Abstract
Spindle assembly required during mitosis depends on microtubule polymerization. We demonstrate that the evolutionarily conserved low-complexity protein, BuGZ, undergoes phase transition or coacervation to promote assembly of both spindles and their associated components. BuGZ forms temperature-dependent liquid droplets alone or on microtubules in physiological buffers. Coacervation in vitro or in spindle and spindle matrix depends on hydrophobic residues in BuGZ. BuGZ coacervation and its binding to microtubules and tubulin are required to promote assembly of spindle and spindle matrix in Xenopus egg extract and in mammalian cells. Since several previously identified spindle-associated components also contain low complexity regions, we propose that coacervating proteins may be a hallmark of proteins that comprise a spindle matrix that functions to promote assembly of spindles by concentrating its building blocks.
Keywords: spindle, spindle matrix, phase transitions, coacervation, tubulin, microtubule, low complexity, unstructured
Introduction
Since the discovery of spindle apparatus in the 1800s (Lukacs, 1981), much attention has focused on how microtubules (MT) interact with chromosomes to ensure equal partitioning of chromosomes into daughter cells. Investigation of the mechanisms by which MTs and MT-associated proteins regulate mitosis (Walczak et al., 2010) is fueled by the ease of visualizing the spindle-shaped MT fibers, the disruption of chromosome segregation and cell division upon MT perturbation, and the discovery of tubulin (Oakley, 2000). In addition to spindle MTs, a set of material that surrounds and permeates spindle MTs have periodically drawn attention (Goldman and Rebhun, 1969 Schibler and Pickett-Heaps, 1980; Scholey et al, 2001; Johansen and Johansen, 2007; Johansen et al., 2011; Leslie et al., 1987; Pickett-Heaps et al., 1984; Pickett-Heaps and Forer, 2009; Schweizer et al., 2014; Wein et al., 1998; Zheng, 2010; Zheng and Tsai, 2006).
Historically this ill-defined spindle-associated material has been referred to as spindle matrix. One vague but generally accepted feature of spindle matrix is that it retains some integrity upon MT disassembly. Based on this criterion, several spindle matrix proteins have been identified and studied in the context of spindle assembly and chromosome segregation. For example among the Drosophila spindle matrix proteins (Fabian et al., 2007; Johansen et al., 2011; Qi et al., 2005; Qi et al., 2004; Rath et al., 2004; Walker et al., 2000; Yao et al., 2012; Yao et al., 2014), Megator regulates spindle assembly checkpoints (SAC) (Lince-Faria et al., 2009). A conserved protein, BuGZ, which was identified as part of the lamin-B (LB) spindle matrix in Xenopus (Tsai et al., 2006; Ma et al., 2009), has recently been shown to facilitate chromosome alignment by controlling both stability and kinetochore loading of the SAC component Bub3 (Jiang et al., 2014; Toledo et al., 2014). Additionally, LB (Tsai et al., 2006) and poly ADP-ribose (Chang et al., 2004), along with other spindle assembly factors (SAFs), such as dynein, Nudel, NuMA, and kinesin Eg5 (Civelekoglu-Scholey et al., 2010; Goodman et al., 2010; Ma et al., 2009; Tsai et al., 2006), may regulate spindle morphogenesis. Despite these studies, the structural nature of the spindle matrix remains undefined and whether it constitutes a cohesive functional unit is unclear. In fact, some modeling and biophysical probing of spindle apparatus have not provided evidence for the existence of spindle matrix (Brugues and Needleman, 2014; Gatlin et al., 2010; Shimamoto et al., 2011). Thus whether spindle matrix is a real structural element of spindle apparatus or a mere artifact induced upon depolymerization of spindle MTs remains an open question.
Unlike membranous organelles, the spindle apparatus is not surrounded by membrane barrier during vertebrate mitosis. However, spindles may need to concentrate many components in order to support spatially and temporally diverse reactions. Consistently, tubulin and some SAFs are shown to be concentrated in the region where nascent spindle begins to assemble in C. elegans embryos (Hayashi et al., 2012). This concentration process is independent of MTs but it requires nuclear envelope permeabilization and RanGTPase, which stimulates spindle assembly (Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999).
Proteins, such as elastin and elastin-like peptides, can undergo liquid-liquid phase transitions or coacervation to form liquid droplets (Yeo et al., 2011). The phase separation has been proposed to promote concentration of molecules into the liquid droplets, which can then facilitate biochemical reactions (Hyman et al., 2014). Indeed, the liquid droplet feature of P granules and nucleoli is consistent with the idea that assembly and function of these non-membranous organelles could be driven by the phase transition of some of their structural components (Brangwynne et al., 2009; Brangwynne et al., 2011). No proteins of these organelles, however, have yet been shown to undergo functionally relevant phase transition. Interestingly, when engineered as multiple tandem repeats, SRC homology 3 (SH3) domains of NCK and proline-rich motif (PRM) of N-WASP form multivalent interactions, which allow the protein mixture to undergo phase transitions to form liquid droplets. These droplets concentrate actin to promote F-actin assembly in vitro (Li et al., 2012). Despite the observed in vitro phase transitions into liquid droplets, no protein has been shown to function in vivo via phase transitions.
Here we examine the spindle regulatory protein BuGZ, which we noted contains evolutionarily conserved low complexity sequence, and demonstrate that it forms a MT-independent structure through temperature- and hydrophobic residue-dependent coacervation. This phase transition property allows the concentration of tubulin along MTs and supports assembly of spindle MTs and of the biochemically defined spindle matrix structure. Based on these results we propose a model and line of investigation for further developing our understanding of observed properties and possible functions of spindle matrix.
Results
BuGZ promotes assembly of spindle apparatus
Our previous studies show that BuGZ binds MTs to promote kinetochore loading of Bub3 and chromosome alignment (Jiang et al., 2014). We noticed that human BuGZ (hBuGZ) depletion in HeLa cells resulted in a more severe disruption of spindle morphology and reduction of MT intensity than those depleted of Bub3, especially when RNAi treatment was extended to 72 h (Figure S1A–B). The more severe spindle defects in hBuGZ-depleted cells were consistent with a stronger chromosome misalignment than those depleted of hBub3 (Figure S1C). This suggests that BuGZ could directly regulate spindle assembly independent of Bub3’s kinetochore function.
Previously we developed a bead-based spindle assembly assay (Tsai and Zheng, 2005) by tethering the mitotic kinase Aurora A to 2.8-μm magnetic beads via antibodies. These beads function as MT organizing centers to induce efficient spindle assembly in CSF Xenopus egg extract (referred to as extract below) in the presence of RanGTP. Since spindles induced by Aurora A beads and RanGTP do not have chromosomes and kinetochores, we can test the kinetochore-independent function of BuGZ in spindle assembly. Immunodepletion of Xenopus BuGZ (xBuGZ) by ~90% (Figure 1A) resulted in a significant reduction of astral MT length and bipolar spindle numbers (Figure 1B–D). Most bipolar spindles formed in the absence of xBuGZ were also significantly shorter than those of controls (Figure 1E). These defects were fully rescued by purified xBuGZ (Figure 1A–E). xBuGZ depletion also disrupted spindle assembly induced by sperm chromatin. Major phenotypes included spindles with MT aggregates surrounding sperm chromatin or spindles with reduced MTs, followed by asters, half spindles, or abnormal spindle shapes with normal MT density (Figure 1F–H), and all of the defects were also rescued by purified xBuGZ (Figure 1F–H). Thus BuGZ promotes spindle assembly independent of its kinetochore function.
Figure 1. BuGZ promotes spindle assembly independent of kinetochores.
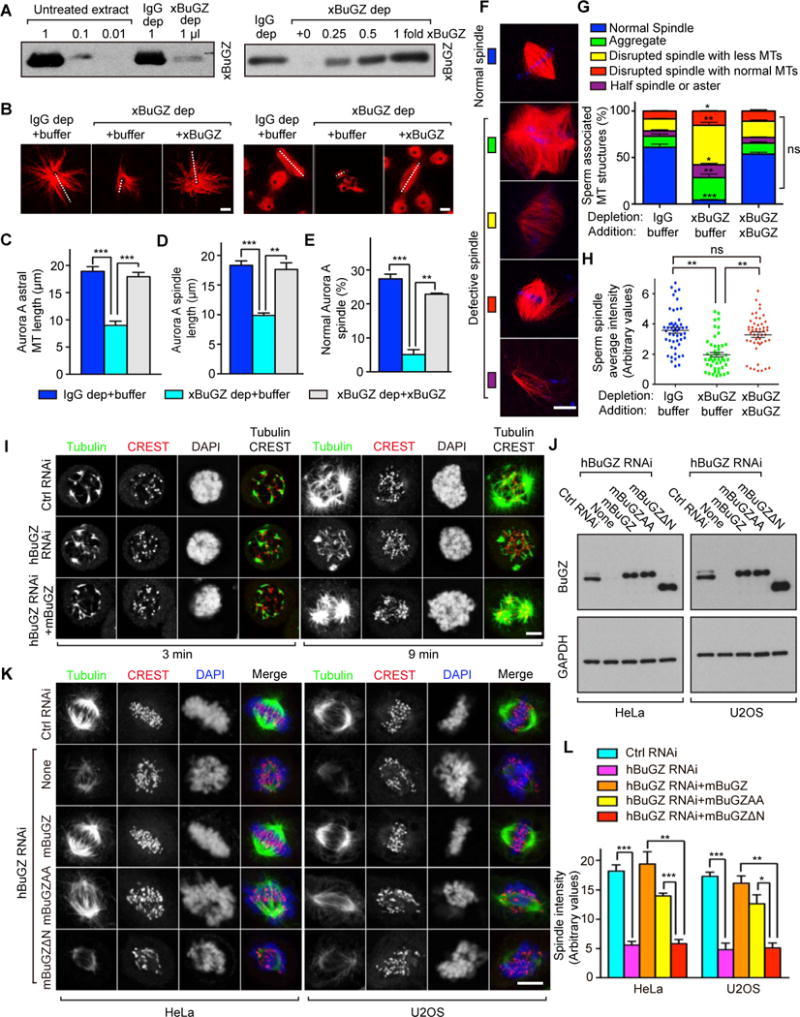
A. Western blotting of xBuGZ depletion (left) and add-back (right) in extracts. xBuGZ depletion (dep) efficiency and xBuGZ addition are shown in titrations.
B–E. Representative images (B) show that xBuGZ depletion reduced astral MT length, bipolar spindle formation and length, which were all rescued by His-xBuGZ. ~50 (C, D) or ~500 (E) structures were measured in each experiment and condition. White dashed lines in B indicate Aurora A spindle length and longest astral MTs measured.
F–H. xBuGZ depletion caused multiple sperm spindle defects (F), which was rescued by His-xBuGZ (G, H). ~500 (G) and ~50 (H) structures were analyzed in each experiment and condition.
I. hBuGZ depletion reduced astral MT re-growth in mitotic HeLa cells, and was rescued by mBuGZ. Cold-treated cells were examined at 3 or 9 min after returning to 37°C. MTs, centromeres, and chromosomes were stained by tubulin antibody, CREST serum, and DAPI, respectively.
J. Western blotting analyses of HeLa and U2OS cells treated by hBuGZ siRNA and transfected with indicated plasmids.
K–L. HeLa or U2OS images (K) show that hBuGZ depletion by 72 h of RNAi diminished MT intensity in spindles, and was rescued fully by mBuGZ, partially by mBuGZAA, but not by mBuGZΔN. Cells were blocked with 10 μM MG132 for 1 h before immunostaining. ~30 cells were measured for each experiment and condition (L). Error bars, standard error of the means (SEM). Student’s t-test: *p<0.05, **p<0.01, ***p<0.001, three independent experiments. Scale bars, 5 μm.
BuGZ-MT interaction promotes spindle assembly
To understand how BuGZ promotes spindle assembly, we treated HeLa cells with control or hBuGZ siRNA and then depolymerized MTs in the cold. MT regrowth was examined after returning cells to 37°C. hBuGZ depletion greatly reduced astral MT regrowth, which was rescued by expressing the RNAi-insensitive wild-type mouse BuGZ (mBuGZ, Figure 1I–J). The N-terminal 92 amino acids of BuGZ bind directly to MTs, while the Gle2-binding sequence (GLEBS) within the C-terminus of BuGZ directly binds and stabilizes Bub3 (Jiang et al., 2014). Replacing the two highly conserved glutamic acids (E) in GLEBS with alanine (A) results in a mutant (mBuGZAA) that fails to bind and stabilize Bub3, while mBuGZΔN lacking the N-terminal 92 amino acids does not bind to spindles in vivo and MTs in vitro (Jiang et al., 2014). The wild-type mBuGZ and mBuGZAA bound to spindle MTs and MTs assembled from pure tubulin (Figure S1D–F). To analyze which of these two known domains in BuGZ promotes assembly of spindle, we depleted endogenous hBuGZ from HeLa or U2OS cells by RNAi. BuGZ RNAi-induced spindle defects, judged by spindle intensity, were rescued fully by wild-type mBuGZ and partially by mBuGZAA, but not by mBuGZΔN (Figure 1J–L). hBuGZ depletion did not alter interphase MT densities (Figure S1G). Thus MT binding of BuGZ promotes spindle MT assembly.
Spindle matrix assembly and stability require BuGZ and a physiological temperature
Since BuGZ was identified as a spindle matrix component, we assayed for spindle matrix by assembling Aurora A-bead spindles in extract and then depolymerizing MTs using nocodazole at room temperature (RT) (Ma et al., 2009; Tsai et al., 2006). The nocodazole-insensitive material that remains on the Aurora A beads, i.e. the spindle matrix, was isolated using a magnet and analyzed by Western blotting or immunostaining probing known spindle matrix markers, lamin-B3 (LB3, the major lamin in extracts), dynein, Eg5, NuMA, and XMAP215 (Ma et al., 2009; Tsai et al., 2006). Depleting xBuGZ greatly diminished recovery of the spindle matrix, but this was rescued by purified xBuGZ (see the Noc, RT panels in Figure 2A–C). Although xBuGZ depletion diminished the recovery of LB3, depleting LB3 did not affect association of xBuGZ with the spindle matrix (Figure S2A). Thus BuGZ may function upstream of LB3 to promote spindle matrix assembly.
Figure 2. Effects of BuGZ and temperature on spindle matrix.
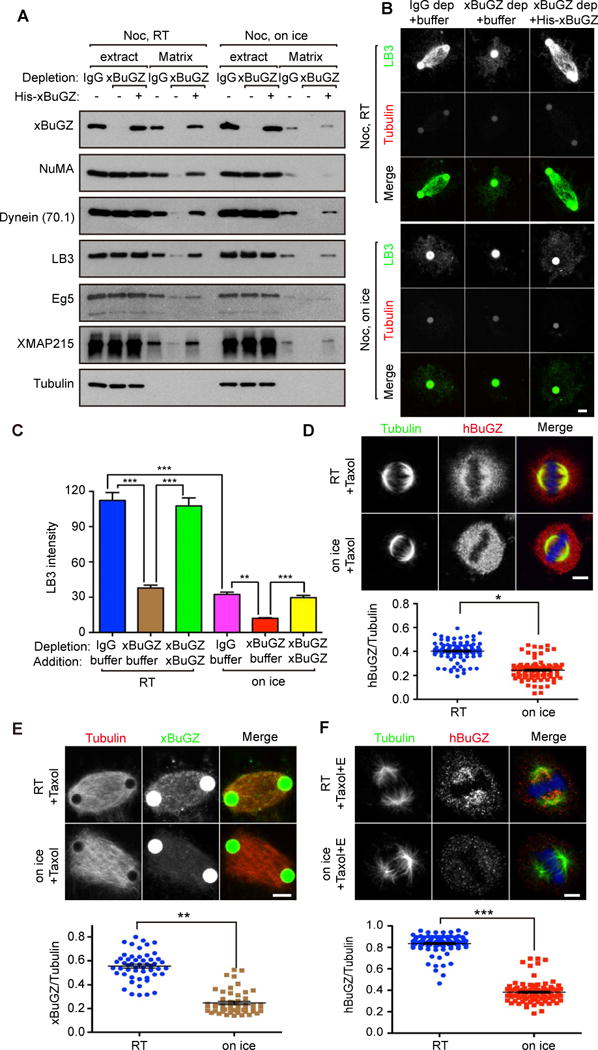
A–C. Both BuGZ and temperature influenced spindle matrix assembly. Spindle matrices were prepared with or without xBuGZ after nocodazole (Noc) treatment at RT or on ice and assayed by Western blotting (A) or immunostaining (B) using indicated markers. Tubulin, negative control. LB3 intensity of ~30 spindle matrices associated with one or two beads was quantified (C). Less spindle matrix was present in the cold than at RT. Green Aurora A beads appear larger than 2.8 μm due to secondary anti-rabbit antibody staining.
D. Taxol treated HeLa cells were incubated at RT or on ice. Metaphase cells were visualized by immunostaining with tubulin (green), hBuGZ (red) antibodies, and DAPI (blue). ~100 cells were quantified for each condition.
E. Treatment of taxol-stabilized Aurora A spindles on ice for 5 min diminished xBuGZ signal on spindles visualized by fluorescein-labeled MTs (red) and xBuGZ immunostaining (green). ~50 spindles were quantified for each condition.
F. Extraction of taxol-stabilized and cold-treated metaphase spindles in HeLa cells diminished hBuGZ signal on spindles compared to RT extraction. Metaphase cells shown were immunostained using tubulin and hBuGZ antibodies and DAPI. ~100 cells were quantified for each condition.
Error bars, SEM. Student’s t-test: p*<0.05, **p<0.01, ***p<0.001, three independent experiments. Scale bar, 5 μm.
When spindle MTs were depolymerized by nocodazole on ice, less matrices were associated with Aurora A beads than those incubated at RT (compare IgG matrix panels in Figure 2A–B). Quantification of LB3 staining revealed a significant reduction of matrices around beads upon cold treatment or upon xBuGZ depletion (Figure 2C). xBuGZ depletion plus cold treatment caused an additional matrix reduction that could be rescued by purified xBuGZ (Figure 2C). Thus spindle matrix assembly and stability require BuGZ and a physiological temperature.
BuGZ exhibits temperature-sensitive binding to spindle MTs
Unlike many MT-associated SAFs that decorate MT fibers densely and brightly, BuGZ appears as a loose ‘haze’ enriched on spindles (Figure S1E) (Jiang et al., 2014). When HeLa cells were incubated at RT or on ice for 5 min followed by immunostaining, we found that cold treatment diminished BuGZ signal on spindles, whether or not the spindle MTs were stabilized with taxol (Figure 2D, S2B). xBuGZ signal was also reduced on cold-treated Aurora A-bead spindles stabilized by taxol (Figure 2E).
We then treated HeLa cells by taxol and collected mitotic cells by shake-off. Detergent extraction of these cells on ice or at RT in the presence of taxol followed by immunoblotting showed that more hBuGZ was extracted in the cold as compared to tubulin or CENP-A controls (Figure S2C). Immunostaining further indicated that spindle-associated hBuGZ was more sensitive to the extraction on ice than at RT (Figure 2F).
BuGZ exhibits temperature-dependent phase transition via conserved phenylalanine and tyrosine
We analyzed vertebrate BuGZ protein sequences using PONDR and SEG, programs designed to predict the disordered (Xue et al., 2010) and low complexity regions (Wootton, 1994) in proteins, respectively. The N-terminus of BuGZ, containing the MT binding domain and zinc fingers, was predicted to have normal amino acid complexity, while the rest of BuGZ was largely unstructured with low amino acid complexity (Figure 3A). Since some disordered and low complexity (DLC) proteins can undergo phase transitions, we examined Sf9 cells expressing YFP-tagged xBuGZ (YFP-xBuGZ) via baculovirus. YFP-xBuGZ formed bright droplet-like spheres in the cytosol, whereas YFP was evenly distributed (Figure 3B).
Figure 3. Temperature- and concentration-dependent phase transition of BuGZ.
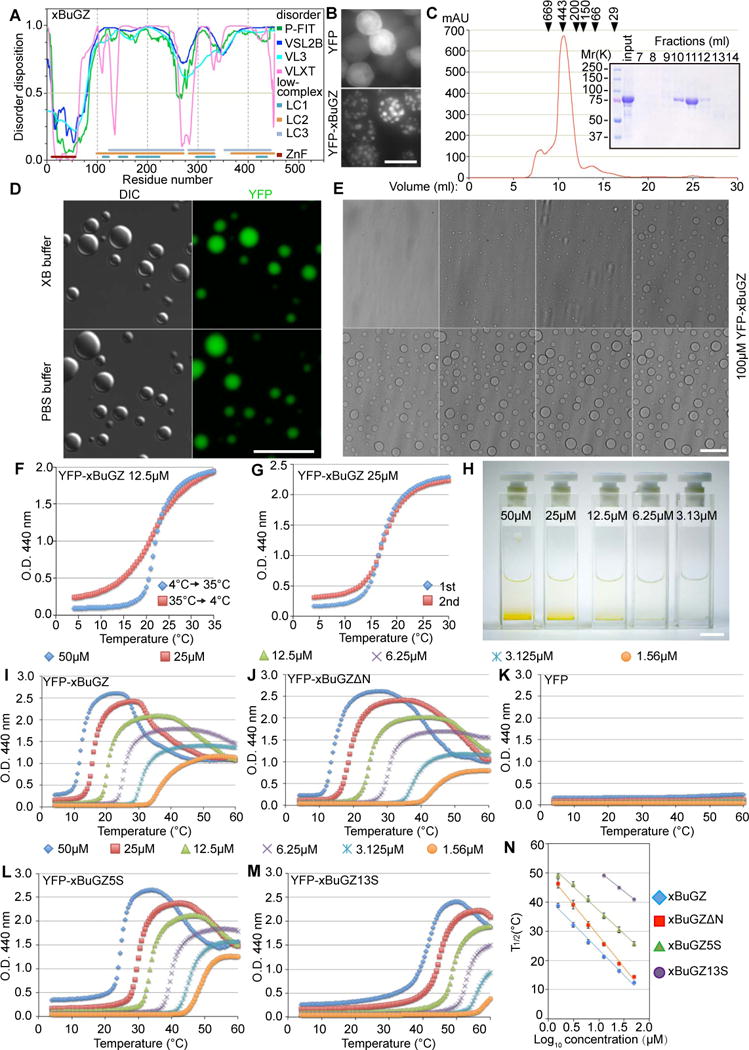
A. Structural features of xBuGZ. The line at 0.5 (y-axis) is the cutoff for disorder (>0.5) and order (<0.5) predictions. P-FIT, VSL2B, VL3, and VLXT, predictors for disordered dispositions. LC1, LC2, and LC3 indicate low complexity regions determined at three stringencies. ZnF, zinc fingers, predicted structured region.
B. YFP-xBuGZ formed spheres in Sf9 cells. YFP served as control. Scale bar, 20 μm.
C. Gel filtration chromatography of YFP-xBuGZ. Arrowheads, positions of size markers (in kDa): thyroglobulin, apoferritin, amylase, alcohol dehydrogenase, albumin, and carbonic anhydrase. Fractions 7–14 were analyzed.
D. YFP-xBuGZ formed droplets in vitro as visualized by DIC and fluorescence microscopy. Scale bar, 20 μm.
E. Temperature-dependent droplet formation by YFP-xBuGZ in XB buffer as visualized by Hoffman modulation contrast microscopy. Temperature ramp, 4 to 20°C at 1°C/min. Scale bar, 20 μm.
F–G. Turbidity assay of reversibility (F) and repeatability (G) of phase transition by YFP-xBuGZ. Increase (4°C to 35°C) and decrease (35°C to 4°C) in temperature in F had the same temperature ramp. The temperature ramp in G was 4°C–30°C. Ramp rate, 0.5°C/min.
H. Concentration-dependent accumulation of YFP-xBuGZ coacervates at the bottom of cuvettes after turbidity measurements. Scale bar, 1 cm.
I–K. Concentration- (color-coded) and temperature-dependent phase transition of YFP-xBuGZ (I) and YFP-xBuGZΔN (J), but not YFP (K), based on the turbidity assay.
L–M. xBuGZ5S (L) and xBuGZ13S (M) coacervation at increasingly higher protein concentrations and temperatures.
N. The temperature at which the turbidity was half (T1/2) of the difference between maximum and 4°C absorbance was plotted against log10 protein concentration. YPF-xBuGZ13S did not reach maximum turbidity at 3.125 and 1.56 μM even at 60°C. Error bars, standard deviation from three independent experiments.
We purified His-, GST-, GFP-, or YFP-tagged xBuGZ or mBuGZ expressed in either Sf9 cells or bacteria (Figure 3C, S3A). Upon warming each protein formed droplets of varying sizes in physiologically relevant buffers and the droplet size increased over time (Figure 3D, S3B–C). When the solutions were cooled on ice, the droplets disintegrated over time, as judged by the disappearance of fluorescence in the droplets (Figure S3D). Under the same conditions and the same or much higher concentrations, purified GST, GFP, YFP, or other SAFs such as GFP-EB1 did not form droplets (Figure S3E). Live imaging showed that at 100 μM of YFP-xBuGZ small droplets became visible at ~10°C and larger droplets formed upon further increase in temperature (Figure 3E).
Turbidity assay showed that purified YFP-xBuGZ underwent an abrupt increase in solution turbidity above a critical temperature and the process was reversible upon cooling to the same temperature (Figure 3F). After dissolution of droplets on ice, BuGZ underwent the same degree of phase transitions upon warming, indicating that the phase separation was repeatable (Figure 3G). Formation of coacervates at high concentrations of YFP-xBuGZ eventually led to large-scale phase separations that become visible to the naked eye due to protein settlement at the bottom of cuvettes (Figure 3H). By varying protein concentrations and temperature, we found that a lower concentration of YFP-xBuGZ required a higher temperature for coacervation (Figure 3I). YFP-xBuGZΔN exhibited similar coacervation properties as YFP-xBuGZ, especially at higher protein concentrations (Figure 3I–J, 3N), indicating that the MT-binding sequence is dispensable for phase transition in vitro. By contrast, YFP did not undergo any noticeable phase transitions at equivalent concentrations and temperatures (Figure 3K).
The above findings suggest that BuGZ could undergo intermolecular interactions mediated by hydrophobic residues. We found that BuGZ orthologs are all abundant in proline (P). For example, mBuGZ and xBuGZ contain 15–19% P, which often occur next to hydrophobic residues (Figure S3F). The hydrophobic and aromatic residues phenylalanine (F) and tyrosine (Y) are implicated in hydrogel formation of some nucleoporins (Frey and Gorlich, 2007), transcription factors, and RNA binding proteins (Kato et al., 2012; Kwon et al., 2013). Since both F and Y are highly conserved among vertebrate BuGZ (Figure S3F), we mutated the last 5 or all 13 conserved Fs and Ys in the predicted DLC region of xBuGZ to serine (S) to create YFP-xBuGZ5S or YFP-xBuGZ13S (Figure S3F). Coacervation of YFP-xBuGZ5S and YFP-xBuGZ13S required increasingly higher concentration and temperature (Figure 3L–N). Thus conserved Fs and Ys are required for BuGZ phase transition.
We found that a fragment of xBuGZ corresponding to amino acids 258–334 (xBuGZ-B) (Figure S3F, black underline), which did not form droplets on its own (Figure S3E), inhibited the phase transition of YFP-xBuGZ when used at high concentrations (Figure S3G). Replacing the 2F and 1Y by S in xBuGZ-B (xBuGZ-B3S, also did not form droplets on its own, Figure S3E) largely disrupted the inhibitory property (Figure S3G–I). At concentrations that did not fully block His-xBuGZ droplet formation, GFP-xBuGZ-B strongly incorporated into the droplets as compared to GFP-xBuGZ-B3S (Figure S3J). Thus, intermolecular BuGZ interactions in droplets may be mediated, in part, by Fs and Ys. Moreover, xBuGZ-B may disrupt droplet formation by blocking proper alignment of full-length xBuGZ molecules critical for coacervation.
BuGZ bundles MTs via MT binding and phase transition
We incubated 0–4 μM YFP-xBuGZ, YFP-xBuGZΔN, YFP-xBuGZ13S, or YFP with rhodamine-labeled and taxol-stabilized short MTs at 37°C. 4 μM BuGZ, but not xBuGZΔN, xBuGZ13S, or YFP, caused prominent MT bundling, although careful inspection showed that some MT bundles were formed even at 1 μM xBuGZ (Figure 4A). The bundled MTs were longer and brighter than the input MT fragments. To quantify the bundling activity, we measured the length and average brightness of individual MTs or MT bundles formed with 2 μM of different xBuGZ proteins and YFP because MT bundles at higher xBuGZ concentrations became a network. xBuGZ increased MT bundle length and average intensity, but xBuGZΔN, xBuGZ13S, or YFP did not (Figure 4B–C). xBuGZ-B inhibited MT bundling induced by xBuGZ, while xBuGZ-B3S was much less effective (Figure 4B–C). Thus MT binding and phase transition of BuGZ promote MT bundling.
Figure 4. Bundling of MTs by BuGZ depends on its MT binding and phase transition.
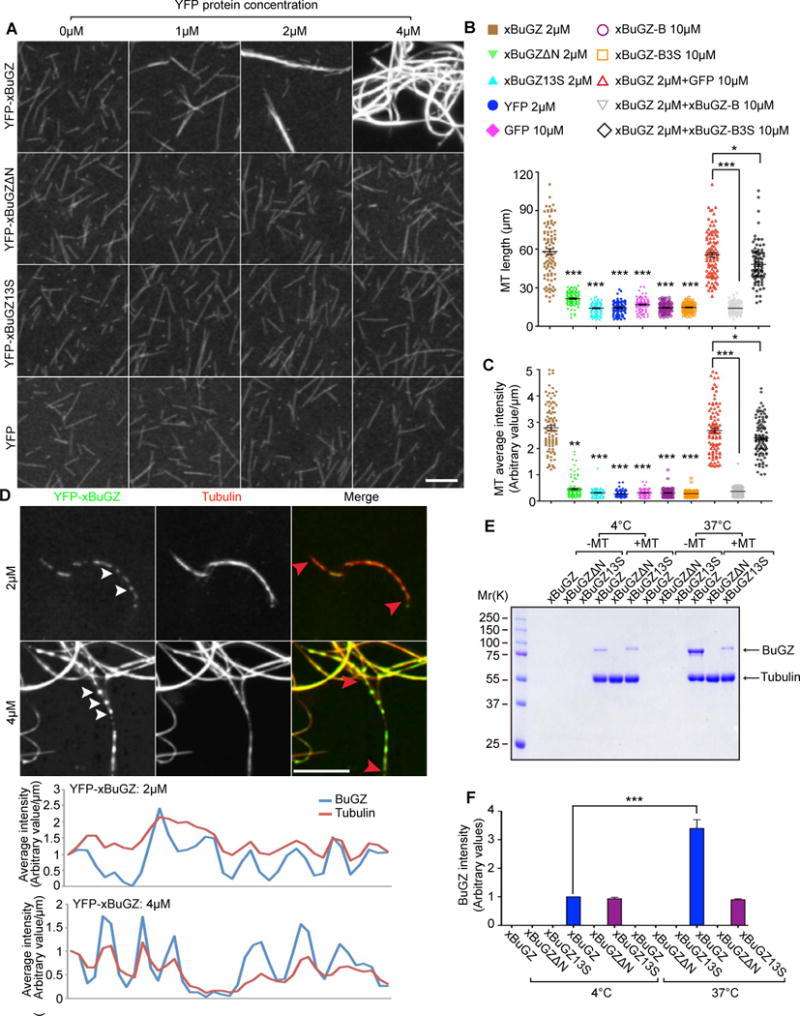
A. YFP-xBuGZ, but not YFP-BuGZΔN, YFP-BuGZ13S, or YFP, induced MT bundling from taxol-stabilized and rhodamine-labeled MT seeds.
B–C. Quantifications of length (B) and average intensity (C) of MTs or MT bundles formed in the presence of indicated proteins and concentrations. MT images were randomly captured under a 63× objective. ~100 individual MTs or MT bundles were measured.
D. YFP-xBuGZ droplets (green, white arrowheads) along some MT bundles (red). Line scans of tubulin and YFP-BuGZ intensity of the indicated segments of MTs (red arrowheads) are shown.
E–F. Purified YFP-xBuGZ, but not YFP-xBuGZ13S, had increased binding to preformed MTs at 37°C compared to 4°C, but YFP-xBuGZΔN fail to bind. Quantification is shown in F.
Error bars, SEM. Student’s t-test: *p<0.05, **p<0.01, ***p<0.001, three independent experiments. Scale bars, 10 μm.
Although 2 or 4 μM YFP-xBuGZ, YFP-xBuGZΔN, YFP-xBuGZ13S, and YFP did not form visible droplets at 37°C (Figure S4A), when incubated with MT seeds, YFP-xBuGZ was enriched on MT bundles and small YFP-xBuGZ droplets were visible along some bundles (arrowheads, Figure 4D, S4B). Line scan showed that the droplets tended to appear at sites of thicker MT bundles flanked by thinner regions (Figure 4D). YFP-xBuGZΔN and YFP-xBuGZ13S failed to form MT-associated droplets, even though YFP-xBuGZ13S showed MT association (Figure S4B). MT pelleting also showed that, despite intact MT binding domain and similar MT binding at 4°C, significantly less YFP-xBuGZ13S bound to MTs at 37°C than did YFP-xBuGZ (Figure 4E–F). When high concentrations (60–100 μM) of YFP-xBuGZ were used to induce MT bundling, large deformed BuGZ droplets extended beyond the associated MT bundles (Figure S4C), suggesting that the MT bundles could flatten the droplets when local BuGZ was not in excess. Thus BuGZ preferably undergoes phase transitions along MTs, while the multivalent MT binding sites in BuGZ coacervates in turn promote MT bundling.
BuGZ phase transition concentrates tubulin and promotes MT polymerization
When different xBuGZ were incubated with rhodamine-tubulin in the presence of nocodazole to prevent MT assembly, tubulin was concentrated in YFP-xBuGZ, but not in YFP-xBuGZΔN, droplets (Figure 5A). When incubated with beads coated with different forms of BuGZ at 4°C in the presence of nocodazole, purified tubulin was pulled down by YFP-xBuGZ or YFP-xBuGZ13S, but not by YFP-xBuGZΔN (Figure 5B).
Figure 5. BuGZ coacervation promotes MT polymerization by concentrating tubulin.
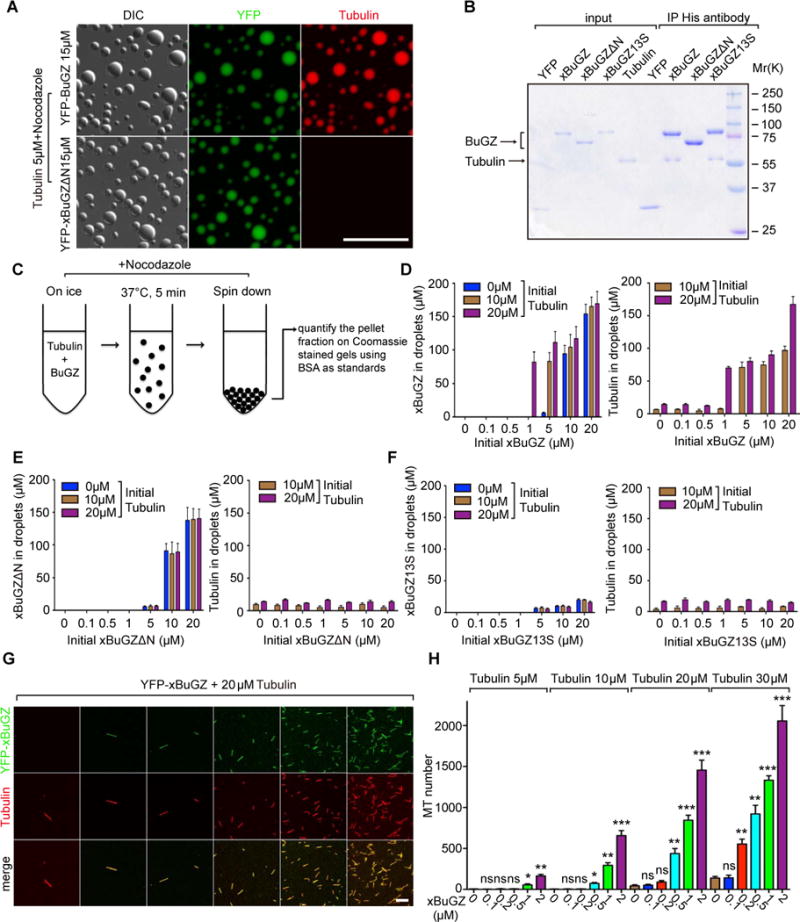
A. Tubulin was concentrated in droplets formed by YFP-xBuGZ, but not by YFP-xBuGZΔN. Scale bar, 20 μm.
B. Anti-His-tag antibody pulled down indicated xBuGZ and tubulin at 4°C.
C. Illustration of the spin down assay.
D. Higher concentrations of YFP-xBuGZ and tubulin are found in droplets (Y-axis) than in initial solution concentrations (X-axis).
E–F. Compared to initial solution concentrations (X-axis), YFP-xBuGZΔN (E) coacervation only concentrated itself, but not tubulin, in droplets (Y-axis), whereas YFP-xBuGZ13S (F) did not coacervate or concentrate itself or tubulin (Y-axis).
G–H. Representative fields of MTs polymerized. MTs were counted in 20 random microscopic fields using a 63× objective. Scale bar, 10 μm.
Error bars, SEM. Student’s t-test: ns, not significant, *p<0.05, **p<0.01, ***p<0.001, three independent experiments.
Next purified BuGZ and tubulin were combined on ice in the presence of nocodazole and incubated at 37°C followed by pelleting the droplets (Figure 5C). Both YFP-xBuGZ and YFP-xBuGZΔN were enriched in droplets by many fold than the initial solution concentrations (Figure 5D–E). Tubulin was greatly enriched in the YFP-xBuGZ, but not the YFP-xBuGZΔN, droplets (Figure 5D–E). Neither YFP-xBuGZ13S nor tubulin was enriched in the pellet fractions (Figure 5F).
Since MT assembly is greatly aided by high tubulin concentrations (Caudron et al., 2000), the concentration of tubulin by BuGZ droplets could lead to enhanced MT assembly. Indeed, in vitro MT assembly assays (Oegema et al., 1999; Zheng et al., 1995) showed that mBuGZ and xBuGZ, but not YFP, YFP-xBuGZΔN, or YFP-xBuGZ13S, stimulated MT assembly even at low concentrations (Figure 5G–H, S5A–C). xBuGZ-B, when present in excess, inhibited the xBuGZ-stimulated MT polymerization, whereas xBuGZ-B3S was much less effective (Figure S5D). Thus the BuGZ-stimulated tubulin concentration and MT polymerization require both MT/tubulin binding and phase transition of BuGZ.
We estimated endogenous xBuGZ in extracts to be ~0.1 μM, and polyethylene glycol, a crowding agent, induced purified xBuGZ at this concentration to undergo phase transition in vitro (Figure S5E–F). Importantly, spindle concentration of xBuGZ was estimated as 0.5–0.86 μM (Figure S5G). Thus endogenous BuGZ could undergo phase transition to promote MT polymerization during spindle assembly.
BuGZ exhibits phase transition property in spindle matrix
If BuGZ in droplets could undergo continuous exchange with solution BuGZ, wild-type and mutant forms of YFP-xBuGZ should have different abilities to exchange into preformed His-xBuGZ droplets. Indeed, YFP-xBuGZ and YFP-xBuGZΔN exchanged into the His-BuGZ droplets efficiently, while YFP-BuGZ5S and YFP-BuGZ13S exhibited weak and background incorporations, respectively (Figure 6A). We also created two additional GFP-tagged fragments, xBuGZ-A and -C, corresponding to 111–187 aa and 376–452 aa in the DLC region (Figure S3F, pink and purple underlines), and their F/Y mutants: xBuGZ-A3S and xBuGZ-C5S, respectively. Similar to xBuGZ-B these fragments did not formed droplets on their own (Figure S6A). GFP-xBuGZ-A, -B, and -C, but not their mutants, exchanged into and disrupted the His-BuGZ droplets (Figure S6B; also see Figure S3G).
Figure 6. Tubulin/MT binding and phase transitions of BuGZ promote spindle matrix assembly and function.
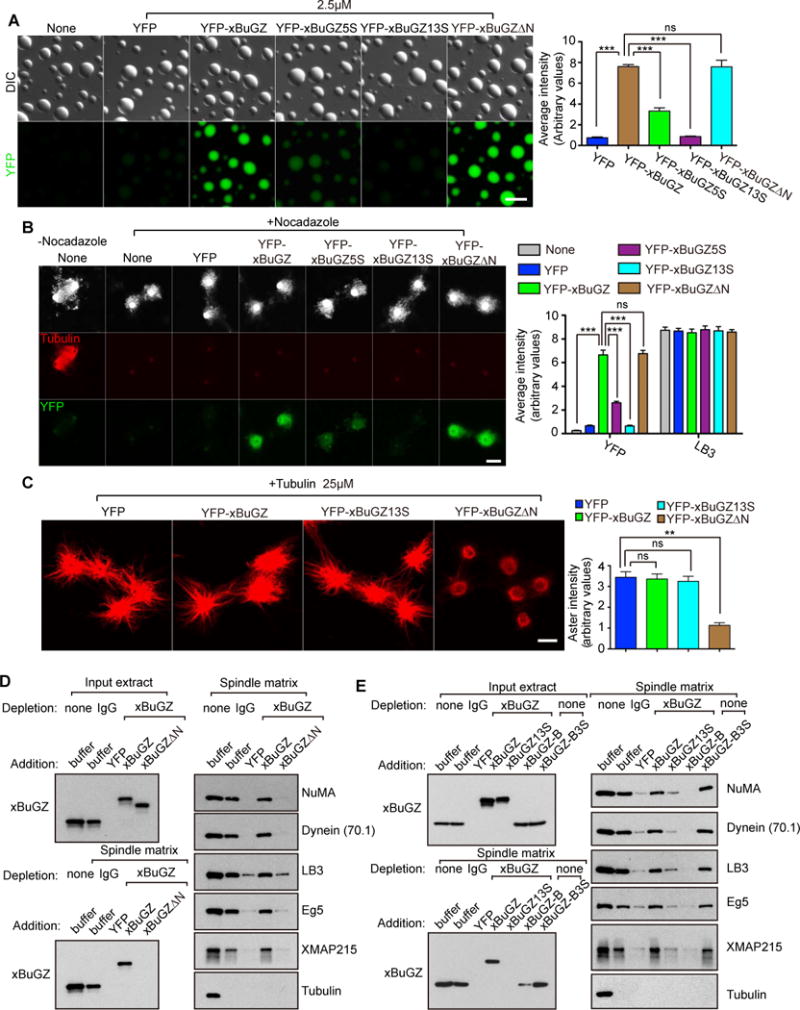
A. Incorporation of YFP-xBuGZ into preformed His-xBuGZ droplets in vitro required Fs and Ys but not the N-terminus of xBuGZ. YFP intensity was quantified in ~50 droplets.
B. Incorporation of 0.1 μM YFP-xBuGZ into isolated spindle matrix required Fs and Ys but not the N-terminus of xBuGZ. ~30 structures were analyzed.
C. Incubation of YFP-xBuGZΔN, but not YFP, YFP-xBuGZ, or YFP-xBuGZ13S, with isolated spindle matrix disrupted matrix-mediated MT assembly. ~30 asters were analyzed.
D. Spindle matrix assembly required MT-binding of BuGZ. YFP-xBuGZ, but not YFP-xBuGZΔN, rescued spindle matrix assembly upon endogenous xBuGZ depletion, as assayed by Western blotting analyses using spindle matrix markers.
E. Spindle matrix assembly required BuGZ coacervation. YFP-xBuGZ, but not YFP-xBuGZ13S, rescued spindle matrix assembly upon endogenous xBuGZ depletion. The addition of GFP-BuGZ-B, but not -BuGZ-B3S, into unperturbed extract disrupted spindle matrix assembly.
Error bars, SEM. Student’s t-test: ns, not significant, **p<0.01, ***p<0.001, three independent experiments. The numbers of structures quantified are for each experiment and condition. Scale bars, 10 μm.
Next we incubated isolated spindle matrices with 0.1 μM wild-type or mutant YFP-xBuGZ. YFP-xBuGZ and YFP-xBuGZΔN exchanged into spindle matrices (marked by LB3) strongly, while YFP-xBuGZ5S exhibited weak exchange (Figure 6B). YFP-xBuGZ13S exhibited only background incorporation into the matrix, similar to the YFP control (Figure 6B). When isolated spindle matrices were incubated with 0.5 μM GFP-xBuGZ-A, -B, or -C to disrupt the coacervation of endogenous xBuGZ, each fragment, but not their mutants, incorporated into and reduced the size of the matrix (Figure S6C). Therefore phase transition is required for both BuGZ incorporation into and the maintenance of preformed spindle matrix, indicating that BuGZ exhibits a phase transition property in the spindle matrix.
Phase transition and tubulin binding of BuGZ promote MT assembly from spindle matrix
The exchange of YFP-xBuGZ or -xBuGZΔN into the spindle matrix replaced most endogenous xBuGZ, while YFP-xBuGZ5S and -xBuGZ13S exhibited increasingly less replacement of endogenous xBuGZ (Figure 6B, S6D). The total BuGZ levels in the matrices, however, remained similar to controls (Figure S6D). After incubating isolated spindle matrices with 0.1 μM of different YFP-xBuGZs followed by addition of 25 μM tubulin with or without nocodozole, the YFP-xBuGZΔN-incorporated spindle matrices had greatly diminished abilities to concentrate tubulin and to promote MT polymerization (Figure 6C, S6E). YFP-xBuGZ13S did not affect such abilities of the matrices (Figure 6C, S6E), consistent with its failure to incorporate (Figure 6B, S6D). Thus phase transition and tubulin-binding of BuGZ are required for the spindle matrix to concentrate tubulin and promote MT assembly.
Phase transition and MT binding of BuGZ promote spindle matrix assembly
To test whether phase transition of xBuGZ is critical for spindle matrix assembly, extract depleted of endogenous xBuGZ was supplemented with purified YFP-xBuGZ, YFP-xBuGZΔN, or YFP-xBuGZ13S. Alternatively, 2 μM of purified BuGZ-B or BuGZ-B3S were added to unperturbed extract. We found that YFP-xBuGZ, but not xBuGZΔN or xBuGZ13S, rescued spindle matrix assembly (Figure 6D–E). xBuGZ-B, but not xBuGZ-B3S, disrupted spindle matrix assembly (Figure 6E). Therefore both MT binding and phase transition of BuGZ are required for spindle matrix assembly.
Phase transition and MT binding of BuGZ promote spindle assembly
To understand whether phase transition of BuGZ is important for spindle assembly, we used GFP-xBuGZ-A, -B, and -C to disrupt phase transition of endogenous xBuGZ in extract. 1 μM each of the fragments disrupted spindle assembly induced by Aurora A beads or sperm, whereas their mutants did not (Figure S7A–D). When isolated spindles were incubated with 0.2 μM GFP-xBuGZ-A, -B, or -C, a concentration that did not disrupt spindle assembly, we found efficient spindle incorporation of these fragments, whereas their mutants incorporated poorly (Figure S7E). Thus these fragments may bind to endogenous BuGZ to disrupt phase transition and spindle assembly.
Next we incubated isolated Aurora A-bead spindles with 0.1 μM YFP-xBuGZ or mutants. YFP-xBuGZ, YFP-xBuGZ5S, and YFP-xBuGZ13S showed a decreasing exchange into spindles (Figure 7A), confirming the importance of BuGZ phase transition in spindle association. YFP-xBuGZΔN exhibited the weakest exchange of all (Figure 7A), consistent with the idea that MT binding of BuGZ facilities its phase transition. We then depleted endogenous xBuGZ in extract and induced spindle formation by Aurora A beads or sperm. Spindle defects due to xBuGZ depletion were fully rescued by purified xBuGZ, but not by xBuGZΔN or YFP-xBuGZ13S (Figure 7B–F).
Figure 7. Tubulin/MT binding and coacervation of BuGZ promotes spindle assembly.
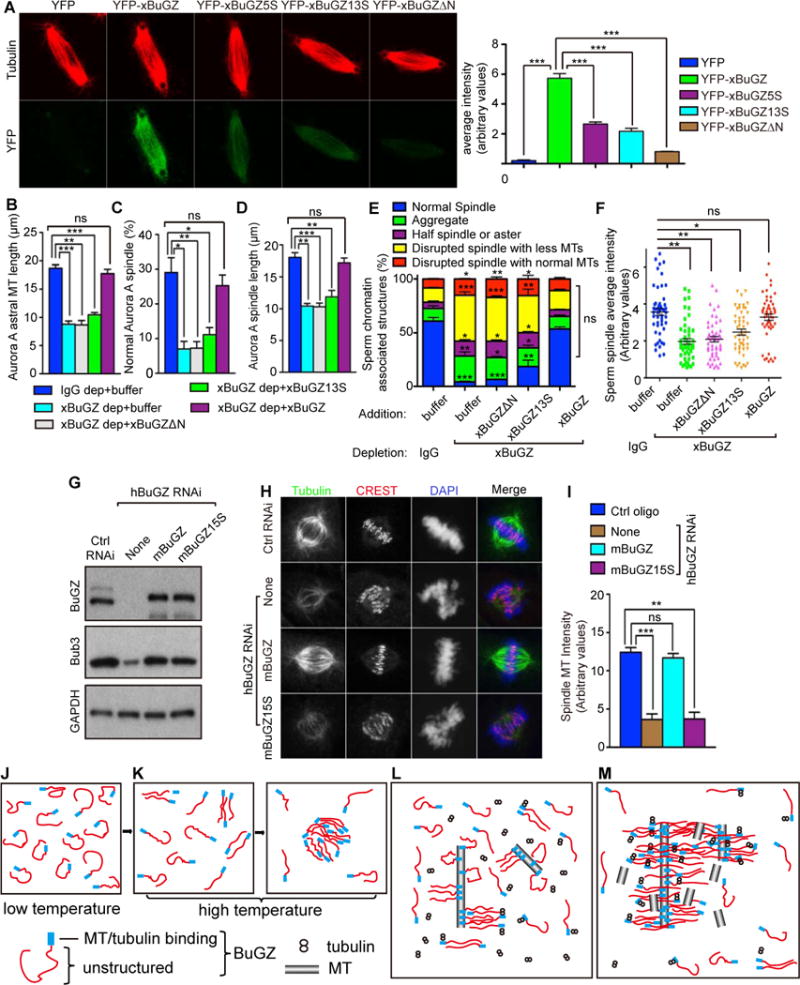
A. Incorporation of 0.1 μM YFP-xBuGZ into isolated spindles required Fs and Ys and MT binding. ~30 spindles were analyzed. Scale bars, 10 μm.
B–D. xBuGZ, but not xBuGZΔN or xBuGZ13S, rescued astral MT length (B), percentages of normal spindles (C), and length of spindles (D) induced by Aurora A beads. ~50 (B, D) or ~500 (C) structures were analyzed.
E–F. Only wild-type xBuGZ, but not xBuGZΔN or xBuGZ13S, rescued defective morphology (E) or MT intensity (F) of spindles in xBuGZ-depleted extracts. ~500 sperm-associated MT structures (E) or ~50 spindles (F) were analyzed.
G–I. Expression of mBuGZ, but not mBuGZ15S, in hBuGZ depleted HeLa cells (G) rescued normal spindle MT intensity (H, I). ~30 spindles were analyzed. Spindles, centromeres, and chromosomes were stained. Scale bars, 5 μm.
J–M. Models for BuGZ phase transition in vitro (J–K) or during spindle assembly (L–M). See explanations in the Discussion.
Error bars, SEM. Student’s t-test: ns, not significant, *p<0.05, **p<0.01, ***p<0.001 from three independent experiments. The numbers of structures analyzed are for each experiment and condition.
In the predicted DLC region, mBuGZ has extra 15 amino acids (including 1Y and 1F) not found in xBuGZ (see Figure S3F). We replaced these two and the other 13 Fs and Ys with S to create mBuGZ15S. HeLa cells were transfected with control or hBuGZ siRNAs followed by expression of RNAi-insensitive Flag-mBuGZ or -mBuGZ15S. Reduction of MT intensity due to hBuGZ RNAi was rescued by wild-type mBuGZ but not by mBuGZ15S (Figure 7G–I). Since xBuGZΔN also failed to rescue MT intensity (Figure 1J–L), both MT binding and phase transition of BuGZ are required to promote spindle assembly in cells.
Discussion
The vague structure-function definitions and uncertain composition of the spindle matrix have made its study both challenging and controversial. Among the studied spindle matrix proteins, LB was initially suggested to be a structural component of the matrix because its depletion resulted in reduced spindle matrix as judged by markers, such as NuMA and Eg5 (Tsai et al., 2006). However, due to the difficulties in studying lamins biochemically, the structure that LB assumes in the spindle matrix remains challenging to decipher. Similarly, despite the identification of several spindle matrix proteins in Drosophia, the assembly mechanism of these proteins remains unknown (Johansen and Johansen, 2007). Through analyses of the spindle matrix component BuGZ, which can be expressed and purified as a soluble protein, we have uncovered its phase transition property in spindle and spindle matrix assembly.
Based on our in vitro studies, we propose that at low temperature the DLC region of BuGZ assumes a variety of quasi-folded states in solution due to weak intra-molecular hydrophobic interactions, which limits intermolecular interactions (Figure 7J). Temperature increase disrupts (or denatures) the quasi-folded BuGZ to allow intermolecular interactions, leading to phase transition (Figure 7K). By studying phase transition of BuGZ in spindle and matrix, we propose that during spindle assembly, the binding of N-terminal BuGZ to MTs limits quasi-folding of BuGZ. By bringing BuGZ molecules close to one another on MTs, intermolecular interactions and phase transition of BuGZ are enhanced (Figure 7L), which in turn bundles MTs and concentrates tubulin (Figure 7M). The elevated tubulin concentration near existing MTs then promotes MT polymerization during spindle assembly.
Our mutational analyses show that BuGZ coacervation depends on highly conserved Fs and Ys in the DLC region of BuGZ. Hydrophobicity-dependent phase transition has been studied in various proteins and polymers. In the well-characterized phase transition of elastin, hydrophobic patches are required for its coacervation, which is critical for subsequent filament assembly (Yeo et al., 2011). Our analyses of the predicted DLC region of BuGZ revealed that highly conserved hydrophobic residues and prolines (P) are enriched in two segments that flank a region with relatively low P and hydrophobicity. This suggests that intermolecular hydrophobic interactions mediated by the hydrophobic patches contribute to BuGZ coacervation. Consistent with this, BuGZ coacervation is dependent on temperature, protein concentration, and Fs and Ys. The aromatic feature of these residues may also mediate phase transitions independent of their hydrophobicity. Inhibition of BuGZ coacervation by the xBuGZ fragments containing F and Y could be caused by disruption of proper alignment of hydrophobic and aromatic residues important for intermolecular BuGZ interactions. BuGZ droplets formed in vitro do not appear to further form filaments based on our preliminary analyses by electron microscopy (unpublished). Although additional studies are required to fully understand the biophysical properties that underline BuGZ phase transition, our findings suggest that unfolding of BuGZ at elevated temperature could drive coacervation through intermolecular hydrophobic interactions.
The temperature-dependent BuGZ phase transition and spindle matrix stability suggest that BuGZ is a structural component of spindle matrix. Indeed, based on known markers of Xenopus spindle matrix, we show that BuGZ in spindle matrix exhibits a phase transition property, which is required for assembly and stability of spindle matrix. The MT-independent and temperature-dependent phase transitions of BuGZ that we uncover here also help to explain why upon MT depolymerization the spindle matrix can retain its structural integrity at RT. Our findings beg the question whether phase transitions represent a key structural property of spindle matrix.
Using PONDR and SEG, we found additional DLC-containing proteins in the proteome of Xenopus spindle matrix (unpublished). Similar analyses revealed the presence of long stretches of DLC regions in several Drosophila spindle matrix proteins, such as Megator, Chromator, EAST, and Skeletor (unpublished). Since multiple proteins can undergo phase transitions together through complicated intermolecular interactions, we speculate that the spindle matrix could be a complex coacervate, whose formation relies on various combinations of proteins depending on organisms and cell types. These coacervates could contain different phases to segregate different biochemical reactions that could communicate with one another. This idea may explain the apparent unrelatedness of some spindle matrix components. Formation of complex coacervates may involve both DLC and structured proteins, which could explain why the largely structured proteins, such as lamin and actin, are found in the spindle and spindle matrix (Zheng and Tsai, 2006; Pickett-Heaps and Forer, 2009). Additionally, proteins participating in phase transition in the spindle and spindle matrix could undergo rapid flux in and out of the structures. The phase transition features may explain the lack of a discrete localization of BuGZ on spindle MTs and a lack of a clearly defined morphology of the spindle matrix upon MT depolymerization.
We show that phase transition of BuGZ does not require MTs, but the MT binding domain of BuGZ helps to not only facilitate BuGZ phase transition along MTs but to concentrate tubulin. Both phase transition and MT binding of BuGZ are required for promoting MT polymerization and bundling in vitro. Importantly, we show that all of these properties are also required for BuGZ to promote spindle assembly. Therefore, one functional consequence of BuGZ-mediated spindle matrix assembly along existing MTs appears to allow efficient polymerization and bundling of MTs in the spindle. By increasing local tubulin concentration, BuGZ could also promote MT nucleation from microtubule nucleators such as γTuRC (Zheng et al., 1995).
Recent studies have shown that MT-dependent branched MT nucleation facilitates spindle assembly (Petry et al., 2013). Since BuGZ could concentrate tubulin along existing MTs, it would be interesting to further study whether BuGZ-mediated phase transitions could help to concentrate other SAFs known to promote branched MT nucleation. Additionally, we have shown that BuGZ interacts with Bub3 to promote the binding of Bub3 to kinetochores in a MT-dependent manner (Jiang et al., 2014). Purified BuGZ-Bub3 complex undergoes more efficient phase transitions than BuGZ alone in vitro (unpublished). It would be interesting to further study whether increasing Bub3 concentration along spindle MTs via phase transition could promote assembly of the Bub3-Bub1-BubR1 complex for its kinetochore loading.
We have shown that nuclear transport receptors such as importin α and β disrupt spindle matrix assembly, which is attenuated by RanGTP (Tsai et al., 2006). Since BuGZ and some other known spindle matrix components are nuclear proteins in interphase, it will be important to further explore whether the RanGTP-importin system regulates phase transitions of BuGZ and other spindle matrix proteins in mitosis. As a conserved protein found in both vertebrate and invertebrate, our study of BuGZ here should open a door to further characterize the structural properties and functions of the spindle matrix in different organisms.
Experimental Procedures
Expression vectors
For all expression constructs see Table S1.
Cell culture and Xenopus egg extract
All cells were grown under standard culturing conditions. CSF egg extracts were prepared as described before and only those that were tested to support spindle assembly were used for further experiments. All assays in egg extracts are detailed in Supplemental Materials.
Immunofluorescence and quantifications
Cells were fixed by 4% paraformaldehyde in PBS for 7 min, followed by extraction in 0.5% Triton in PBS for 10 min. Xenopus MT structures were fixed by ice cold methanol for 5 min. Samples were then blocked in 4% BSA in PBS for >1 h followed by primary antibody incubation overnight at 4°C. Nikon ECLIPSE E800 or Leica SP5 microscopes were used for imaging. To quantify spindle MT intensity or the ratio of BuGZ and MT immunostaining intensities, metaphase spindles in cells, Aurora A, or sperm MT structures were captured at the same exposure using a 63× objective on Nikon ECLIPSE E800. Two 15×15 pixel regions, corresponding to the brightest areas of each half spindle was chosen based on tubulin or BuGZ intensity and the average intensities in these areas were determined. The background fluorescence was subtracted using the intensity measured in areas away from the spindle.
Protein expression, purification, and interaction
Proteins were purified using Glutathione agarose (Sigma) or Ni-NTA agarose (QIAGEN) according to manufacturer’s protocols. Some proteins were further purified by gel filtration. To study tubulin and xBuGZ interaction, cycled tubulin was added to beads coated with purified His-tagged YFP-xBuGZ, YFP-BuGZΔN, or YFP via antibody to 6His and incubated. See details in Supplemental Materials.
BuGZ phase transitions
Purified mBuGZ, xBuGZ, or control proteins were thawed on ice and diluted into ice-cold buffers on ice followed by incubation at 37°C for 5 min and DIC or fluorescence microscopy. For turbidity assay, purified proteins were diluted in XB buffer on ice (300 μl final) and then loaded into 750-μl cuvettes (28F-Q-10, Starna Cells, CA) in a cold room. Turbidity was measured at 440 nm using a Cary 300 UV-VIS spectrophotometer with a Peltier-Thermostatted Multi-cell Holder (Agilent Technologies). Temperature ramp rate, 0.5°C/min from 4 to 60°C. To estimate protein concentrations in BuGZ droplets, 100 μl of protein samples were incubated at 37°C for 5 min followed by centrifugation at 2000 rpm in a microfuge (Eppendorf E5430) for 5 min at RT. Proteins in pellet fractions were quantified by Coomassie blue staining using BSA as standard. See details in Supplemental Materials.
MT polymerization, bundling, and binding
Purified xBuGZ and cycled tubulin were mixed on ice. MT polymerization was performed at 37°C for 5 min. After fixation, MTs were counted in 20 random microscopy fields using a 63× objective. The same amount of rhodamine-MT seeds were mixed with different purified xBuGZ proteins and incubated at 37°C for 5 min and imaged immediately. MT length and intensity were measured based on micrographs. To visualize the binding of wild-type and mutant xBuGZ to MTs, rhodamine-labeled and taxol-stabilized MTs were mixed with different versions of YFP-xBuGZ. See details in Supplemental Materials.
Sequence analysis for protein disorder and low complexity
The PONDR (http://www.disprot.org/index.php) (Xue et al., 2010) and SEG programs (http://mendel.imp.ac.at/METHODS/seg.server.html) (Wootton, 1994) were used to analyze the DLC regions of BuGZ and other spindle matrix proteins at default settings. See details in Supplemental materials.
Supplementary Material
Acknowledgments
We thank Dr. Andreas Merdes for the NuMA antibody, Ona Martin for proof reading, Ona Martin and Lynne Hugendubler for technical support, Dr. Pengcheng Zhang for help with the turbidity assay, and the Zheng lab for critical reading. Supported by the Chinese Academy of Sciences (XDA01010107 to X.Z.), Ministry of Science and Technology of China (2014CB964803 to X.Z.), National Science Foundation of China (31010103910 to X.Z. and Y.Z.), and GM056312 and GM06023 (to Y.Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
Conceptualization, YZ; Methodology, H.J., S.W., Y.Z., H.C., and X.Z.; Investigation, H.J., S.W., X.H., and Y.H.; Writing-Original draft, Y.Z., H.J., and S.W.; Writing-Review & Editing, Y.Z., X.Z., H.C., S.W., and H.J.; Funding Acquisition, Y.Z. and X.Z. Resources, Y.Z., X.Z. and H.C.; Supervision, Y.Z. and X.Z.
References
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugues J, Needleman D. Physical basis of spindle self-organization. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18496–18500. doi: 10.1073/pnas.1409404111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron N, Valiron O, Usson Y, Valiron P, Job D. A reassessment of the factors affecting microtubule assembly and disassembly in vitro. Journal of molecular biology. 2000;297:211–220. doi: 10.1006/jmbi.2000.3554. [DOI] [PubMed] [Google Scholar]
- Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Tao L, Brust-Mascher I, Wollman R, Scholey JM. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. The Journal of cell biology. 2010;188:49–68. doi: 10.1083/jcb.200908150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian L, Xia X, Venkitaramani DV, Johansen KM, Johansen J, Andrew DJ, Forer A. Titin in insect spermatocyte spindle fibers associates with microtubules, actin, myosin and the matrix proteins skeletor, megator and chromator. Journal of cell science. 2007;120:2190–2204. doi: 10.1242/jcs.03465. [DOI] [PubMed] [Google Scholar]
- Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Gatlin JC, Matov A, Danuser G, Mitchison TJ, Salmon ED. Directly probing the mechanical properties of the spindle and its matrix. The Journal of cell biology. 2010;188:481–489. doi: 10.1083/jcb.200907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Rebhun LI. The structure and some properties of the isolated mitotic apparatus. Journal of cell science. 1969;4:179–209. doi: 10.1242/jcs.4.1.179. [DOI] [PubMed] [Google Scholar]
- Goodman B, Channels W, Qiu M, Iglesias P, Yang G, Zheng Y. Lamin B counteracts the kinesin Eg5 to restrain spindle pole separation during spindle assembly. The Journal of biological chemistry. 2010;285:35238–35244. doi: 10.1074/jbc.M110.140749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Kimura K, Kimura A. Localized accumulation of tubulin during semi-open mitosis in the Caenorhabditis elegans embryo. Molecular biology of the cell. 2012;23:1688–1699. doi: 10.1091/mbc.E11-09-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annual review of cell and developmental biology. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Jiang H, He X, Wang S, Jia J, Wan Y, Wang Y, Zeng R, Yates J, 3rd, Zhu X, Zheng Y. A microtubule-associated zinc finger protein, BuGZ, regulates mitotic chromosome alignment by ensuring Bub3 stability and kinetochore targeting. Developmental cell. 2014;28:268–281. doi: 10.1016/j.devcel.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen KM, Forer A, Yao C, Girton J, Johansen J. Do nuclear envelope and intranuclear proteins reorganize during mitosis to form an elastic, hydrogel-like spindle matrix? Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2011;19:345–365. doi: 10.1007/s10577-011-9187-6. [DOI] [PubMed] [Google Scholar]
- Johansen KM, Johansen J. Cell and molecular biology of the spindle matrix. International review of cytology. 2007;263:155–206. doi: 10.1016/S0074-7696(07)63004-6. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Current biology. 1999;CB 9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie RJ, Hird RB, Wilson L, McIntosh JR, Scholey JM. Kinesin is associated with a nonmicrotubule component of sea urchin mitotic spindles. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:2771–2775. doi: 10.1073/pnas.84.9.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lince-Faria M, Maffini S, Orr B, Ding Y, Claudia F, Sunkel CE, Tavares A, Johansen J, Johansen KM, Maiato H. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. The Journal of cell biology. 2009;184:647–657. doi: 10.1083/jcb.200811012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs D. [Walter Flemming, discoverer of chromatin and mitotic cell division] Orvosi hetilap. 1981;122:349–350. [PubMed] [Google Scholar]
- Ma L, Tsai MY, Wang S, Lu B, Chen R, Iii JR, Zhu X, Zheng Y. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nature cell biology. 2009;11:247–256. doi: 10.1038/ncb1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR. An abundance of tubulins. Trends in cell biology. 2000;10:537–542. doi: 10.1016/s0962-8924(00)01857-2. [DOI] [PubMed] [Google Scholar]
- Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, Zheng Y. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. The Journal of cell biology. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell. 2013;152:768–777. doi: 10.1016/j.cell.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps J, Spurck T, Tippit D. Chromosome motion and the spindle matrix. The Journal of cell biology. 1984;99:137s–143s. doi: 10.1083/jcb.99.1.137s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps J, Forer A. Mitosis: spindle evolution and the matrix model. Protoplasma. 2009;235:91–99. doi: 10.1007/s00709-009-0030-2. [DOI] [PubMed] [Google Scholar]
- Qi H, Rath U, Ding Y, Ji Y, Blacketer MJ, Girton J, Johansen J, Johansen KM. EAST interacts with Megator and localizes to the putative spindle matrix during mitosis in Drosophila. Journal of cellular biochemistry. 2005;95:1284–1291. doi: 10.1002/jcb.20495. [DOI] [PubMed] [Google Scholar]
- Qi H, Rath U, Wang D, Xu YZ, Ding Y, Zhang W, Blacketer MJ, Paddy MR, Girton J, Johansen J, et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Molecular biology of the cell. 2004;15:4854–4865. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath U, Wang D, Ding Y, Xu YZ, Qi H, Blacketer MJ, Girton J, Johansen J, Johansen KM. Chromator, a novel and essential chromodomain protein interacts directly with the putative spindle matrix protein skeletor. Journal of cellular biochemistry. 2004;93:1033–1047. doi: 10.1002/jcb.20243. [DOI] [PubMed] [Google Scholar]
- Schibler MJ, Pickett-Heaps JD. Mitosis in Oedogonium: spindle microfilaments and the origin of the kinetochore fiber. European journal of cell biology. 1980;22:687–698. [PubMed] [Google Scholar]
- Scholey JM, Rogers GC, David JS. Mitosis, microtubules, and the matrix. Journal of Cell Biology. 2001;154:261–266. doi: 10.1083/jcb.200101097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer N, Weiss M, Maiato H. The dynamic spindle matrix. Current opinion in cell biology. 2014;28:1–7. doi: 10.1016/j.ceb.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Shimamoto Y, Maeda YT, Ishiwata S, Libchaber AJ, Kapoor TM. Insights into the micromechanical properties of the metaphase spindle. Cell. 2011;145:1062–1074. doi: 10.1016/j.cell.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo CM, Herman JA, Olsen JB, Ding Y, Corrin P, Girard EJ, Olson JM, Emili A, DeLuca JG, Paddison PJ. BuGZ is required for Bub3 stability, Bub1 kinetochore function, and chromosome alignment. Developmental cell. 2014;28:282–294. doi: 10.1016/j.devcel.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Zheng Y. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Current biology. 2005;CB 15:2156–2163. doi: 10.1016/j.cub.2005.10.054. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Cai S, Khodjakov A. Mechanisms of chromosome behaviour during mitosis. Nature reviews Molecular cell biology. 2010;11:91–102. doi: 10.1038/nrm2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DL, Wang D, Jin Y, Rath U, Wang Y, Johansen J, Johansen KM. Skeletor, a novel chromosomal protein that redistributes during mitosis provides evidence for the formation of a spindle matrix. The Journal of cell biology. 2000;151:1401–1412. doi: 10.1083/jcb.151.7.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein H, Bass HW, Cande WZ. DSK1, a kinesin-related protein involved in anaphase spindle elongation, is a component of a mitotic spindle matrix. Cell motility and the cytoskeleton. 1998;41:214–224. doi: 10.1002/(SICI)1097-0169(1998)41:3<214::AID-CM3>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Computers & chemistry. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochimica et biophysica acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Rath U, Maiato H, Sharp D, Girton J, Johansen KM, Johansen J. A nuclear-derived proteinaceous matrix embeds the microtubule spindle apparatus during mitosis. Molecular biology of the cell. 2012;23:3532–3541. doi: 10.1091/mbc.E12-06-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Wang C, Li Y, Ding Y, Rath U, Sengupta S, Girton J, Johansen KM, Johansen J. The spindle matrix protein, Chromator, is a novel tubulin binding protein that can interact with both microtubules and free tubulin. PloS one. 2014;9:e103855. doi: 10.1371/journal.pone.0103855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GC, Keeley FW, Weiss AS. Coacervation of tropoelastin. Advances in colloid and interface science. 2011;167:94–103. doi: 10.1016/j.cis.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Zheng Y. A membranous spindle matrix orchestrates cell division. Nature reviews Molecular cell biology. 2010;11:529–535. doi: 10.1038/nrm2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Tsai MY. The mitotic spindle matrix: a fibro-membranous lamin connection. Cell cycle. 2006;5:2345–2347. doi: 10.4161/cc.5.20.3365. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


