Acute lymphoblastic leukemia (ALL) is the most common malignancy in children, occurring in 32 per million children every year in the United States (1). Racial differences in the incidence of ALL between blacks and whites have been well recognized (2). Based on SEER data from 2000 to 2006, the ALL incidence in blacks was 16 per million person-years (1). This was lower than the estimated incidence of 33 per million person-years in non-Hispanic whites, a finding similar to that reported by McNeil (2).
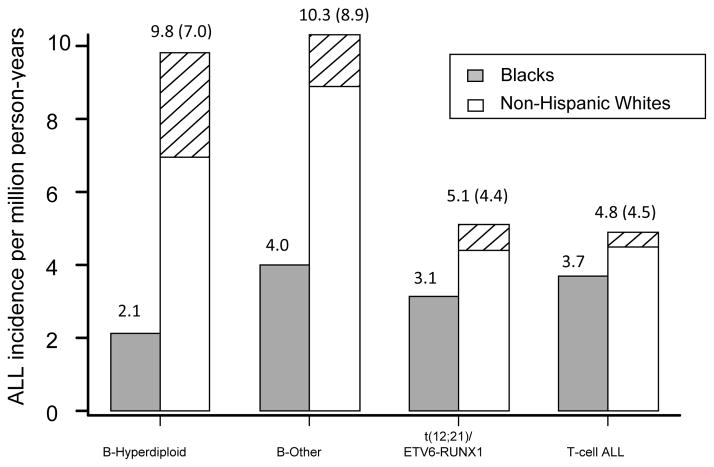
ALL is also a heterogeneous disease including multiple subtypes (3). The distribution of ALL subtypes also differs between blacks and whites (4). A higher percentage of blacks with ALL have T-cell ALL or B-lineage ALL with the TCF3-PBX1 fusion, whereas a higher percentage of whites have B-lineage hyperdiploid ALL (4). In about 70% of cases, SEER distinguished T-cell ALL from B-lineage ALL, but there was not information on ploidy or chromosomal translocations. Therefore, we estimated the subtype-specific incidence in blacks and whites based on the overall incidence of ALL using SEER data (1) and the distribution of ALL subtypes that we observed in 560 patients with newly diagnosed ALL enrolled on St Jude trials, including 112 black patients and 448 non-Hispanic white patients (Figure 1). We focused on four major non-overlapping ALL subtypes with sufficient numbers of patients, including T-cell ALL, B-lineage hyperdiploid ALL with ≥50 chromosomes, B-lineage ALL bearing the t(12;21)/ETV6-RUNX1 fusion, and B-lineage ALL without defined genetic or chromosomal abnormalities (i.e. without hyperdiploidy, hypodiploidy, TCF3-PBX1, BCR-ABL, MLL/AF4, or ETV6-RUNX1). We estimated that the incidence of T-cell ALL does not differ between blacks and whites (3.7 per million vs. 4.8 per million, p = 0.17), and that the incidence of B-lineage ALL was lower in blacks (9.3 per million person-years) than in whites (25.2 per million person-years, p < 1 × 10−6). According to those SEER data that were evaluable, the incidence of T-cell ALL was also similar in blacks and in whites (both at 3 per million person-years), whereas the incidence of B-lineage ALL was about 8 per million in blacks vs. 21 per million in whites, comparable with what we had estimated. In addition, our data showed that the incidence of B-hyperdiploid ALL differed significantly between whites and blacks (9.8 per million vs. 2.1 per million person-years, respectively, relative risk [RR] = 4.6 for whites compared to blacks), as did B-lineage ALL without defined genetic or chromosomal abnormalities (RR = 2.6) and B-lineage ALL with the t(12;21)/ETV6-RUNX1 fusion (RR = 1.6).
Figure 1.
The subtype-specific U.S. incidence of ALL in blacks and non-Hispanic whites. The hatched areas indicate cases in whites that can be attributed to a higher frequency of the risk allele at rs10821936 in whites compared to blacks. Numbers in parentheses are the estimated incidence of ALL in whites if the frequency of high-risk genotypes at rs10821936 were the same in whites as in blacks. B-other includes B-lineage ALL without defined genetic or chromosomal abnormalities.
We hypothesized that the racial differences in ALL incidence could partly be attributed to differences in allele frequencies between populations of African and European ancestries at disease-associating loci. We have recently identified that germline diversity in the ARID5B gene (SNP rs10821936) was significantly associated with childhood ALL risk in non-Hispanic whites based on a genome-wide association analysis (5). An ARID5B SNP in high linkage disequilibrium with rs10821936 also associated with ALL risk in an independent study (6). The high risk genotypes at rs10821936 particularly increased the risk of developing the B-hyperdiploid ALL subtype. Here we performed a case-control analysis to investigate whether genotypes at rs10821936 was also associated with the risk of developing childhood ALL in blacks.
Patients included 93 black children with newly diagnosed ALL enrolled on St Jude trials Total XIIIB and Total XV between 1994 and 2007. Controls included 112 unrelated subjects from the International HapMap Project (www.hapmap.org), including 52 unrelated subjects of African Ancestry in SouthWest USA (ASW) and 60 Yuruba (YRI) samples. Patient genotypes were assessed by Affymetrix 500K or SNP6.0 Mapping array and confirmed by an independent genotyping method as described (5). Genotypes of controls were downloaded from Hapmap version 29. Logistic regression and Fisher’s Exact test were used to compare the differences in allele and genotype frequencies at the ARID5B SNP rs10821936 between patients with ALL and controls. The frequency of the C risk allele at rs10821936 was significantly higher in black children with ALL (minor allele frequency = 0.33) than in controls of African ancestry (0.163 in ASW and 0.20 in YRI, odds ratio [OR] = 2.08; 95% C.I. 1.32 – 3.27, p = 0.0015, Table 1). Black patients carrying one or two copies of the C allele at rs10821936 had a higher risk of developing higher B-hyperdiploid ALL compared to controls (OR=6.62; 2.0 – 21.9; p=0.0001), similar to our previous findings in whites. The odds ratios relating this ARID5B genotype to the other subtypes of ALL were moderate, ranging from 2.01 in patients with t(12;21)/ETV6-RUNX1 fusion to 2.80 in T-cell ALL. The odds ratios relating ARID5B SNPs to subtype-specific ALL risk were similar in magnitude between blacks and whites (Table 2).
Table 1.
Genotype and allele distribution of ARID5B SNP rs10821936 in patients (by ALL subtypes and controls) in blacks
| rs10821936
|
|||||
|---|---|---|---|---|---|
| CC | CT | TT | Allele freq (C) | ||
| Patients | Total | 10 | 41 | 42 | 0.33 |
| B-hyperdiploid | 1 | 11 | 4 | 0.40 | |
| B-other | 1 | 13 | 15 | 0.26 | |
| t(12;21)/ETV6-RUNX1 | 3 | 8 | 12 | 0.30 | |
| T-cell ALL | 5 | 9 | 11 | 0.38 | |
|
| |||||
| Controls | Total | 6 | 29 | 77 | 0.18 |
| YRI* | 5 | 14 | 41 | 0.20 | |
| ASW** | 1 | 15 | 36 | 0.16 | |
YRI, HapMap samples from Yoruba in Ibadan, Nigeria
ASW, HapMap samples from a population of African Ancestry in SouthWest United States
Table 2.
Odds ratios comparing genotypes of rs10821936 between patients and controls in non-Hispanic whites and blacks
| Non-Hispanic Whites | Blacks | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| n | P-value | Odds ratio* (95% C.I) | n | P-value | Odds ratio (95% C.I) | ||
| C vs. T | All Patients | 317 | 1.4×10−15 | 1.91 (1.60 – 2.20) | 93 | 0.0015 | 2.09 (1.31 – 3.30) |
|
| |||||||
| CC/CT vs. TT | All Patients | 317 | 4.0×10−10 | 2.16 (1.67 – 2.82) | 93 | 0.0008 | 2.67 (1.50 – 4.76) |
| B-Hyperdiploid | 108 | 1.3×10−10 | 4.63 (2.68 – 8.48) | 16 | 0.0021 | 6.62 (2.0 – 21.9) | |
| B-Other | 121 | 0.0026 | 1.83 (1.22 – 2.79) | 29 | 0.09 | 2.05 (0.89 – 4.72) | |
| t(12;21)/ETV6-RUNX1 | 45 | 0.09 | 1.78 (0.92 – 3.64) | 23 | 0.13 | 2.01 (0.82 – 4.96) | |
| T-cell ALL | 43 | 0.36 | 1.36 (0.70 – 2.71) | 25 | 0.023 | 2.80 (1.16 – 6.77) | |
Odds ratio is estimated using logistic regression comparing allele frequencies or genotype frequencies between ALL cases and controls. Controls for whites included individuals as previously described (5); controls for blacks included 112 subjects as described in text.
It is notable that the frequency of the C risk allele was lower in the control populations of African ancestry (0.163 in ASW, 0.20 in YRI) than in populations of European Ancestry (0.33)(5). The question then naturally arises as to whether this allele frequency difference in the general population might contribute to racial differences in the incidence of ALL. To this end, we calculated the absolute risk of developing ALL in whites with different rs10821936 genotypes, based on the average subtype-specific incidence and the relative risks between genotypes. Since ALL is a rare disease, the afore-estimated odds ratios could be used equivalently as relative risks. If the frequency of the risk allele in whites were 0.18 (the same frequency as in blacks), instead of the observed 0.33, we would expect only 7 per million person-years to develop B-hyperdiploid ALL, compared to the observed 9.8 per million person-years observed in whites in the U.S. Therefore, 2.8 per million person-years of B-hyperdiploid cases among whites could be explained by the greater frequency of the high risk rs10821936 genotypes in whites compared to blacks (hatched area in Figure 1). Combining the four major ALL subtypes, a total of 5.2 per million person-years (30% of the observed racial difference in ALL incidence) could be attributed to the higher frequency of the risk allele at rs10821936 in whites compared to blacks.
Our data show that inherited variation at rs10821936 in the ARID5B gene is associated with the risk of developing childhood ALL in blacks, as we previously described in non-Hispanic American whites (5). This further indicates the importance of the ARID5B gene in the etiology of childhood ALL. Further work is needed to identify the causal SNPs and elucidate the mechanism. It is remarkable that this single locus explains approximately 30% of the racial difference in ALL incidence between blacks and whites. Nevertheless, even after accounting for the racial differences in frequency of the risk allele at rs10821936, there remains a lower incidence of B-lineage ALL without known genetic abnormalities in blacks. These unexplained racial differences could be due to racial differences in allele frequencies at additional inherited loci, in environmental exposure to leukemogens, or in immune function (7). In addition, B-lineage ALL without known cytogenetic abnormalities could include a heterogeneous group of cryptic genomic variations such as chromosomal gains or losses (8). It is possible that germline polymorphisms are associated with subgroups of ALL cases with cryptic gains or losses. Future systematic cataloging of race-specific lesions in ALL patients should shed new light on the etiology of this disease.
Acknowledgments
We thank our protocol co-investigators, clinical and research staff, particularly Pamela McGill, Natalie Lowery, Sean Freeman, Shaherah Amos, Nancy Kornegay and Dr. Jennifer Pauley, as well as patients and families for their participation. We thank the staff from the Hartwell Center for Bioinformatics and Biotechnology.
Footnotes
Conflicts of interest statement
The authors of this submitted manuscript have no conflicts to disclose.
References
- 1.Surveillance Epidemiology and End Results Program (SEER) Stat Database: Incidence - SEER 17 Regs Limited-use, Katrina/Rita Population Adjustment. 2009 [cited April 2009; Available from: www.seer.cancer.gov.
- 2.McNeil DE, Cote TR, Clegg L, Mauer A. SEER update of incidence and trends in pediatric malignancies: acute lymphoblastic leukemia. Med Pediatr Oncol. 2002 Dec;39(6):554–557. doi: 10.1002/mpo.10161. discussion 552–553. [DOI] [PubMed] [Google Scholar]
- 3.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004 Apr 8;350(15):1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 4.Pui CH, Sandlund JT, Pei D, Rivera GK, Howard SC, Ribeiro RC, et al. Results of therapy for acute lymphoblastic leukemia in black and white children. JAMA. 2003;290(15):2001–2007. doi: 10.1001/jama.290.15.2001. [DOI] [PubMed] [Google Scholar]
- 5.Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009 Sep;41(9):1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009 Sep;41(9):1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006 Mar;6(3):193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 8.Mullighan CG, Downing JR. Genome-wide profiling of genetic alterations in acute lymphoblastic leukemia: recent insights and future directions. Leukemia. 2009 Jul;23(7):1209–1218. doi: 10.1038/leu.2009.18. [DOI] [PubMed] [Google Scholar]