Abstract
Objectives:
Leptospirosis is one of the most widespread zoonotic infectious diseases affecting humans and animals. Several animal species, including cattle, can act as potential asymptomatic carriers facilitating zoonotic transmission of Leptospira. This study was conducted to assess the occurrence of asymptomatic renal Leptospira carriers among cattle slaughtered in southeastern Georgia, United States.
Methods:
A battery of diagnostic tests, including dark field microscopy, direct fluorescent antibody staining, polymerase chain reaction, and culture, were performed on a set of bovine kidneys (n = 37) collected from an abattoir in southeastern Georgia, United States. Virulence of a field isolate obtained from this study was tested in a hamster experimental model.
Results:
Motile spirochete-like structures were observed by dark field microscopy in 23 (59%) out of 37 kidney samples tested. In all, 29 samples (78%) were positive by direct fluorescent antibody staining. Only 11 (29.7%) samples by polymerase chain reaction and 3 (8.1%) by culture were positive for Leptospira sp. The isolates obtained by culture were confirmed as Leptospira borgpetersenii. Hamsters experimentally infected with one of the Leptospira field isolates obtained from this study did not show clinical signs but developed renal infection with interstitial nephritis and tubular necrosis.
Conclusions:
This study confirms that asymptomatic Leptospira renal infection is present among cattle in the region. Our findings underscore the need for future studies to assess the potential environmental contamination and transmission to humans in contact with infected cattle.
Keywords: Leptospira, cattle, diagnosis, public health
Introduction
Leptospirosis is a reemerging zoonotic disease affecting both humans and animals and is caused by various serovars of Leptospira.1 Asymptomatic renal infection occurs in many domestic and wild animal species. Exposure to urine from infected animals and the contaminated environment results in infection in susceptible humans and animals leading to subclinical mild illness to serious life-threatening disease. Severe and untreated infection may result in medically expensive complications due to renal failure, hepatic disease, pulmonary hemorrhage, and mortality.1–3 Diagnosis of clinical leptospirosis can be challenging due to its protean manifestations resembling other febrile diseases such as influenza, dengue fever, and rickettsial infections.
The incidence of leptospirosis in humans is increasing globally.2,3 Although the disease was initially recognized as an occupational disease, it is now considered an important reemerging zoonotic disease affecting a broader category of the population. Growth and spread of human populations, deforestation and urbanization, global climate change, ecotourism, and the increase in wildlife and pet trade might favor the increased incidence of the disease.3–5 In the United States, most of the recent outbreaks were associated with water-related recreational events.5–7 Abundance of animal reservoirs such as cattle and an ecosystem favoring the maintenance of organisms in the environment will likely increase the exposure and infection rate in susceptible hosts.
Leptospirosis was first reported in cattle in United States in 1946 as outbreaks of severe febrile illness.8 It is an economically significant disease in terms of cattle production, as the main manifestations include abortions, stillbirth, infertility, and loss of milk production.9–11Leptospira borgpetersenii (L. borgpetersenii) serovar Hardjo is the most common host-adapted Leptospira serovar of cattle in North America.9,10 Many recent studies on prevalence of Leptospira infection in cattle are solely based on the seroprevalence.9–11 Abattoir-based studies on Leptospira renal infection date back to the 1990s.12,13 Serological evaluations do not provide accurate estimates of active infection or carrier status. Therefore, a preliminary study was conducted to evaluate the presence of asymptomatic Leptospira renal infection in cattle using kidney samples obtained from a local abattoir using a combination of diagnostic methods.
Materials and methods
Fresh kidneys were collected from cattle slaughtered in a local abattoir. Only 1–2 fresh bovine kidney samples were obtained at a time and processed within 2–4 h of collection in order to improve the detection and isolation rate. Cattle slaughtered in this abattoir came from small independent farms located in southeastern counties in the state of Georgia and from northern Florida. Data on sex, age, and county information were recorded when available. Of the 37 kidney samples, 24 (64.9%) were collected from steers, 5 (13.5%) were collected from bulls, 3 (8.1%) were collected from heifers, and the sex of 5 (13.5%) animals was unknown. In all, 21 (56.8%) cattle were <30 months old, 6 (16.2%) were 30 months or older, and the age of 10 (27.0%) animals was unknown. With respect to the breed, 6 (16.2%) animals were Angus, 1 (2.7%) was Holstein, 1 (2.7%) was Jersey, 1 (2.7%) was a Texas Longhorn, and the breed of 28 (75.7%) animals was unknown. The samples were tested by dark field microscopy (DFM), direct fluorescent antibody staining (DFA), polymerase chain reaction (PCR), and culture. Out of the 37 samples, 3 samples were not examined by DFM and 2 samples were not examined by DFA.
Approximately a 2-inch wedge-shaped sections of kidney encompassing portions of cortex and medulla were processed using aseptic techniques. After removing the capsule, the edges were trimmed and the samples were lightly seared on the surface. Samples were cut into small pieces in a sterile petri dish using a sterile scalpel blade and were mixed with 10 mL of Ellinghausen-McCullough-Johnson-Harris (EMJH) liquid media (Becton and Dickinson Microbiology Systems, Sparks, MD). The homogenate was allowed to settle for 2–3 m. Supernatant of 15 µL was placed on a glass slide under a coverslip and immediately examined by DFM. Samples containing organisms with morphology and motility compatible with Leptospira were recorded as positive. A total of 15 µL of the sample was placed on circles of a glass slide and processed for DFA as described previously.14 Briefly, smears were allowed to air-dry, fixed in chilled acetone for 15 m and treated with anti-Leptospira polyclonal antibody conjugated with fluorescein isothiocyanate (National Veterinary Services Laboratory, Ames, IA) for 1.5 h at 37 °C. The slides were washed three times in phosphate buffered saline (PBS) and air-dried. The slides were examined under 40 × objective of a fluorescent microscope after applying the mounting fluid and a coverslip. The samples with typical morphology of Leptospira and positive fluorescence were recorded as positive.
Culture was performed as described previously with modifications.15 The kidney samples were homogenized as described above and 500 µL of the supernatant from the kidney homogenate was added to 9.5 mL P80-BA liquid oleic media.(National Veterinary Services Laboratory, Ames, IA).. Three serial 10-fold dilutions (10−2, 10−3, and 10−4) were prepared, and 100 µL from the 10−2 dilution was inoculated into two semisolid P-80-BA liquid oleic media tubes with 5-fluorouracil and two without 5-fluorouracil.The tubes were incubated at 29 °C up to 6 months. The samples were examined for the presence of Leptospira at 2-week intervals by DFM. An aliquot of the homogenate was stored in a −70 °C freezer for PCR. DNA was extracted in an automated DNA extraction unit (BioSprint-96, Qiagen Inc, Valencia, CA) using MagMAXTM total nucleic acid isolation kit (Applied Biosystems, Foster City, CA) using manufacturer’s recommendations. Real-time PCR was conducted using a previously described protocol.16
Leptospira isolates obtained by culture were subcultured into semisolid and liquid P-80-BA liquid oleic media. DNA was isolated using QIAamp DNA Mini Kit (QIAGEN Inc, Valencia, CA). In order to further characterize the isolates, PCR was performed targeting Lip L32 (present in all pathogenic Leptospira spp.), IS1500 (specific for L. interrogans), and IS1533(specific for L. borgpetersenii) gene sequences.17–19
To evaluate the virulence of the Leptospira isolates, a preliminary experimental inoculation study was conducted in hamsters using one of the field isolates as described previously.20 Animals were kept following institutional animal care and protocols approved by the University of Georgia’s Institutional Animal Care and Use Committee (IACUC). Four hamsters were inoculated intraperitoneally with 1 mL of Leptospira isolate (BK6) containing 1 × 107organisms. One control hamster was inoculated with P-80-BA liquid oleic media.The hamsters were observed twice daily for the development of clinical signs. After 28 days postinoculation, the hamsters were humanely euthanized, necropsy was conducted, and the organs were collected in 10% neutral buffered formalin and processed for routine histopathology using standard protocols. The kidney samples were cultured on P-80-BA liquid oleic media. Microscopic agglutination testing (MAT) on the hamster serum was performed against six Leptospira serovars.
Results
Motile spirochete-like structures were observed by DFM in 23 out of 34 (59%) kidney samples examined. In all, 29 (78%) samples out of 35 tested were positive by DFA. Of 37 kidney samples tested, 11 (29.7%) samples were positive by PCR and 3 (8.1%) samples were positive by culture. A summary of the results for each test is shown in Table 1. Two Leptospira isolates were obtained from steers under 30 months of age, and the age and sex of the animal that contributed to the third isolate were unavailable. The origin of one isolate is unknown. The cycle threshold (Ct) values for 11 real-time PCR positive samples ranged from 31.63 to 39.64 with an average Ct value of 35.20. The Leptospira isolates obtained were also positive for the screening PCR used in the study. PCR for the Lip L32 gene, which is present in all pathogenic Leptospira, was positive for all three isolates (Figure 1(a)). PCR for insertion sequences IS1500 and IS1533 was performed to differentiate L. borgpetersenii and L. interrogans. All three isolates were positive for IS533 (Figure 1(b)) and negative for IS1500 (Figure 1(c)) confirming the isolates as L. borgpetersenii.
Table 1.
Summary of Leptospira testing results from 37 bovine kidney samples obtained from a local abattoir in southeastern Georgia, United States.
| Test | Positive | Negative | Not evaluated | Total |
|---|---|---|---|---|
| DFM | 23 (62.2%) | 11 (29.7%) | 3 (8.1%) | 37 (100%) |
| DFA | 30 (81.1%) | 5 (13.5%) | 2 (5.4%) | 37 (100%) |
| PCR | 11 (29.7%) | 26 (70.3%) | 0 (0.0%) | 37 (100%) |
| Culture | 3 (8.1%) | 34 (91.9%) | 0 (0.0%) | 37 (100%) |
DFM: dark field microscopy; DFA: direct fluorescent antibody test; PCR: polymerase chain reaction.
Figure 1.
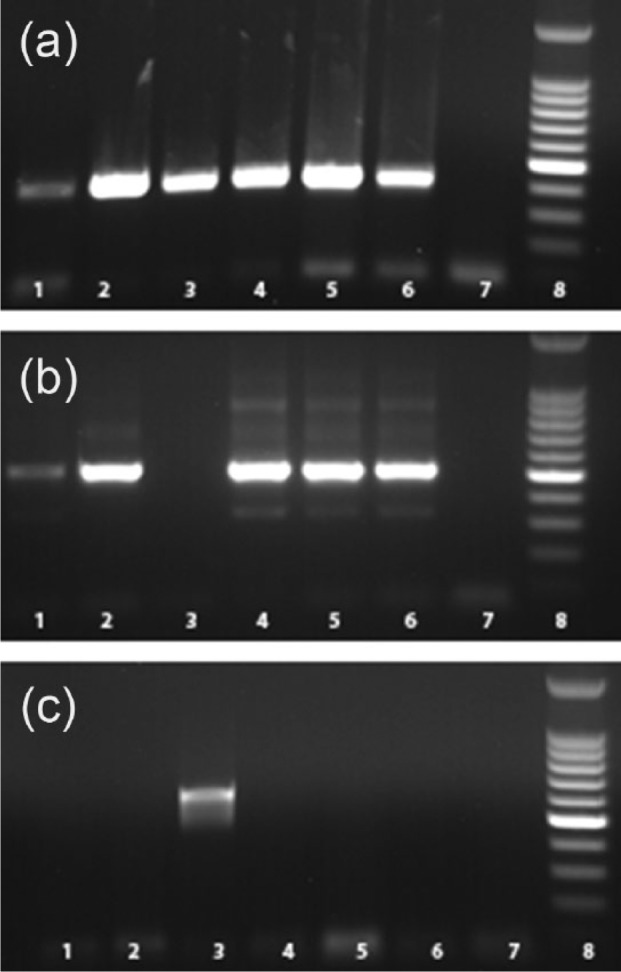
Polymerase chain reaction using field isolates: (a)Lip L32 gene, (b) IS1533 gene, (c) IS1500 gene. Lane 1: L. borgpetersenii serovar Hardjo type A (NVSL S 1343); Lane 2: L. borgpetersenii serovar Hardjo type B (NVSL S 818); Lane 3: L. interrogans serovar Hardjo; Lane 4: field isolate BK6; Lane 5: field isolate BK9; Lane 6: field isolate BK30; Lane 7: field isolate BK6; Lane 8: negative control; Lane 9: molecular weight markers.
NVSL: National Veterinary Services Laboratory.
Experimental inoculation of hamsters with one of the isolates did not induce any clinical signs. The hamsters remained asymptomatic during the entire duration of the study. No gross lesions were observed at necropsy 28 days postinoculation. Histopathological examination revealed multifocal renal tubular ectasia, necrosis, and mild interstitial nephritis in all inoculated hamsters (Figures 2(a) and (b)). The control hamster did not develop any renal lesions. The inoculated hamsters developed an antibody response (MAT = 1/400) only to the serovar Hardjo among the six routinely tested serovars in our laboratory. The Leptospira organisms were recovered from kidneys of all infected hamsters by culture.
Figure 2.
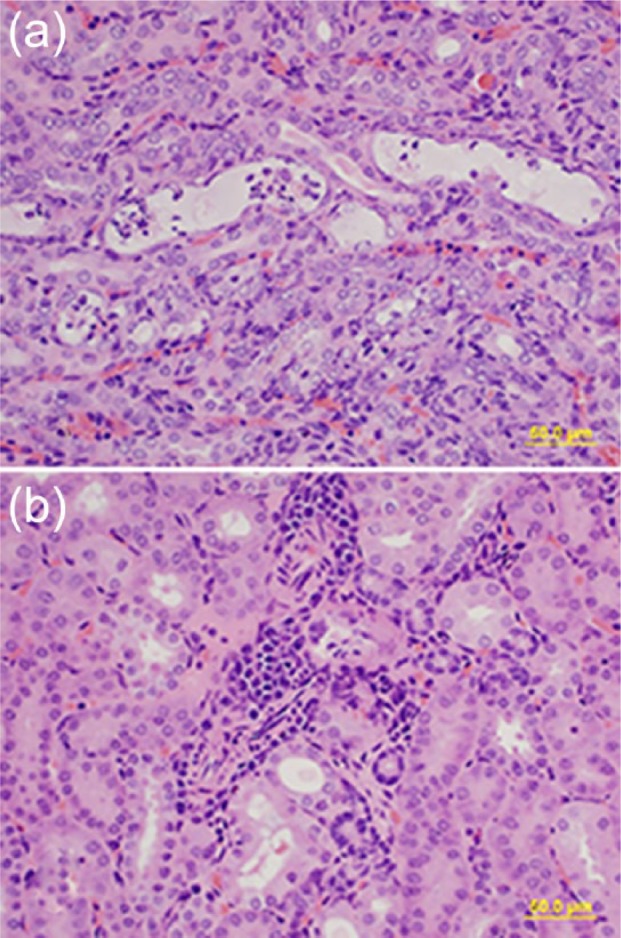
Representative hematoxylin and eosin stained sections of hamster kidney infected with field isolate BK6, 400× magnifications. Note (a) the tubular ectasia and necrosis and (b) interstitial nephritis, bar = 100 µm
Discussion
This study emphasizes that Leptospira borgpetersenii serovar Hardjo infection is prevalent in cattle in the study area. A combination of diagnostic tests was applied to the kidney samples in this study to improve the sensitivity of detection. We observed more samples positive by DFA and DFM than culture and PCR combined. It is important to note that the DFM and DFA are not specific for the detection of the pathogenic Leptospira, and the possibility of false-positive results cannot be excluded.
Three L. borgpetersenii serovar Hardjo isolates were obtained from this study. Leptospira culture is not generally attempted in diagnostic laboratories due to its laborious nature, long periods of incubation, and contamination with other fast-growing bacteria. L. borgpetersenii serovar Hardjo is a very slow-growing and hard to maintain species, and special media (PA-80-BA liquid oleic media) were needed to grow and maintain cultures. The diagnostic complexity due to the presence of large number of serovars and animal reservoirs emphasizes the need of culture to obtain Leptospira isolates for future epidemiologic evaluations and strategic implementation of preventive measures. L. borgpetersenii serovar Hardjo types A and B are reported in the North American cattle population.21,22Leptospira isolates belonging to serovars Pomona and Grippotyphosa have also been isolated from cattle.12,13 In a nationwide abattoir-based study conducted in 1990, the majority of the isolates were L. borgpetersenii serovar Hardjo type A based on restriction endonuclease analysis.12,13 Most of the type B L. borgpetersenii serovar Hardjo isolates from cattle were isolated from the southeastern United States, while type A did not have any particular pattern of distribution. In a recent study which compared virulence of L. borgpetersenii serovar Hardjo A and B types in hamsters, the type A isolate established chronic renal colonization, whereas the type B isolate induced severe debilitating illness.23 The field isolate tested in our study resulted in renal infection in hamsters without inducing clinical signs as described with the type A isolates. Characterization of these L. borgpetersenii serovar Hardjo isolates is ongoing. Genomic sequencing data have suggested that this L. borgpetersenii serovar Hardjo has undergone a considerable genome reduction resulting in reduced indirect transmission potential.24 However, it is important to note that L. borgpetersenii serovar Hardjo was the most commonly diagnosed serovar from notified cases of human leptospirosis in New Zealand.25,26
Leptospirosis is an important occupational disease in New Zealand among abattoir workers.27 Elimination or reduction of L. borgpetersenii serovar Hardjo infection in cattle populations is a desirable goal due to its economic and public health impact, but currently available vaccines do not provide sterile immunity or prevent establishment of renal carriers.23 Many aspects of cattle L. borgpetersenii serovar Hardjo infection, including the establishment of infection, variation in disease patterns, immune response generated, the mode of transmission and maintenance of infection, and the impact of various strains in relation to ecology and management systems remain largely unknown. In a previous study, we conducted in dairy cattle in this region, 7 out of 10 dairy herds in the region tested positive for Leptospira in urine by DFA, and all the herds tested had at least one cow with antibody titers (by MAT) ≥100 for one or more Leptospira serovars.14 Life-threatening disease and mortality due to leptospirosis in dogs are reported in this region.28 Moreover, kidneys of multiple wild animal species (bobcats, coyotes, and opossums) tested were PCR positive for Leptospira indicating widespread distribution of animal reservoirs (S. Rajeev, unpublished data).
Our study indicates an endemic distribution of L. borgpetersenii serovar Hardjo in cattle populations in the region. Most of Georgia, especially the southeastern region has a subtropical, humid climate with moderate to heavy precipitation rate promoting an environmental maintenance of Leptospira. Additionally, there are numerous water bodies that are frequently used for recreation and irrigation. In humans, leptospirosis is a treatable illness if diagnosed early; however, accurate diagnosis is challenging due to its confounding clinical presentations, lack of awareness and clinical suspicion among patients and physicians, and limited diagnostic expertise at the point of care. As the majority of Leptospira cases are mild and may get misdiagnosed as febrile illness such as influenza, it is likely that its incidence in humans is underreported. Considering the abundance of animal reservoirs as shown in our data, changing climatic conditions of the region and the overall global trends in human leptospirosis incidence, studies to explore the distribution and extent of human exposure to Leptospira spp. in the southeastern United States are warranted.
Acknowledgments
We would like to thank Karina Sorensen, Jill Johnson, Lisa Whittington, and other technical staff of bacteriology, serology, and histology sections of Tifton Veterinary Diagnostic and Investigational Laboratory for their valuable technical support.
Footnotes
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: The authors wish to thank the Southeastern Milk Check Off for funding the project.
References
- 1. Levett PN. Leptospirosis. Clin Microbiol Rev 2001; 14: 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McBride AJ, Athanazio DA, Reis MG, et al. Leptospirosis. Curr Opin Infect Dis 2005; 18: 376–386. [DOI] [PubMed] [Google Scholar]
- 3. Guerra MA. Leptospirosis: public health perspectives. Biologicals 2013; 41: 295–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vinetz JM, Glass GE, Flexner CE, et al. Sporadic urban leptospirosis. Ann Intern Med 1996; 125: 794–798. [DOI] [PubMed] [Google Scholar]
- 5. Morgan J, Bornstein SL, Karpati AM, et al. Outbreak of leptospirosis among triathlon participants and community residents in Springfield, Illinois, 1998. Clin Infect Dis 2002; 34: 1593–1599. [DOI] [PubMed] [Google Scholar]
- 6. Stern EJ, Galloway R, Shadomy SV, et al. Outbreak of leptospirosis among Adventure Race participants in Florida, 2005. Clin Infect Dis 2010; 50: 843–849. [DOI] [PubMed] [Google Scholar]
- 7. Meites E, Jay MT, Deresinski S, et al. Reemerging leptospirosis, California. Emerg Infect Dis 2004; 10: 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mathews FP. A contagious disease of cattle associated with Leptospira. Am J Vet Res 1946; 7: 78–93. [PubMed] [Google Scholar]
- 9. Grooms DL, Bolin CA. Diagnosis of fetal loss caused by bovine viral diarrhea virus and Leptospira spp. Vet Clin North Am Food Anim Pract 2005; 21: 463–472. [DOI] [PubMed] [Google Scholar]
- 10. Grooms DL. Reproductive losses caused by bovine viral diarrhea virus and leptospirosis. Theriogenology 2006; 66: 624–628. [DOI] [PubMed] [Google Scholar]
- 11. Guitian J, Thurmond MC, Hietala SK. Infertility and abortion among first-lactation dairy cows seropositive or seronegative for Leptospira interrogans serovar hardjo. J Am Vet Med Assoc 1999; 215: 515–518. [PubMed] [Google Scholar]
- 12. Miller DA, Wilson MA, Beran GW. Relationships between prevalence of Leptospira interrogans in cattle, and regional, climatic, and seasonal factors. Am J Vet Res 1991; 52: 1766–1768. [PubMed] [Google Scholar]
- 13. Miller DA, Wilson MA, Beran GW. Survey to estimate prevalence of Leptospira interrogans infection in mature cattle in the United States. Am J Vet Res 1991; 52: 1761–1765. [PubMed] [Google Scholar]
- 14. Rajeev S, Berghaus RD, Overton MW, et al. Comparison of fluorescent antibody and microscopic agglutination testing for Leptospira in pregnant and nonpregnant cows. J Vet Diagn Invest 2010; 22: 51–54. [DOI] [PubMed] [Google Scholar]
- 15. Zuerner RL. Laboratory maintenance of pathogenic Leptospira. Curr Protoc Microbiol 2005; Chapter 12: Unit 12E.1. [DOI] [PubMed] [Google Scholar]
- 16. Smythe LD, Smith IL, Smith GA, et al. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect Dis 2002; 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levett PN, Morey RE, Galloway RL, et al. Detection of pathogenic leptospires by real-time quantitative PCR. J Med Microbiol 2005; 54: 45–49. [DOI] [PubMed] [Google Scholar]
- 18. Zuerner RL, Alt D, Bolin CA. IS1533-based PCR assay for identification of Leptospira interrogans sensu lato serovars. J Clin Microbiol 1995; 33: 3284–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zuerner RL, Bolin CA. Differentiation of Leptospira interrogans isolates by IS1500 hybridization and PCR assays. J Clin Microbiol 1997; 35: 2612–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haake DA. Hamster model of leptospirosis. Curr Protoc Microbiol 2006; Chapter 12: Unit 12E.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ellis WA, Thiermann AB, Montgomery J, et al. Restriction endonuclease analysis of Leptospira interrogans serovar hardjo isolates from cattle. Res Vet Sci 1988; 44: 375–379. [PubMed] [Google Scholar]
- 22. Marshall RB, Winter PJ, Thiermann AB, et al. Genotypes of Leptospira interrogans serovar hardjo in cattle in the UK. Vet Rec 1985; 117: 669–670. [DOI] [PubMed] [Google Scholar]
- 23. Zuerner RL, Alt DP, Palmer MV. Development of chronic and acute golden Syrian hamster infection models with Leptospira borgpetersenii serovar Hardjo. Vet Pathol 2012; 49: 403–411. [DOI] [PubMed] [Google Scholar]
- 24. Bulach DM, Zuerner RL, Wilson P, et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc Natl Acad Sci U S A 2006; 103: 14560–14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benschop J, Heuer C, Jaros P, et al. Sero-prevalence of leptospirosis in workers at a New Zealand slaughterhouse. N Z Med J 2009; 122: 39–47. [PubMed] [Google Scholar]
- 26. Dreyfus A, Benschop J, Collins-Emerson J, et al. Sero-prevalence and risk factors for leptospirosis in abattoir workers in New Zealand. Int J Environ Res Public Health 2014; 11: 1756–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mansell C, Benschop J. Leptospirosis is an important multi-species zoonotic disease in New Zealand. N Z Med J 2014; 127: 5–8. [PubMed] [Google Scholar]
- 28. Rajeev S, Woldemeskel MW, Westmoreland DS. Pathology in practice. Leptospirosis. J Am Vet Med Assoc 2012; 240: 957–959. [DOI] [PubMed] [Google Scholar]


