Abstract
Although the field of pharmacogenetics has existed for decades, the implementation of, pharmacogenetic testing in clinical care has been slow. There are numerous publications, describing the barriers to clinical implementation of pharmacogenetics. Recently, several freely, available resources have been developed to help address these barriers. In this review we, discuss current programs that use preemptive genotyping to optimize the pharmacotherapy of, patients. Array-based preemptive testing includes a large number of relevant pharmacogenes, that impact multiple high-risk drugs. Using a preemptive approach allows genotyping results to, be available prior to any prescribing decision so that genomic variation may be considered as, an inherent patient characteristic in the planning of therapy. This review describes the common, elements among programs that have implemented preemptive genotyping and highlights key, processes for implementation, including clinical decision support.
Keywords: Pharmacogenomics, Precision medicine, Individualized medicine, Personalized medicine, Clinical decision support
Introduction
Clinical pharmacogenetics research has the goal of defining what genetic variation is important for influencing interpatient variability in drug response. The field has existed since the 1950s, but as the laboratory tools for interrogating genomic variation have continued to evolve uncovering rare variants and multigenic effects, the need for large clinical studies to generate solid evidence and accompanying laboratory mechanistic studies have become increasingly important. However, there are some gene/drug pairs for which evidence is compelling and that meet the threshold for clinical implementation. For these select gene/drug pairs, there is already sufficient research to justify some degree of clinical implementation; yet, implementation of pharmacogenetic testing in the clinic is rare. In this review we discuss the use of pharmacogenetic testing as a preemptive clinical tool and focus not on whether to implement pharmacogenetics, but how to do so.
As has been noted (1-6), focusing on pharmacogenetics as an early area for clinical implementation has several advantages over other areas of clinical genomics, including avoiding some ethical/insurability issues, with an initial focus on genetic variants that have importance for drug dosing but that have little “incidental” importance for disease risk. A growing number of clinically actionable pharmacogenetic variants exist (7-21). Incorporating pharmacogenetic testing into patient care has the potential to improve clinical outcomes, decrease length of treatment, decrease cost of treatment, and decrease adverse effects from drug therapy (22).
Many health care providers who have implemented pharmacogenetics have done so on a gene-by-gene basis. The decision to perform a genetic test in this way is based on the likelihood that a “high-risk” drug (one substantially influenced by a specific genetic variation) will be prescribed for a given patient or group of patients. An advantage of this method is the increased likelihood that the genetic test result is applied by the clinician, because the prescribing decision is linked to the performance of the genetic test. Pharmacogenetic test results may then be used as one characteristic along with others to dose medications, as exemplified by warfarin-dosing algorithms that use both genetic and non-genetic factors to individualize warfarin doses (23-26). However, this “per gene” reactive testing has disadvantages such as high expense; a slow turn-around time, which may be too slow to be useful for initial prescribing decisions; and a substantial knowledge base needed for clinicians to be aware of important gene/drug relations to prompt ordering each genetic test. Indeed, although such single-gene tests have been available for many years from clinical laboratories, uptake in the clinic has been slow, therefore, high-risk medications are often given to patients at high risk for drug failure or adverse drug effects due to lack of pharmacogenetic testing.
To be practical for use in prescribing decisions, pharmacogenetic test results should ideally be available preemptively. By preemptive, we mean that the test result is available in the medical record as a pre-prescription patient characteristic: the test result has not been ordered because a specific pharmacogenetically high-risk drug is being contemplated, but the result is available because a broad screening of multiple genes has already been performed. This preemptive approach may counteract many of the disadvantages of reactive pharmacogenetic testing (27-29). The recent availability of high-quality genotyping arrays and other multiplex approaches that are oriented to pharmacogenetics and reasonably priced makes preemptive genotyping financially feasible. Unlike pharmacogenetic testing for individual genes, array-based preemptive testing can include a large number of relevant pharmacogenes that cover most, if not all, pharmacogenetically high-risk drugs. The test results are then available prior to any prescribing decision involving these high-risk drugs, consistent with the vision that in every prescribing decision, patient genomic variation will be considered as an inherent patient characteristic (30), as are age, weight, renal function, and allergy status. A recent study reviewed the prescription history of 52,942 patients at Vanderbilt University Medical Center, and considering six medications with adverse effects linked to pharmacogenes, the researchers found that a preemptive genotyping program would potentially have avoided 398 adverse events (31).
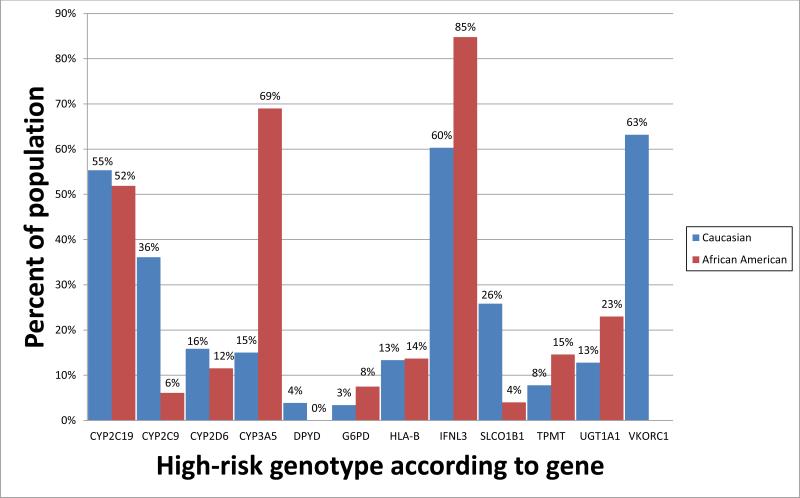
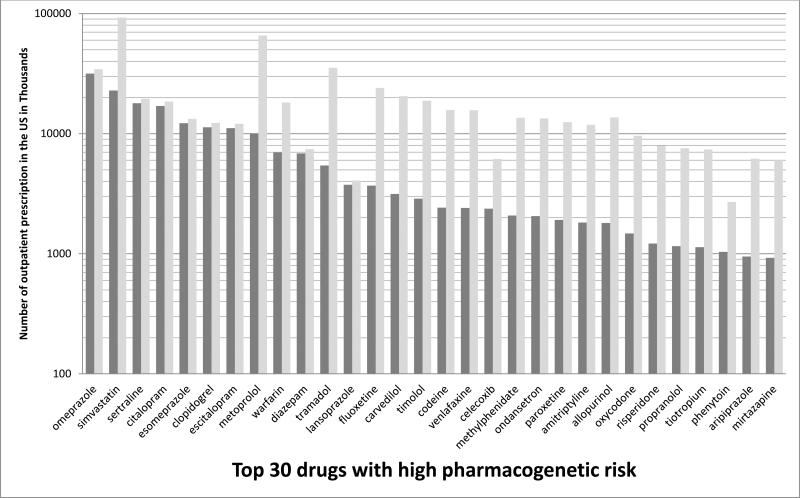
It should be acknowledged that only a fraction of commonly used medications are candidates for clinical action based on pharmacogenetics now. Approximately 1000 drugs are approved by the United States Food and Drug Administration, and of these, fewer than 100 are candidates for pharmacogenetic testing and clinical action (excluding cancer drugs whose prescribing may be influenced by somatically acquired genomic variants). Actionable drug prescribing is most strongly linked to approximately 12 commonly tested genes (Supplement Table 1, www.pharmgkb.org/cpic/pairs). The estimated proportion of whites and blacks harboring high-risk genomic variants for 12 genes with at least one known actionable inherited variant varies substantially based on self-declared race (Figure 1). Assuming each of these genes is inherited independently, 98.5 % of whites and 99.1% of blacks in the United States have at least one high-risk diplotype. For pharmacogenetics to impact the outcome of a patient, the patient must have received a pharmacogenetically high-risk drug and had an underlying actionable pharmacogenetic phenotype. In the United States, the 30 most commonly prescribed pharmacogenetically high-risk drugs accounted for 738 million prescriptions (32) (Figure 2). The number of prescriptions dispensed in the United States for each high-risk drug in 2013 can be found in Supplemental Table 2. At St. Jude, 2023 of 4245 (48%) pediatric patients received at least one pharmacogenetically high-risk drug in a one-year period, comparable to the 54% of adult patients reported by Vanderbilt Medical Center (31).
Figure 1.
Percentage of individuals expected to have a high-risk diplotype for 12 genes identified by the Clinical Pharmacogenetics Implementation Consortium (CPIC) to have at least one known actionable inherited variant plotted by self-reported race category [white (red bars) or black (blue bars)]. For the genes CYP2C9, CYP2C19, CYP2D6, CYP3A5, SLCO1B1, TPMT, UGT1A1, and VKORC1, diplotype frequencies were obtained from the St. Jude Children's Research Hospital PG4KDS study, based on data from 624 black patients and 732 white patients. Due to small sample size in other race categories, other race categories were omitted from the figure. For the genes for which validated diplotype data were not yet available from the PG4KDS study, (DPYD, G6PD, HLA-B, and IFNL3), high-risk diplotype frequencies were estimated using published allele frequency data (7; 8; 13; 15; 48).
High risk diplotypes were considered as follows: for CYP2C19, diplotypes containing a *2 , *3 or *17 allele; for CYP2C9, diplotypes containing a *2 or *3 allele; for CYP2D6, diplotypes resulting in activity scores of < 1 or > 2; for CYP3A5, diplotypes containing a *1 allele; for DPYD, diplotypes containing *2A, *3, or the rs67376798 A variant; for G6PD, those with G6PD deficient phenotypes; for HLA-B, diplotypes containing a *5701 or *5801 allele; for IFNL3, diplotypes containing an rs12979860 T allele; for SLCO1B1, diplotypes containing a *5 or *15 allele; for TPMT, diplotypes containing a *2, *3A, *3B, *3C, *4 or *8 allele; for UGT1A1, diplotypes containing two copies of the *28 allele; and for VKORC1, diplotypes containing an rs9923231 A allele. High-risk diplotypes for blacks were not calculated for VKORC1 due to the low predictive value of genotype-driven warfarin dosing in this population.
Figure 2.
Number of outpatient prescriptions dispensed in the United States for the calendar year 2013 for top 30 drugs with high pharmacogenetic risk plotted by drug and diplotype risk category. The dark grey portion of each bar represents prescriptions potentially prescribed to blacks or whites with a high-risk diplotype for the applicable gene(s); the light grey portion represents those prescribed to those without a high-risk diplotype. Total number of prescriptions for each drug was collected from the IMS Health (IMS) National Prescription Audit proprietary prescription database (32). This database contains all retail prescriptions filled from a representative sample of 35,000 (73% of the approximately 50,000) U.S.-based retail pharmacies. IMS then proportionately extrapolates their data on the basis of populations served by the included pharmacies to provide weekly estimates of all prescriptions filled in the United States for these drugs. The National Prescription Audit database does not track prescriptions filled by in-hospital pharmacies.
The number of prescriptions potentially prescribed to black or white patients with a high-risk diplotype per drug was calculated as follows: (total number of prescriptions for a drug) * (percent of Americans with Caucasian ancestry (74.8%)) * (percent of high-risk diplotypes in whites for each corresponding gene as shown in Figure 1) + (total number of prescriptions for a drug) * (percent of Americans with African American ancestry (12.6%)) * (percent of high-risk diplotypes in blacks for each corresponding gene as shown in Figure 1), where the percent of Caucasians and African Americans was derived from the 2010 U.S. census (http://www.census.gov/2010census/). For warfarin, only whites with a high-risk diplotype were used in the calculation. If the drug is affected by two genes, the presence of a high-risk diplotype for either gene was considered as the presence of a high-risk diplotype for that drug.
Barriers
The challenges to implementation of pharmacogenetics have been reviewed elsewhere (33-43) and can be broadly grouped into lack of guidance for how to use pharmacogenetic data in the clinic, absence of infrastructure to handle genetic data, clinicians’ resistance to use genetic data in clinical practice, and concerns about costs and reimbursement. Even with the increasing availability of clinical guidelines for specific gene/drug pairs that clearly indicate how prescribing should be modified based on test results, (7-21; 44; 45) actual implementation of pharmacogenetically-guided prescribing remains a challenge. The result is that pharmacogenetically high-risk drugs continue to be prescribed to patients whose relevant genotype is unknown.
Another barrier to the implementation of pharmacogenetics into routine clinical practice is the deficit of support systems and infrastructure to handle large amounts of genomic data that can be generated from an array-based assay. Pharmacogenetic data are most useful when the electronic health record (EHR) contains systems that provide the data in a manner and time that render them readily available for use in the clinic: the data should be apparent to the prescriber at the time of prescribing and they must be available throughout the lifetime of the patient. The several days delay in turn-around time on a send-out pharmacogenetic test result may render the test less useful, as is the case for warfarin (17). The lack of infrastructure to handle genomic data is particularly problematic due to fragmentation of the United States healthcare system. Between 2000 and 2002, United States Medicare beneficiaries saw a median of seven physicians in four different offices (46). Each provider has their own EHR and rarely can their software systems communicate with each other. This fragmentation causes problems for having data available preemptively, as well as for consistent interpretation and application of pharmacogenetic test results. An advantage of genomic data is that they need to be generated only once for the lifetime of a patient. However, the lack of a single EHR in which laboratory and medication data are integrated reduces the utility of genomic data. As a patient moves from one provider to the next, his/her genomic data, which are relevant over a lifetime, do not necessarily follow.
Clinician resistance to widespread implementation of these tests may arise from gaps in education about what tests are available and how to order, interpret, and incorporate pharmacogenetic tests in the context of other clinical variables. Many clinicians completed their training in the pre-genomics era or otherwise did not have formal education in the field of pharmacogenetics. Recent surveys of clinicians in the U.S. reported that as few as 29% had received education in pharmacogenetics (47; 48). Pharmacists and physicians who reported feeling well-informed about the availability and applications of testing and had received pharmacogenetics education were more often early adopters of pharmacogenetic testing (47; 49; 50).
Another major barrier may be difficulty in receiving reimbursement for these clinical tests. A 2013 study analyzing barriers to reimbursement using a one drug/one test model, found that payors’ reimbursement varied greatly in terms of gene/drug pairs and amount reimbursed (51). FDA recommendations for testing before using the drug had little effect on utilization of genetic testing; cost of the test as well as lack of clinical evidence were cited as some of the most significant reasons for limited reimbursement. New models for reimbursement are required that focus on the interpretation of multiple results arising from array-based tests, instead of a single result from a single test (37), and for reimbursement of preemptive tests. With costs of whole exome sequencing now less than $1000, and array-based genotyping less than that, the expense of genomic testing (a once-in-a-lifetime test), will soon be trivial compared to medical procedures that are performed repeatedly to diagnose and monitor various conditions (e.g. echocardiograms, MRI scans, physical exams). However, there will be expenses involved for personnel to interpret the test results, to produce reports for clinical use, and to oversee the EHR technology. But in health care systems that reimburse for sick care, as opposed to preventing adverse outcomes, there is still a barrier as to which entity bears the cost of preemptive genotyping that will serve the patient on a life-long basis, and this has hindered adoption of multi-gene testing.
Resources
The Pharmacogenomic Knowledgebase, PharmGKB (www.pharmgkb.org), offers a repository for pharmacogenetic information with both a clinical and research oriented focus (52). Originally created in 1999, this website-based tool is used by researchers and clinicians alike to address pharmacogenetic questions. The website can be searched by using numerous terms including genes, variants, drugs, and diseases. As of April 2013, over 5000 genetic variants had been annotated covering over 900 drug-related genes and over 600 drugs (52). The knowledgebase includes “Very Important Pharmacogene” summaries, pharmacologic pathways, and tables with clinical annotations and appropriate references. PharmGKB is home to summaries of clinical pharmacogenetic dosing guidelines, including those developed by the Clinical Pharmacogenetics Implementation Consortium (CPIC).
CPIC was established in 2009 as a joint effort between PharmGKB and the NIH's Pharmacogenomics Research Network to address the major barriers of how to use pharmacogenetic information in the clinical setting (48). The consortium consists of clinicians and scientists with expertise in pharmacogenetics, pharmacogenomics, and laboratory medicine. One of CPIC's major goals is to provide peer-reviewed, evidence based, freely available gene/drug based guidelines to facilitate clinical adoption of pharmacogenetics. CPIC guidelines address the most commonly cited barrier to clinical implementation of pharmacogenetics, namely, how to interpret genotypes and how to use that information to alter prescribing (48). These guidelines provide clear and specific therapeutic recommendations for use of pharmacogenetic tests. Written with the assumption that the genomic data are already available, the guidelines answer the question of how to use genetic test results, rather than whether to collect them. The evidence for each recommendation is graded on quality and each recommendation is graded on strength using standardized criteria. The guidelines adhere to most of the Institute of Medicine's Standards for Developing Trustworthy Clinical Practice Guidelines and are updated regularly to reflect changes in evidence (53). As of March 2014, 12 guidelines have been published covering 10 genes and 24 drugs (http://www.pharmgkb.org/view/dosing-guidelines.do?source=CPIC#). The guidelines are regularly updated and are supported with supplementary tables to facilitate translation of the guidelines into machine-readable EHR content. The list of planned CPIC guidelines is also updated regularly (www.pharmgkb.org/cpic/pairs).
The Royal Dutch Association for the Advancement of Pharmacy created the Pharmacogenetic Working Group in 2005 to meet the need for evidence-based therapeutic pharmacogenetic recommendations. The group consists of 15 members from the Netherlands including pharmacists, physicians, chemists, pharmacologists, epidemiologists, and toxicologists. Recommendations from the group have been published in two papers in 2008 and 2011 (44; 45). The recommendations do not indicate who should be tested, but rather what to do with the test results including drug avoidance and dose manipulations. To date, recommendations for 53 drugs and 11 genes have been published (45). The recommendations are specific to the drug, gene, and genotype/phenotype. The evidence for each recommendation is graded on quality of evidence and clinical relevance. Detailed analyses of the supporting data are available in Dutch from the Working Group. Their recommendations are posted on PharmGKB and several members of the Working Group also participate in CPIC.
The gene/drug guidelines discussed above provide recommendations on how to use pharmacogenetic results, not whether to order genetic tests. There are numerous other clinical pharmacogenetic guidelines that have been released for individual genetic tests or for individual drugs (54-58). However, most of these guidelines deal with the issue of whether and when to order genetic tests, and secondarily, how to use the results. For the most part, a preemptive approach to genotyping obviates the decision about whether to test.
The United States Food and Drug Administration maintains a website listing over 100 drugs available in the United States which have pharmacogenetic data in their package labeling (http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm). The placement of pharmacogenetic data in the label differs among drugs, sometimes appearing in sections on clinical response, adverse events, dosing, warnings, or mechanisms of drug action. Actionability of the product labeling varies greatly depending on the drug, but specific clinical recommendations on dosing and use of the drugs based on specific pharmacogenetic test results are relatively rare. Of note, eight drugs contain boxed warnings regarding pharmacogenetic data (i.e., arsenic trioxide, rasburicase, valproic acid, abacavir, clopidogrel, lenalidomide, carbamazepine, and codeine), indicating that the FDA considers the drug to carry a significant risk of serious or even life-threatening adverse effects.
The Genetic Testing Registry provides a clearinghouse for information regarding laboratories offering clinical genetic tests (http://www.ncbi.nlm.nih.gov/gtr/) (59). Laboratories voluntarily submit information about the tests they offer. This website includes details for each test such as purpose, methods, clinical validity, utility, and contact information.
Current programs for preemptive pharmacogenetic testing
Only a limited number of sites have published on their experience to routinely use preemptive pharmacogenetic testing to guide prescribing. In response to the slow integration of pharmacogenetics into clinical practice, the Pharmacogenomics Research Network formed the Translational Pharmacogenetic Program (TPP) to implement routine pharmacogenetically based prescribing within diverse health care systems (28). The TPP originally comprised six sites (Mayo Clinic, Ohio State University, St. Jude Children's Research Hospital, University of Florida, University of Maryland, and Vanderbilt University Medical Center), each charged with developing methods to implement pharmacogenetics into clinical care at their respective site. The program has recently expanded to include the University of Chicago and Brigham and Women's Hospital. Models using a preemptive approach and on-demand/point-of-care testing are being investigated. TPP is among the first groups to identify and overcome real-world barriers to adoption of evidence-based pharmacogenetics in diverse health care settings and to provide practical knowledge for broad dissemination. During this process, logistic barriers to implementation of pharmacogenetics are identified and resolved and the solutions are disseminated.
A preemptive model for implementing pharmacogenetics presents a unique information technology challenge: to provide genomic data to clinicians when the data are most useful and to provide readily available access to the data over the lifetime of the patient. Of the eight sites, several sites have developed programs using a preemptive pharmacogenetic approach (Table 1).
Table 1.
Summary of the genotyping platform used by 5 U.S. institutions to implement array-based preemptive pharmacogenetic testing
| Institution | Genotyping platform | Number of genes assayed |
|---|---|---|
| Mayo Clinic (43) | PGRN-Seq | 84 |
| Mount Sinai Medical Center (42) | PGRN-Seq | 84 |
| St. Jude Children's Research Hospital (65) | Affymetrix DMET Plus array | 230 |
| University of Florida and Shands Hospital (35) | Life Technologies Quant Studio Open Array | 120 |
| Vanderbilt University Medical Center (69) | VeraCode ADME Core Panel | 34 |
St. Jude Children's Research Hospital
After years of using single-gene tests, at St. Jude Children's Research Hospital (60-62), we have gained experience in preemptive implementation of pharmacogenetic tests in the clinic by using array-based tests, which overcomes many of the barriers described above (63). Our group has used single-gene tests, primarily for TPMT and CYP2D6, to guide clinical prescribing decisions for thiopurines and codeine, respectively (60-62). In May 2011, a clinical research protocol, PG4KDS, was opened with the goal of selectively migrating array-based pharmacogenetic tests into routine patient care, so that results would be available preemptively (64). Genotyping is performed in a CLIA-approved laboratory, using the Affymetrix DMET Plus array supplemented with a CYP2D6 copy number determination using a quantitative PCR assay. The array interrogates 1936 variants in 230 genes (65). The genotype data are stored as individual, patient-specific files for each gene in a database separate from the EHR.
The Pharmacogenetics Oversight Committee (POC), a subcommittee of the Pharmacy and Therapeutics (P&T) committee, evaluates the available evidence of a gene/drug pair for migration into the EHR, with CPIC guidelines playing a critical role in the evaluation (66). Gene/drug pairs with adequate evidence and clinical decision support (CDS) are moved from research databases into the EHR and are then available for clinical purposes for all past and future enrolled patients (67). Once a gene has been selected for migration to the EHR, multiple steps must be completed so that adequate CDS and EHR infrastructure is available for the genetic tests to be optimally used. The passive and active CDS are utilized to inform prescribers of a patient's pharmacogenetic results.
Translation tables relate each diplotype result to a drug-related phenotype. These phenotypes are assigned a clinical priority status, if the clinical priority is high-risk or actionable, then a problem list entry for the EHR is also assigned (66). A high-risk phenotype is one for which changes in usual prescribing would be recommended for at least one drug, for example, codeine prescribed to a CYP2D6 ultra-rapid metabolizer. Custom codified problem list entries were created because currently available coding systems, such as SNOMED, ICD-9, or ICD-10, do not have the specificity needed to accurately trigger clinically actionable CDS (67).
For each gene result, basic quality control steps are taken to minimize sample mix-up, to identify inconsistencies with prior genotypes, and to resolve any discrepancies with prior data. For each gene, we build translation tables (based on CPIC guidelines) to link the diplotype to phenotype, priority status, a clinically relevant interpretation of the phenotype, and, if applicable, the actionable pharmacogenetic phenotype designation (66; 67). Assigning the diplotype is key; this drives the assignment of phenotype, the interpretation of the test results, and the downstream interruptive CDS. The interpretations are provided in a written pharmacogenetic consultation linked in the EHR to the test result. We created a web-based tool (Consult Builder) to use formatted sentences to build a templated interpretation for each diplotype, using consistent language across genes where possible (66). The clinician approving the interpretive consultation has the option of using the template or customizing the interpretation based on patient-specific information on drug exposure or effects. These interpretive written consultations represent passive CDS necessary to successfully implement preemptive pharmacogenetics (66). The diplotype and accompanying pharmacogenetic consultation are viewable in a pharmacogenetic tab in the patient's EHR (64).
Active interruptive CDS takes two forms: pre-test and post-test alerts. Pre-test alerts fire to the prescriber when an order is placed for a drug linked to a gene/drug pair if the patient does not already have a documented genotype (67). Until the widespread adoption of preemptive multi-gene genotyping, these alerts will be necessary to prompt the prescriber to consider ordering a genotype test prior to the first dose. The prescriber also has the option to cancel or continue with the current order without ordering a genotype test. As preemptive genotyping becomes more widespread, more post-test alerts will fire, which notify prescribers who have ordered an affected drug for a patient who has a high-risk phenotype of the gene/drug interaction. The alert describes the risk to the patient, offers a therapeutic recommendation, and requires the prescriber to modify the order, cancel the order, or acknowledge the alert and continue with the current order (64). Examples of screenshots and wording of these CDS alerts have previously been published (61), (66; 67).
Those patients who consent to individualized communication (more than 97% of patients thus far enrolled) are informed of their genetic results via standardized formatted letters which are mailed to the address of their choice every time a new gene test result is placed in their EHR. The letters are patient specific and include the patient's genotype result, phenotype, and a clinical interpretative report written in lay language. Each letter is posted in the patient's St. Jude EHR for viewing by clinicians and the personalized letters are suitable for patients to share with outside providers, allowing those providers the potential to use the test results to inform their prescribing as well.
As of January 2014, four genes and 12 drugs have been migrated into the EHR (64). A total of 56 active CDS alerts have been created. For the 1016 patients with pharmacogenetic results in their EHR, 3,776 genotypes have been entered into the EHR. With the gene/drug pairs thus far implemented, 792 patients (78%) have at least one actionable phenotype.
The goal for St. Jude's preemptive pharmacogenetics program is to implement in the clinic all CPIC gene/drug pairs. Although this program is currently implemented via a research protocol, in the future, the goal is for such an approach to become standard-of-care throughout medicine.
Vanderbilt University Medical Center
Vanderbilt University Medical Center began a preemptive pharmacogenetics implementation project, called Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment (PREDICT), in September 2010 (39). The objective of the project is to develop the infrastructure and framework for incorporating genomic test results into the medical record and making these data available preemptively to practitioners. The program's initial focus was on antiplatelet therapy following placement of a cardiovascular stent. Providers can enroll any patient on the program, but the enrollment focus is on groups of patients with anticipated cardiac catheterization with coronary artery stenting or a risk score estimating likelihood of exposure to pharmacogenomically relevant medications (68). Using the VeraCode ADME Core Panel, patients were genotyped for 184 variants in 34 genes related to drug response (69). This panel produced high quality data for most genes reported, with the notable exception of CYP2D6 and copy number variations. The genotype results are stored in a database separate from the EHR (39). Genotypes are moved into the EHR as the evidence for their clinical use is determined to be sufficient. The process for moving a gene into the EHR begins with a subcommittee of the P&T committee reviewing and evaluating the literature and deciding if a gene is actionable. This review is then presented to the P&T committee, which approves or denies the gene as actionable.
CYP2C19 *2/*2 was the first genotype determined to be actionable in Vanderbilt's program (68). Genotype data are viewable in a section of the EHR specific for genetic information. Each genotype is displayed in standard star (*) allele notation and is accompanied by a phenotype interpretation. Decision support was developed for actionable genes and integrated into inpatient and outpatient EHR applications for medication orders (68). The decision support for CYP2C19 was coupled with clopidogrel and recommended the use of prasugrel as an alternative in patients with an actionable genotype (68). As of November 2013, 10,000 patients had been genotyped in the PREDICT program (68). CDS for several gene/drug pairs had been implemented (CYP2C19-clopidogrel, SLCO1B1-simvastatin, CYP2C9 and VKORC1-warfarin, CYP3A5-tacrolimus, and TPMT-thiopurines) (68). With these gene/drug pairs, 91% of patients had at least one actionable genotype (68). Using this preemptive array-based method, the investigators report that over 5,000 genetic tests were eliminated compared to what would have been ordered had reactive single gene tests been used (68)
University of Florida and Shands Hospital
The University of Florida and Shands Hospital launched a clinical implementation program in 2011 called the Personalized Medicine Program (38). The program uses chip-based genotyping (Life Technologies Quant Studio Open Array) to generate potentially multiple clinically actionable genotypes. This custom array interrogates 256 single nucleotide polymorphisms (SNPs) (35). The focus of the program is to have genotyping data available preemptively (38). The hospital's P&T committee and a Personalized Medicine Program subcommittee are used to regulate which pharmacogenetic data are actionable and should be entered into the medical record. This regulation process includes evaluating the literature, establishing genotype-phenotype relationships, determining recommendations on alternative therapies, and approving wording for clinical decision support tools. Once this process is complete, relevant genotypes may be added to patient's medical records. CYP2C19 and clopidogrel was the first gene/drug pair implemented in June 2012.
As of March 2013, about 800 patients had genotype results in the EHR. Eight of the 256 SNPs were used to determine a patient's CYP2C19 diplotype. CYP2C19 phenotypes of intermediate and poor metabolizer were considered actionable for clopidogrel. Obtaining a CYP2C19 genotype result is considered a part of standard clinical care at Shands Hospital and is covered by a clinical consent, rather than by research informed consent. Electronic CDS alerts physicians ordering clopidogrel for patients undergoing percutaneous coronary intervention (PCI) who have an actionable CYP2C19 genotype and recommends an alternative antiplatelet agent. Clinical pharmacists are also alerted to patients undergoing PCI. This allows pharmacists to follow up with patients who were discharged before their genotype results were available. Patients must agree to a research informed consent to have the remaining 248 SNPs data moved into the EHR in the future to preemptively inform prescribing as other gene/drug pairs are approved by the P&T committee. Stanford University Medical Center will serve as a replication site for this program (35).
Mayo Clinic
The Mayo Clinic, with assistance from the Pharmacogenomic Research Network and the Electronic Medical Records and Genomics Network (eMERGE), developed a protocol to create a best practice for clinical implementation of genetic results to improve patient outcomes (43). The protocol is named “Right Drug, Right Dose, Right Time-Using Genomic Data to Individualize Treatment” (RIGHT protocol). The three main objectives of the protocol's pilot study were to: 1) identify patients who would likely benefit from pharmacogenetic driven intervention, 2) use next generation sequencing (NGS), PGRNseq, and a CYP2D6 assay to obtain genotype results for 85 pharmacogenes and integrate the results into the EHR, and 3) develop and implement point of care CDS for high-risk pharmacogenetic results. Patients were selected for enrollment if they were determined to have a high-likelihood of receiving a high-risk drug, based on a predictive model using chronic disease states and demographic information. A combination of genotyping assays was required due to technical challenges of sequencing CYP2D6 using NGS. High-risk pharmacogenetic results were displayed as molecular diagnostic laboratory test results using standard notation and accompanied by an interpretation. These results served as triggers for CDS alerts to fire when high-risk drugs were ordered for these patients.
A Mayo Clinic pharmacogenomics task force provided initial oversight for clinical implementation including selection of gene/drug pairs and CDS development. Gene/drug pairs were selected based on information available from sources such as the FDA, PharmGKB, and CPIC. Before their implementation, CDS had to be approved by several institutional groups including the P&T committee, pharmaceutical formulary committee, and relevant disease-oriented task forces. Prescribers were educated about gene/drug pairs using a web-based “just-in-time” system. Patients were able to access their genetic results using the Mayo Clinical Online Patient Services account. A total of 1013 patients were enrolled on the protocol. As of July 2013, four gene/drug pairs (HLA-B*1502-carbamazepine, HLA-B*5701-abacavir, TPMT-thiopurines, and IFNL3-interferon) were approved for implementation and several more pairs (CYP2D6-codeine, tamoxifen, and tramadol; CYP2C19-clopidogrel; and SLCO1B1-simvastatin) were in the CDS development process.
Mount Sinai Medical Center
Mount Sinai Medical Center, a member of eMERGE, initiated the CLIPMERGE PGx program in February 2013 (42). The goal of the program is to establish an infrastructure for the clinical implementation of pharmacogenetic data. Using an existing DNA biobank “BioMe” as a cohort, patients likely to receive a drug with pharmacogenetically relevant interactions and who receive medical care at Mount Sinai Internal Medicine Associates are targeted for enrollment. Investigators for CLIPMERGE PGx consider clopidogrel, warfarin, simvastatin, and several types of antidepressants as having pharmacogenetically relevant interactions, because pharmacogenetics practice guidelines, such as CPIC guidelines, are available. Using these guidelines as a template, active CDS is developed to deliver alerts to clinicians at the point of care. Specific training sessions are provided to clinicians, who will receive the alerts. To facilitate development and management of the CDS, a data management system, which is separate from and interfaces with Mount Sinai's EPIC EHR, is used. Several gene/drug pairs have been implemented (CYP2C19-clopidogrel, CYP2C9 and VKORC1-warfarin, SLCO1B1-simvastatin, CYP2D6 and CYP2C19-TCAs, and CYP2D6-SSRIs).
Developing a process for implementation
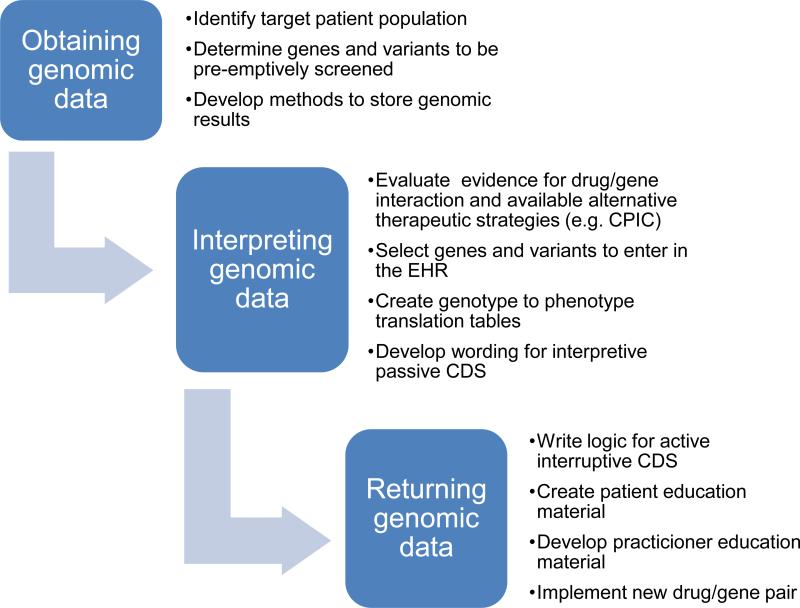
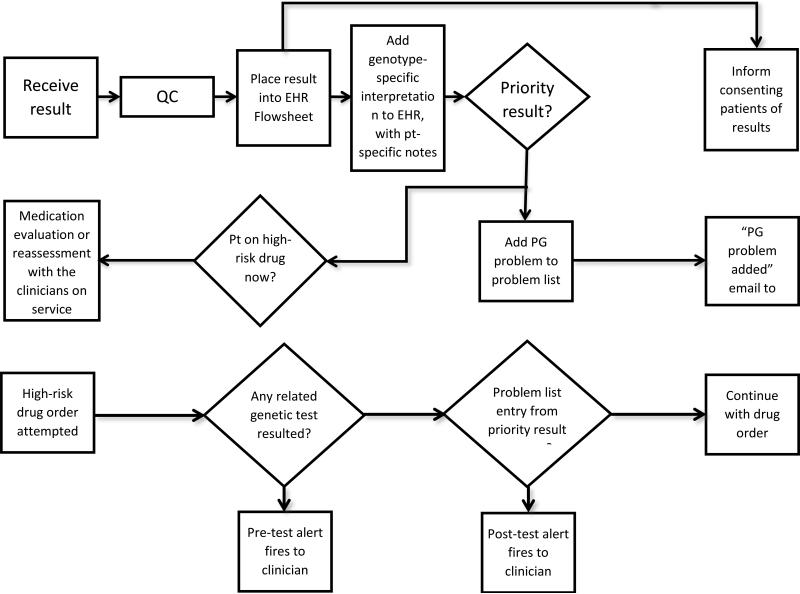
There are some common elements to the programs which have implemented preemptive pharmacogenetics testing (Figure 3). Each has a systematic process for genotyping and for migrating test results into the EHR (Figure 4). An institutional infrastructure is present to support clinical implementation. Each of the programs discussed here has a governing committee, which is a subcommittee of the P&T committee, to provide oversight for which gene test results are placed in the EHR and which drugs are the subject of CDS alerts. Each program has a process for evaluating the evidence for implementing each gene/drug pair in clinical practice, which has been facilitated by the development of peer-reviewed guidelines (7-21). Multi-gene arrays are chosen to include pharmacogenes likely to be used in the EHR. Adequate CDS for each gene/drug pair is developed and implemented, with some customization for each practice site. Each program has established a process to add relevant genotypes to the EHR over time. Each site has determined what level of patient consent is required for clinical testing, with some sites deciding that no special informed consent is needed.
Figure 3.
Steps required to implement preemptive pharmacogenetics. CDS= clinical decision support
Figure 4.
Steps needed to translate a genotype result into a clinically useful action
Education
Because of rapidly changing evidence for gene/drug pairs, successful clinical implementation requires ongoing clinician education (47; 49). These education efforts may focus on general education for all clinicians and specific advanced education for those clinicians who will be making genetic test interpretations and recommendations, which may include pharmacists or prescribers. With every new gene/drug pair implemented, clinicians may need to be provided educational material through various methods to allow adequate learning about the new gene/drug pair. Also, information on dosing recommendations according to pharmacogenetic status may be added to the institution's formulary, so that it is available to clinicians without having to link it to a specific patient's results. The delivery of pharmacogenetic education may be as on-demand services or as formal didactic sessions. On-demand services include websites such as www.pharmgkb.org, videos or formulary references whereas formal education sessions may include presentations on pharmacogenetics at conferences within the organization, or a structured education and competency process tailored to the individual's job functions. St. Jude's preemptive pharmacogenetic educational efforts include all of these aspects, and many elements are freely available (www.stjude.org/pg4kds http://www.ashp.org/menu/PracticePolicy/ResourceCenters/Emerging-Sciences/Pharmacogenomics.aspx http://www.g-2-c-2.org/). Because no amount of education will be sufficient to deal with the ever-growing number of clinically relevant gene/drug pairs, CDS built into the EHR is essential to alert clinicians to the need to further investigate specific gene/drug pairs at the time of prescribing.
Electronic clinical decision support
An essential component of implementing preemptive clinical pharmacogenetics is an electronic EHR with the ability to customize CDS (64; 70-72). Indeed, it is the very ability to create customized CDS that can be activated at the time of ordering a high-risk drug that makes preemptive pharmacogenetic testing plausible. Genetic test results differ from other laboratory test results because they remain relevant over a patient's entire lifetime. Because preemptively determined test results are placed in the EHR potentially months or years before a related drug may be ordered, the ability to actively deliver patient and drug-specific information based on existing genetic test results through decision support alerts to clinicians at the point of care is crucial (67; 73). Without effective active CDS, which includes interruptive alerts that are delivered through drug-specific order entry or dispensing functions of the EHR, pharmacogenetic results previously collected could easily be forgotten or lost within a patient's medical record and an actionable phenotype may not be considered in the decision to prescribe a high-risk drug affected by that phenotype. Passive CDS includes genetic test results and their interpretation, which is essential to communicate results to clinicians and provide guidance that is available at any time through the EHR (66; 74).
Conclusion
Millions of prescriptions are dispensed every year for pharmacogenetically high-risk medications. When 12 pharmacogenes with at least one known actionable inherited variant are considered, over 97% of the U.S. population has at least one high-risk diplotype. A preemptive approach to clinical implementation of pharmacogenetic testing, made possible by the use of EHRs and multi-gene testing platforms, provides a mechanism for harnessing genetic testing to improve drug prescribing in the clinic. This preemptive approach has now been adopted in a few early adopter health system settings. The process for clinical implementation, as well as the content underlying clinical decision support for actions based on genetics, is being established in different clinical settings for both children and adults, and the content needed to act on pharmacogenetic test results is being developed and shared so that other sites will not have to recapitulate all of the effort needed to implement at these early adopter sites. By improving standardization and interoperability of health care record systems, education of clinicians, and communication among clinicians and with patients, implementation of genomics to improve medication therapy can be more fully realized.
Supplementary Material
Acknowledgements
Funding Supported by NCI grants CA 21765, NIH/NIGMS
Pharmacogenomics Research Network (U01 GM92666, U19 GM61388, U01 HL105918, R24 GM61374), and by the American Lebanese Syrian Associated Charities (ALSAC).
References
- 1.Meyer UA. Pharmacogenetics - five decades of therapeutic lessons from genetic diversity. Nat Rev Genet. 2004;5:669–76. doi: 10.1038/nrg1428. [DOI] [PubMed] [Google Scholar]
- 2.Veenstra DL, Roth JA, Garrison LP, Jr., Ramsey SD, Burke W. A formal risk-benefit framework for genomic tests: facilitating the appropriate translation of genomics into clinical practice. Genet Med. 2010;12:686–93. doi: 10.1097/GIM.0b013e3181eff533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–13. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 4.Manolio TA, Green ED. Genomics reaches the clinic: from basic discoveries to clinical impact. Cell. 2011;147:14–6. doi: 10.1016/j.cell.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Voora D, Ginsburg GS. A hub for bench-to-bedside pharmacogenomic-based research. Pharmacogenomics. 2011;12:1095–8. doi: 10.2217/pgs.11.62. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364:1144–53. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muir AJ, Gong L, Johnson SG, Lee MT, Williams MS, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for IFNL3 (IL28B) Genotype and PEG Interferon-alpha-Based Regimens. Clinical pharmacology and therapeutics. 2013;2:141–6. doi: 10.1038/clpt.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caudle KE, Thorn CF, Klein TE, Swen JJ, McLeod HL, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing. Clinical pharmacology and therapeutics. 2013;94:640–5. doi: 10.1038/clpt.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, et al. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clinical pharmacology and therapeutics. 2013;93:324–5. doi: 10.1038/clpt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clinical pharmacology and therapeutics. 2013;94:317–23. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leckband SG, Kelsoe JR, Dunnenberger HM, George AL, Jr., Tran E, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for HLA-B genotype and carbamazepine dosing. Clinical pharmacology and therapeutics. 2013;94:324–8. doi: 10.1038/clpt.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hicks JK, Swen JJ, Thorn CF, Sangkuhl K, Kharasch ED, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clinical pharmacology and therapeutics. 2013;93:402–8. doi: 10.1038/clpt.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hershfield MS, Callaghan JT, Tassaneeyakul W, Mushiroda T, Thorn CF, et al. Clinical pharmacogenetics implementation consortium guidelines for human leukocyte antigen-B genotype and allopurinol dosing. Clinical pharmacology and therapeutics. 2013;93:153–8. doi: 10.1038/clpt.2012.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilke RA, Ramsey LB, Johnson SG, Maxwell WD, McLeod HL, et al. The Clinical Pharmacogenomics Implementation Consortium: CPIC Guideline for SLCO1B1 and Simvastatin-Induced Myopathy. Clinical pharmacology and therapeutics. 2012;92:112–7. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin MA, Klein TE, Dong BJ, Pirmohamed M, Haas DW, Kroetz DL. Clinical pharmacogenetics implementation consortium guidelines for HLA-B genotype and abacavir dosing. Clinical pharmacology and therapeutics. 2012;91:734–8. doi: 10.1038/clpt.2011.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crews KR, Gaedigk A, Dunnenberger HM, Klein TE, Shen DD, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for Codeine Therapy in the Context of Cytochrome P450 2D6 (CYP2D6) Genotype. Clinical pharmacology and therapeutics. 2012;91:321–6. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JA, Gong L, Whirl-Carrillo M, Gage BF, Scott SA, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clinical pharmacology and therapeutics. 2011;90:625–9. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott SA, Sangkuhl K, Gardner EE, Stein CM, Hulot JS, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clinical pharmacology and therapeutics. 2011;90:328–32. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clinical pharmacology and therapeutics. 2011;89:387–91. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crews KR, Gaedigk A, Dunnenberger HM, Leeder JS, Klein TE, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Cytochrome P450 2D6 Genotype and Codeine Therapy: 2014 Update. Clinical pharmacology and therapeutics. 2014;4(376):82. doi: 10.1038/clpt.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Relling MV, McDonagh EM, Chang T, Caudle KE, McLeod HL, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for Rasburicase Therapy in the context of G6PD Deficiency Genotype. Clinical pharmacology and therapeutics. 2014 May 2; doi: 10.1038/clpt.2014.97. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu AC, Fuhlbrigge AL. Economic evaluation of pharmacogenetic tests. Clinical pharmacology and therapeutics. 2008;84:272–4. doi: 10.1038/clpt.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International Warfarin Pharmacogenetics C, Klein TE, Altman RB, Eriksson N, Gage BF, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–64. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clinical pharmacology and therapeutics. 2008;84:326–31. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HS, Lee SS, Oh M, Jang YJ, Kim EY, et al. Effect of CYP2C9 and VKORC1 genotypes on early-phase and steady-state warfarin dosing in Korean patients with mechanical heart valve replacement. Pharmacogenetics and genomics. 2009;19:103–12. doi: 10.1097/FPC.0b013e32831a9ae3. [DOI] [PubMed] [Google Scholar]
- 26.Lenzini P, Wadelius M, Kimmel S, Anderson JL, Jorgensen AL, et al. Integration of genetic, clinical, and INR data to refine warfarin dosing. Clinical pharmacology and therapeutics. 2010;87:572–8. doi: 10.1038/clpt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altman RB. Personal genomic measurements: the opportunity for information integration. Clinical pharmacology and therapeutics. 2013;93:21–3. doi: 10.1038/clpt.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuldiner AR, Relling MV, Peterson JF, Hicks K, Freimuth RR, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Overcoming Challenges of Real-World Implementation. Clinical pharmacology and therapeutics. 2013;2:207–10. doi: 10.1038/clpt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altman RB, Whirl-Carrillo M, Klein TE. Challenges in the pharmacogenomic annotation of whole genomes. Clinical pharmacology and therapeutics. 2013;94:211–3. doi: 10.1038/clpt.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamburg MA, Collins FS. The path to personalized medicine. The New England journal of medicine. 2010;363:301–4. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 31.Schildcrout JS, Denny JC, Bowton E, Gregg W, Pulley JM, et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clinical pharmacology and therapeutics. 2012;92:235–42. doi: 10.1038/clpt.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.National Prescription Audit. IMS Health; Danbury, CT USA: [Google Scholar]
- 33.O'Donnell PH, Bush A, Spitz J, Danahey K, Saner D, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clinical pharmacology and therapeutics. 2012;92:446–9. doi: 10.1038/clpt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira NL, Weinshilboum RM. The impact of pharmacogenomics on the management of cardiac disease. Clinical pharmacology and therapeutics. 2011;90:493–5. doi: 10.1038/clpt.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson JA, Burkley BM, Langaee TY, Clare-Salzler MJ, Klein TE, Altman RB. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clinical pharmacology and therapeutics. 2012;92:437–9. doi: 10.1038/clpt.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrugia G, Weinshilboum RM. Challenges in implementing genomic medicine: the Mayo Clinic Center for Individualized Medicine. Clinical pharmacology and therapeutics. 2013;94:204–6. doi: 10.1038/clpt.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JA. Pharmacogenetics in clinical practice: how far have we come and where are we going? Pharmacogenomics. 2013;14:835–43. doi: 10.2217/pgs.13.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson JA, Elsey AR, Clare-Salzler MJ, Nessl D, Conlon M, Nelson DR. Institutional profile: University of Florida and Shands Hospital Personalized Medicine Program: clinical implementation of pharmacogenetics. Pharmacogenomics. 2013;14:723–6. doi: 10.2217/pgs.13.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, et al. Operational Implementation of Prospective Genotyping for Personalized Medicine: The Design of the Vanderbilt PREDICT Project. Clinical pharmacology and therapeutics. 2012;1:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Relling MV, Altman RB, Goetz MP, Evans WE. Clinical implementation of pharmacogenomics: overcoming genetic exceptionalism. Lancet Oncol. 2010;11:507–9. doi: 10.1016/S1470-2045(10)70097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott SA. Personalizing medicine with clinical pharmacogenetics. Genet Med. 2011;13:987–95. doi: 10.1097/GIM.0b013e318238b38c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gottesman O, Scott SA, Ellis SB, Overby CL, Ludtke A, et al. The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clinical pharmacology and therapeutics. 2013;94:214–7. doi: 10.1038/clpt.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clinic proceedings. 2014;89:25–33. doi: 10.1016/j.mayocp.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swen JJ, Wilting I, de Goede AL, Grandia L, Mulder H, et al. Pharmacogenetics: from bench to byte. Clinical pharmacology and therapeutics. 2008;83:781–7. doi: 10.1038/sj.clpt.6100507. [DOI] [PubMed] [Google Scholar]
- 45.Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clinical pharmacology and therapeutics. 2011;89:662–73. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 46.Pham HH, Schrag D, O'Malley AS, Wu B, Bach PB. Care patterns in Medicare and their implications for pay for performance. N Engl J Med. 2007;356:1130–9. doi: 10.1056/NEJMsa063979. [DOI] [PubMed] [Google Scholar]
- 47.Stanek EJ, Sanders CL, Taber KA, Khalid M, Patel A, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clinical pharmacology and therapeutics. 2012;91:450–8. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 48.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clinical pharmacology and therapeutics. 2011;89:464–7. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCullough KB, Formea CM, Berg KD, Burzynski JA, Cunningham JL, et al. Assessment of the pharmacogenomics educational needs of pharmacists. Am J Pharm Educ. 2011;75:51. doi: 10.5688/ajpe75351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shields AE, Lerman C. Anticipating clinical integration of pharmacogenetic treatment strategies for addiction: are primary care physicians ready? Clinical pharmacology and therapeutics. 2008;83:635–9. doi: 10.1038/clpt.2008.4. [DOI] [PubMed] [Google Scholar]
- 51.Cohen J, Wilson A, Manzolillo K. Clinical and economic challenges facing pharmacogenomics. The pharmacogenomics journal. 2013;13:378–88. doi: 10.1038/tpj.2011.63. [DOI] [PubMed] [Google Scholar]
- 52.Thorn CF, Klein TE, Altman RB. PharmGKB: the Pharmacogenomics Knowledge Base. Methods Mol Biol. 2013;1015:311–20. doi: 10.1007/978-1-62703-435-7_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caudle KE, Klein TE, Hoffman JM, Muller DJ, Whirl-Carrillo M, et al. Incorporation of Pharmacogenomics into Routine Clinical Practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline Development Process. Current drug metabolism. 2014;2:209–17. doi: 10.2174/1389200215666140130124910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Recommendations from the EGAPP Working Group: can UGT1A1 genotyping reduce morbidity and mortality in patients with metastatic colorectal cancer treated with irinotecan? Genet Med. 2009;11:15–20. doi: 10.1097/GIM.0b013e31818efd9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valdes R, Payne D, Linder M. Laboratory analysis and application of pharmacogenetics to clinical practice. Laboratory medicine practice guidelines. 2010 [Google Scholar]
- 56.Recommendations from the EGAPP Working Group: routine testing for Factor V Leiden (R506Q) and prothrombin (20210G>A) mutations in adults with a history of idiopathic venous thromboembolism and their adult family members. Genet Med. 2011;13:67–76. doi: 10.1097/GIM.0b013e3181fbe46f. [DOI] [PubMed] [Google Scholar]
- 57.Manns MP, Czaja AJ, Gorham JD, Krawitt EL, Mieli-Vergani G, et al. Diagnosis and management of autoimmune hepatitis. Hepatology. 2010;51:2193–213. doi: 10.1002/hep.23584. [DOI] [PubMed] [Google Scholar]
- 58.Meggitt SJ, Anstey AV, Mohd Mustapa MF, Reynolds NJ, Wakelin S. British Association of Dermatologists' guidelines for the safe and effective prescribing of azathioprine 2011. The British journal of dermatology. 2011;165:711–34. doi: 10.1111/j.1365-2133.2011.10575.x. [DOI] [PubMed] [Google Scholar]
- 59.Rubinstein WS, Maglott DR, Lee JM, Kattman BL, Malheiro AJ, et al. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic acids research. 2013;41:D925–35. doi: 10.1093/nar/gks1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Relling MV, Pui CH, Cheng C, Evans WE. Thiopurine methyltransferase in acute lymphoblastic leukemia. Blood. 2006;107:843–4. doi: 10.1182/blood-2005-08-3379. [DOI] [PubMed] [Google Scholar]
- 61.Crews KR, Cross SJ, McCormick JN, Baker DK, Molinelli AR, et al. Development and implementation of a pharmacist-managed clinical pharmacogenetics service. Am J Health Syst Pharm. 2011;68:143–50. doi: 10.2146/ajhp100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stocco G, Cheok MH, Crews KR, Dervieux T, French D, et al. Genetic polymorphism of inosine triphosphate pyrophosphatase is a determinant of mercaptopurine metabolism and toxicity during treatment for acute lymphoblastic leukemia. Clinical pharmacology and therapeutics. 2009;85:164–72. doi: 10.1038/clpt.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crews KR, Hicks JK, Pui CH, Relling MV, Evans WE. Pharmacogenomics and individualized medicine: translating science into practice. Clinical pharmacology and therapeutics. 2012;92:467–75. doi: 10.1038/clpt.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoffman JM, Haidar CE, Wilkinson MR, Crews KR, Baker DK, et al. PG4KDS: A Model for the Clinical Implementation of Pre-emptive Pharmacogenetics. American journal of medical genetics. Part C, Seminars in medical genetics. 2014;1:45–55. doi: 10.1002/ajmg.c.31391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernandez CA, Smith C, Yang W, Lorier R, Crews KR, et al. Concordance of DMET plus genotyping results with those of orthogonal genotyping methods. Clinical pharmacology and therapeutics. 2012;92:360–5. doi: 10.1038/clpt.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hicks JK, Crews KR, Hoffman JM, Kornegay NM, Wilkinson MR, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clinical pharmacology and therapeutics. 2012;92:563–6. doi: 10.1038/clpt.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bell GC, Crews KR, Wilkinson MR, Haidar CE, Hicks JK, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. Journal of the American Medical Informatics Association : JAMIA e1:e93-9. 2013 doi: 10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Driest SL, Shi Y, Bowton EA, Schildcrout JS, Peterson JF, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clinical pharmacology and therapeutics. 2013 doi: 10.1038/clpt.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oetjens MT, Denny JC, Ritchie MD, Gillani NB, Richardson DM, et al. Assessment of a pharmacogenomic marker panel in a polypharmacy population identified from electronic medical records. Pharmacogenomics. 2013;14:735–44. doi: 10.2217/pgs.13.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Overby CL, Kohane I, Kannry JL, Williams MS, Starren J, et al. Opportunities for genomic clinical decision support interventions. Genet Med. 2013;15:817–23. doi: 10.1038/gim.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goldspiel BR, Flegel WA, Dipatrizio G, Sissung T, Adams SD, et al. Integrating pharmacogenetic information and clinical decision support into the electronic health record. Journal of the American Medical Informatics Association : JAMIA. 2013;3:522–8. doi: 10.1136/amiajnl-2013-001873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Welch B, Kawamoto K. The Need for Clinical Decision Support Integrated with the Electronic Health Record for the Clinical Application of Whole Genome Sequencing Information. Journal of Personalized Medicine. 2013;3:306–25. doi: 10.3390/jpm3040306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilke RA, Xu H, Denny JC, Roden DM, Krauss RM, et al. The emerging role of electronic medical records in pharmacogenomics. Clinical pharmacology and therapeutics. 2011;89:379–86. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaspers MW, Smeulers M, Vermeulen H, Peute LW. Effects of clinical decision-support systems on practitioner performance and patient outcomes: a synthesis of high-quality systematic review findings. Journal of the American Medical Informatics Association : JAMIA. 2011;18:327–34. doi: 10.1136/amiajnl-2011-000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.