. Genetic regulation of bone mass and susceptibility to osteoporosis. Genes Dev 2006; 2006(20): 2492–2506. [DOI] [PubMed] [Google Scholar]
- 7. Bae S-C, Lee YH. Phosphorylation, acetylation and ubiquitination: the molecular basis of RUNX regulation. Gene 2006; 366: 58–66. [DOI] [PubMed] [Google Scholar]
- 8. Napierala D, Garcia-Rojas X, Sam K, et al. Mutations and promoter SNPs in RUNX2, a transcriptional regulator of bone formation. Mol Genet Metab 2005; 86: 257–268. [DOI] [PubMed] [Google Scholar]
- 9. Lee H-J, Koh J-M, Hwang J-Y, et al. Association of a RUNX2 promoter polymorphism with bone mineral density in postmenopausal Korean women. Calcif Tissue Int 2009; 84: 439–445. [DOI] [PubMed] [Google Scholar]
- 10. Pineda B, Hermenegildo C, Laporta P, et al. Common polymorphisms rather than rare genetic variants of the Runx2 gene are associated with femoral neck BMD in Spanish women. J Bone Miner Metab 2010; 28: 696–705. [DOI] [PubMed] [Google Scholar]
- 11. Vaughan T, Reid DM, Morrison NA, et al. RUNX2 alleles associated with BMD in Scottish women: interaction of RUNX2 alleles with menopausal status and body mass index. Bone 2004; 34: 1029–1036. [DOI] [PubMed] [Google Scholar]
- 12. Karasik D, Dupuis J, Cho K, et al. Refined QTLs of osteoporosis-related traits by linkage analysis with genome-wide SNPs: Framingham SHARe. Bone 2010; 46: 1114–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baniwal SK, Shah PK, Shi Y, et al. Runx2 promotes both osteoblastogenesis and novel osteoclastogenic signals in ST2 mesenchymal progenitor cells. Osteoporos Int 2012; 23: 1399–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doecke JD, Day CJ, Stephens AS, et al. Association of functionally different RUNX2 P2 promoter alleles with BMD. J Bone Miner Res 2006; 2006(21): 265–273. [DOI] [PubMed] [Google Scholar]
- 15. Ermakov S, Malkin I, Keter M, et al. Family-based association study of polymorphisms in the RUNX2 locus with hand bone length and hand BMD. Ann Hum Genet 2008; 72: 510–518. [DOI] [PubMed] [Google Scholar]
- 16. Alcantara EH, Shin M-Y, Sohn H-Y, et al. Diosgenin stimulates osteogenic activity by increasing bone matrix protein synthesis and bone-specific transcription factor Runx2 in osteoblastic MC3T3-E1 cells. J Nutr Biochem 2011; 22: 1055–1063. [DOI] [PubMed] [Google Scholar]
- 17. Centrella M, McCarthy TL. Estrogen receptor dependent gene expression by osteoblasts – direct, indirect, circumspect and speculative effects. Steroids 2012; 77: 174–184. [DOI] [PubMed] [Google Scholar]
- 18. Elefteriou F, Yang X. Genetic mouse models for bone studies – strengths and limitations. Bone 2011; 49: 1242–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gallet M, Vanacker J-M. ERR receptor as potential targets in osteoporosis. Trends Endocrinol Metab 2010; 21: 637–641. [DOI] [PubMed] [Google Scholar]
- 20. Hie M, Iitsuka N, Otsuka T, et al. Zinc deficiency decreases osteoblasts and osteoclasts associated with the reduced expression of Runx2 and RANK. Bone 2011; 49: 1152–1159. [DOI] [PubMed] [Google Scholar]
- 21. Jang WG, Kim EJ, Bae I-H, et al. Metformin induces osteoblast differentiation via orphan nuclear receptor SHP-mediated transactivation of Runx2. Bone 2011; 48: 885–893. [DOI] [PubMed] [Google Scholar]
- 22. Lee C-H, Huang Y-L, Liao J-F, et al. Ugonin K promotes osteoblastic differentiation and mineralization by activation of p38 MAPK- and ERK-mediated expression of Runx2 and osterix. Eur J Pharmacol 2011; 668: 383–389. [DOI] [PubMed] [Google Scholar]