Abstract
Objectives
We investigated the frequency of MYC and TERC increased gene copy number (GCN) in early-stage non–small cell lung cancer (NSCLC) and evaluated the correlation of these genomic imbalances with clinicopathologic parameters and outcome.
Materials and Methods
Tumor tissues were obtained from 113 resected NSCLCs. MYC and TERC GCNs were tested by fluorescence in situ hybridization (FISH) according to the University of Colorado Cancer Center (UCCC) criteria and based on the receiver operating characteristic (ROC) classification.
Results
When UCCC criteria were applied, 41 (36%) cases for MYC and 41 (36%) cases for TERC were considered FISH-positive. MYC and TERC concurrent FISH-positive was observed in 12 cases (11%): 2 (17%) cases with gene amplification and 10 (83%) with high polysomy. By using the ROC analysis, high MYC (mean ≥2.83 copies/cell) and TERC (mean ≥2.65 copies/cell) GCNs were observed in 60 (53.1%) cases and 58 (51.3%) cases, respectively. High TERC GCN was associated with squamous cell carcinoma (SCC) histology (P = 0.001). In univariate analysis, increased MYC GCN was associated with shorter overall survival (P = 0.032 [UCCC criteria] or P = 0.02 [ROC classification]), whereas high TERC GCN showed no association. In multivariate analysis including stage and age, high MYC GCN remained significantly associated with worse overall survival using both the UCCC criteria (P = 0.02) and the ROC classification (P = 0.008).
Conclusions
Our results confirm MYC as frequently amplified in early-stage NSCLC and increased MYC GCN as a strong predictor of worse survival. Increased TERC GCN does not have prognostic impact but has strong association with squamous histology.
Keywords: MYC, TERC, FISH, non–small cell lung cancer, prognosis
Amplification of genes of the MYC family (MYC, MYCN, and MYCL1) has been described in a variety of cancer cell lines and tumor specimens including lung cancer.1–4 The MYC gene localized at 8q24.1 is a well-characterized oncogene involved in cell growth, differentiation, metabolism, and apoptosis.5
The prognostic role of MYC amplification has been explored both in small cell lung cancer (SCLC) and non–small cell lung cancer (NSCLC). Lockwood et al6 have investigated 104 cancer cell lines from different tumor tissue origins by array comparative genomic hybridization to identify amplified chromosomal segments. Among 53 lung cell lines, of which 36 were originated from NSCLC, 16 from SCLC, and 1 from Mesothelioma, MYC was the most frequently amplified gene.
Some studies on resected NSCLC reported that MYC was associated with tumor progression,7,8 a worse prognosis, and its overexpression was related to metastasis of lung cancer.9 Kubokura et al10 showed that MYC amplification correlated with lymph node metastasis, suggesting a possible negative effect on survival. Iwakawa et al11 showed that MYC amplification was associated with poor prognosis in patients both with small-sized (≤2 cm in greatest dimension) and early-stage I lung adenocarcinoma (ADC). Furthermore, MYC was expressed in large numbers of NSCLCs12 and was amplified or overexpressed in SCC and ADC of the lung.13–15 The MYC increased gene copy number (GCN) leads to overexpression of the MYC protein through Max heterodimer transcription factors that alter gene expression in large part by recruiting histone-modifying enzymes.15
The gain of sequences on the long arm of chromosome 3 (3q) is also a frequent event in many human malignant diseases, including lung cancer.2 Human telomerase gene (TERC), localized on the chromosome 3q26, encodes the RNA component of human telomerase, a ribonucleoprotein enzyme, which acts as a template for the addition of telomeric repeat sequences 5′-TTAGGG-3′16 required for the stability and complete replication of chromosome ends, and it has been associated with cell immortality and the development of cancers.
Recently, Fan et al17 reported significantly higher percentages of cells with amplification of TERC in NSCLC than in SCLC, and the phenomenon occurred more frequently in SCC than in ADC.18 Using FISH methodology, Pelosi et al19 studied the 3q26 amplification in preneoplastic/preinvasive squamous cell lesions of the bronchial mucosa and in 2 subsets of lung SCC, the early hilar (EHSCC), and the parenchyma-infiltrating SCC (PISCC). The authors concluded that 3q26 amplification was likely a late event in the development of SCC of the lung and it is more prevalent in EHSCC than in PISCC, suggesting different pathogenesis for these tumor subtypes.
Foster et al20 showed that the 3q26 amplification was a common feature of pulmonary SCC, confirming its key role in the transition from high-grade preinvasive neoplasia to invasive carcinoma, as also documented in uterine cervix21 and head-and-neck SCC.22 Yan et al23 reported that 3q and 8q amplifications were significantly higher in smoker than those in nonsmoker SCC patients and was associated with tumorigenesis and/or progression of the disease.
In summary, only a few studies are available on MYC and TERC GCN in NSCLC, and no consensus criteria exist on how to assess the status of these genes as prognostic value. This study aimed to evaluate the MYC and TERC gene copy status in NSCLC by FISH using 2 criteria for interpretation, namely, the University of Colorado Cancer Center (UCCC) scoring system proposed for EGFR in lung cancer24 and the receiver operating characteristics (ROC) scoring system. We also evaluated the correlation of these genomic imbalances with clinicopathologic parameters and outcome in resected NSCLC patients.
MATERIAL AND METHODS
Patient Selection
This retrospective study was conducted in a cohort of 113 NSCLC patients who received a radical resection for primary NSCLC at the Thoracic Surgery Unit of the Perugia University at S. Maria della Misericordia Hospital, Italy, between 2002 and 2006. Histologic subtypes and grade of differentiation were determined according to the World Health Organization classification.25 The only criteria used for patient selection was availability of tumor tissue from primary lung cancer and survival data. Neither chemotherapy nor radiotherapy was administered before surgery. A follow-up, including a chest x-ray at 3-month intervals alternated with a total body computed tomography (CT) scan every 6 months, was scheduled for all patients for the first 2 years. Subsequently, the patients underwent a CT scan/y.
Recurrences were detected by imaging techniques and when necessary, confirmed by histologic sampling. The study was reviewed and approved by the local institution’s Ethics Committee, and written informed consent was obtained from each patient for surgical specimen analyses.
FISH Assay
FISH assays were carried out on 4 μm (± 1 μm) thick sections from formalin-fixed, paraffin-embedded tissue blocks from surgically resected tumor specimens of NSCLC patients. The 2-color (TERC-MYC) FISH probe was prepared combining LSI TERC Spectrum Gold and LSI c-MYC Spectrum Aqua, both reagents from Abbott Molecular. Slides were incubated at 56 ± 2°C overnight and incubated in CitriSolv 2 times for 10 minutes each and air dried. Thereafter, specimens were dehydrated in 100% ethanol twice, for 5 minutes each, air dried, and subsequently incubated in 2 × saline sodium citrate (SSC) at 75°C for 9 to 25 minutes, 0.25 mg/mL proteinase K at 45°C for 10 to 25 minutes, and 2 − SSC at room temperature for 5 minutes, and dehydrated in 70%, 85%, and 100% ethanol series for 2 minutes each, and air dried. The probe mixture was applied to the target areas of each slide, which were covered with glass coverslips and sealed with rubber cement. Slides were incubated at 85 ± 1°C in a dry oven for 15 minutes to codenature probe and target DNAs and incubated in moisture chamber at 37 ± 1°C for 16 hours. Coverslips were then removed and the slides were immersed twice in 2 × SSC/0.3% NP-40 at 73 ± 1°C for 2 minute each, then washed in 2 × SSC at room temperature, dehydrated in 70%, 85% and 100% ethanol, and air dried. Vectashield DAPI in mounting medium (14 mL of 0.3 ug/mL) was applied and the areas were covered with 22 × 50 mm coverslips.
Analysis was performed using fluorescence microscope (Zeiss AxioImager). For documentation, images were captured using a charge-coupled device camera (CoolSnap, Photometrics) and merged using dedicated software (CytoVision, Leica Micro- systems). The scoring was carried out in 100 nonoverlapping tumor cell nuclei per patient from 4 representative tumor areas. According to the Colorado criteria for EGFR,24 the GCN for each gene was classified as increased (FISH-positive) when displaying gene amplification ( > 10% of tumor cells with >15 copies of the signals or gene clusters [ > 4 gene copies per cluster] or innumerable tight gene clusters) and high polysomy (≥40% of cells displaying ≥4 copies of the specific gene signal).
Statistical Analysis
The primary end point was to assess whether MYC and TERC GCN affected survival of surgically resected NSCLC. We used the ROC classification system as a continuous variable to determine a cutoff point for MYC and TERC GCN.26 Sensitivity and specificity were expressed in terms of percentage, and the highest value has been chosen as the best cutoff point in discriminating patients who survived compared with those who died. Overall survival (OS) was defined as the time from surgery to the date of death from any cause; patients who were not reported as having died at the time of the analysis were censored at the date they were last known to be alive. Disease-free survival (DFS) was defined as the time from surgery to first local, regional, or distant recurrence; second primary malignancy; or death from any cause, whichever came first. Patients who were alive and did not experience recurrence at the time of the analysis were censored at the last disease assessment date. OS, DFS, and the 95% confidence intervals (CIs) were evaluated by the Kaplan-Meier method comparing the different groups by log-rank test. The Cox proportional hazards model was used to evaluate the prognostic role of each single studied parameter on OS and DFS, in univariate and multivariate analyses. The χ2 test was used to assess the association between clinical features and the MYC and TERC GCN. Unless otherwise specified, all tests are with 1 df. A probability value of <0.05 was considered as statistically significant. Statistical analysis was carried out using Matlab software (the MathWorks version 7.2.0.232).
RESULTS
Patient Characteristics
The clinical characteristics of all 113 patients are summarized in Table 1. The vast majority of patients were men (84%) and in early pathologic stages (78%; stage I to II). Median age was 66.4 years (range, 40 to 84 years). Most of the patients were diagnosed with either SCC (n = 58, 51.3%) or ADC/bronchioloalveolar carcinomas (n = 38, 33.6%). Patients with ADC were younger than those with SCC (P = 0.05). Former or current smokers represented 92% of all patients and were more common in SCC than in ADC (P = 0.001). Well- differentiated grading was found more frequently in ADC than in SCC patients (P < 0.0001). Median OS was 49.5 months (range, 0.5 to 99.7 mo), and median DFS was 41.6 months (range, 0.49 to 99.7 mo). As expected, mean OS and mean DFS were much longer for patients in stage I to II than in stage III (mean ± SD, 53.9 ± 3.0 mo vs. 34.2 ± 5.9 mo; P = 0.0016 and 47.2 ± 3.4 vs. 22.1 ± 5.6 mo; P = 0.0003, respectively).
TABLE 1.
Clinicopathologic Characteristics of NSCLC Patients According to Histologic Types
| SCC (n = 58) | ADC (n = 38)* | Others (n = 17)† | P | |
|---|---|---|---|---|
| Age (y) | ||||
| Median | 69 | 65 | 68 | 0.05 |
| Mean ± SD | 67.4 ± 7.5 | 64.5 ± 9.6 | 67 ± 7.3 | |
| Sex, n (%) | ||||
| Male | 53 (91.4) | 28 (73.7) | 14 (82.4) | 0.05 |
| Female | 5 (8.6) | 10 (26.3) | 3 (17.6) | |
| Smoking status, n (%) | ||||
| Former/current | 58 (100.0) | 31 (81.6) | 15 (88.2) | 0.001 |
| Never | 0 (0.0) | 7 (18.4) | 2 (11.8) | |
| pTNM stage, n (%) | ||||
| I | 34 (58.6) | 28 (73.7) | 8 (47.1) | 0.11 |
| II | 13 (22.4) | 2 (5.3) | 3 (17.7) | |
| III | 11 (19.0) | 8 (21.0) | 6 (35.3) | |
| Grading, n (%) | ||||
| Well differentiated | 4 (6.9) | 8 (21.0) | 0 (0.0) | < 0.0001 |
| Moderately differentiated | 31 (53.5) | 20 (52.6) | 0 (0.0) | |
| Poorly differentiated | 23 (39.6) | 10 (26.4) | 17 (100.0) |
ADC category also includes bronchioloalveolar carcinoma.
Other histology include large cell carcinoma (n = 9) and mixed histology (n = 8).
ADC indicates adenocarcinoma; SCC, squamous cell carcinoma.
Following surgery, 5 patients (4.4%) were treated with chemotherapy, 10 (8.9%) with radiotherapy, 1 (0.9%) with chemoradiotherapy, and the remaining 97 (85.8%) received no adjuvant treatment.
MYC and TERC FISH
Increase in the MYC GCN was measured using FISH and analyzed according to the Colorado scoring system and the ROC classification. When Colorado criteria were applied, 41 (36.2%) cases in total were considered MYC FISH-positive. Specifically, 32 (78%) patients showed high polysomy and 9 (22%) showed gene amplification (GA) (Fig. 2B). MYC high polysomy patients included 18 (56.2%) SCC, 9 (28.1%) ADC, 4 (12.5%) large cell carcinoma, and 1 (3.2%) mixed histology; gene amplification patients included 5 (55.6%) SCC, 2 (22.2%) ADC, and 2 (22.2%) mixed histology. MYC FISH status was not associated with sex, smoking history, histologic types, grading, or pathologic stage (Table 2). Using the ROC classification, a cutoff of MYC copy number per cell at 2.83 identified an area under the curve of 0.62 with sensitivity of 0.61 and specificity of 0.60 (Fig. 1A). Using this cutoff, high MYC GCN (≥2.83 copies per cell) was observed in 60 (53.1%) cases, and no significant differences were observed in sex, smoking status, histology, grading, or pathologic stage between patients with and without high MYC GCN (Table 2).
FIGURE 2.
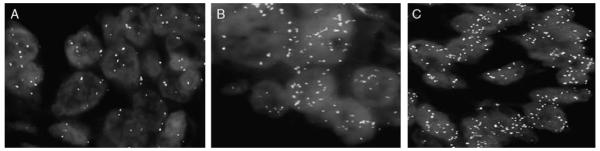
Non–small cell lung cancer sections hybridized with the MYC (Spectrum Aqua)/TERC (Spectrum Gold) probe set showing low copy numbers (A), high copy numbers for MYC (B), and high copy numbers for TERC (C).
TABLE 2.
Clinicopathologic Variables of Patients According to MYC and TERC Gene Copy Status Based on the UCCC Criteria and ROC Classification
| UCCC Criteria |
ROC Classification |
UCCC Criteria |
ROC Classification |
|||||
|---|---|---|---|---|---|---|---|---|
|
MYC FISH + (n = 41, 36.2%) |
MYC FISH − (n = 72, 63.8%) |
MYC≥2.83 (n = 60, 53.1%) |
MYC<2.83 (n = 53, 46.9%) |
TERC FISH + (n = 41, 36.2%) |
TERC FISH − (n = 72, 63.8%) |
TERC≥2.65 (n = 58, 51.3%) |
TERC<2.65 (n = 55, 48.7%) |
|
| Sex, n (%) | ||||||||
| Female | 6 (14.6) | 12 (16.7) | 9 (50.0) | 9 (50.0) | 4 (9.8) | 14 (19.4) | 7 (38.9) | 11 (66.1) |
| Male | 35 (85.4) | 60 (83.3) | 51 (53.6) | 44 (46.3) | 37 (90.2) | 58 (80.6) | 51 (53.7) | 44 (46.3) |
| P | 1.0 | 0.77 | 0.28 | 0.25 | ||||
| Smoking, n (%) | ||||||||
| Former/ current |
40 (97.6) | 64 (88.9) | 56 (53.8) | 48 (46.3) | 40 (97.6) | 64 (88.9) | 56 (53.8) | 48 (46.2) |
| Never | 1 (2.4) | 8 (11.1) | 4 (44.4) | 5 (55.6) | 1 (2.4) | 8 (11.1) | 2 (22.2) | 7 (77.8) |
| P | 0.15 | 0.58 | 0.15 | 0.06 | ||||
| pTNM stage, n (%) | ||||||||
| I | 25 (61.0) | 45 (62.5) | 39 (55.7) | 31 (44.2) | 24 (58.5) | 46 (63.9) | 36 (51.4) | 34 (48.6) |
| II | 9 (22.0) | 9 (12.5) | 10 (55.6) | 8 (44.8) | 7 (17.1) | 11 (15.3) | 10 (55.6) | 8 (44.4) |
| III | 7 (17.0) | 18 (25.0) | 11 (44.0) | 14 (56.0) | 10 (24.4) | 15 (20.8) | 12 (48.0) | 13 (52.0) |
| P | 0.32 | 0.58 | 0.81 | 0.88 | ||||
| Histology, n (%) | ||||||||
| Squamous | 23 (56.1) | 35 (48.1) | 32 (55.2) | 26 (44.8) | 31 (75.6) | 27 (37.5) | 42 (72.4) | 16 (27.6) |
| Non | 18 (43.9) | 37 (51.4) | 28 (50.9) | 27 (40.1) | 10 (24.4) | 45 (62.5) | 16 (29.1) | 39 (20.9) |
| Squamous | ||||||||
| P | 0.56 | 0.65 | 0.0001 | < 0.0001 | ||||
| Grading, n (%) | ||||||||
| Well | 2 (4.9) | 10 (13.9) | 6 (50.0) | 6 (50.0) | 3 (7.3) | 9 (12.5) | 6 (50.0) | 6 (50.0) |
| Moderately | 19 (46.3) | 32 (44.4) | 27 (53.0) | 24 (47.0) | 20 (48.8) | 31 (43.0) | 24 (47.1) | 27 (52.9) |
| Poorly | 20 (48.8) | 30 (41.7) | 27 (54.0) | 23 (46.0) | 18 (43.9) | 32 (44.5) | 25 (50.0) | 25 (50.0) |
| P | 0.34 | 0.56 | 0.72 | 0.95 | ||||
FISH indicates fluorescence in situ hybridization; ROC, receiver operating characteristic classification; UCCC, The University of Colorado Cancer Center criteria.
FIGURE 1.
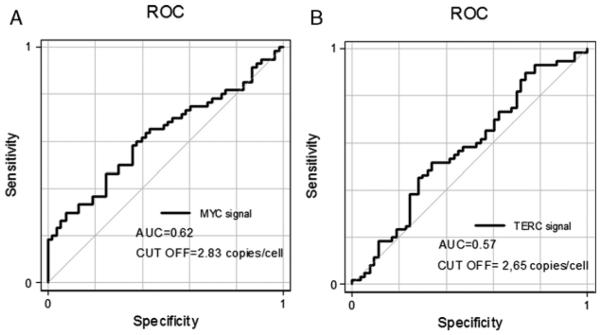
Receiver operating characteristic (ROC) curves for MYC (A) and TERC (B).
TERC GCN was similarly evaluated according to both the Colorado scoring system and ROC classification. When Colorado scoring was applied, 41 (36.2%) cases were considered TERC FISH-positive, of which 26 (23.0%) showed high polysomy and 15 (13.3%) showed gene amplification (Fig. 2C). TERC high polysomy patients included 20 (76.9%) SCC, 2 (7.7%) ADC, 1 (3.9%) large cell carcinoma, and 3 (11.5%) mixed histology; gene amplification patients included 11 (73.4%) SCC, 3 (20%) ADC, and 1 (6.6%) large cell carcinoma. TERC FISH-positive was associated with SCC histology (75.6% vs. 24.4% in non-SCC; P = 0.0001) but no significant differences were observed with sex, smoking history, grading, or pathologic stage (Table 2). The ROC curve classification identified a cutoff of TERC copy number per cell at 2.65 for an area under the curve of 0.57, with sensitivity of 0.58 and specificity of 0.52 (Fig. 1B). Using this cutoff, high TERC GCN was observed in 58 (51.3%) cases (mean per cell 2.65 copies). TERC FISH-positive was associated with SCC histology (72.4% vs. 29.1% in non-SCC; P < 0.0001) but was not associated with other clinicopathologic features (Table 2).
Concurrent MYC and TERC FISH-positive patterns were observed in 12 cases (11%): 2 (17%) cases with GA and 10 (83%) cases with high polysomy for both genes.
Survival Analysis
At a median follow-up time of 53.9 months, 55 patients (48.6%) had died: 41 (74.5%) deaths were due to disease recurrence and 14 (25.5%) to unrelated causes. Eleven (18.9%) of the 58 patients still on follow-up experienced recurrence: local recurrence was observed in 7 patients (63.6%) and recurrence in lung and other sites in 4 patients (36.4%).
We first analyzed patient survival according to MYC and TERC FISH status based on the UCCC criteria. In the univariate analysis summarized in Table 3, MYC FISH-positive showed a shorter DFS and OS than MYC FISH-negative NSCLC patients (median DFS, not reached vs. 26.5 mo; P = 0.032, Fig. 3A; median OS, not reached vs. 45.5 mo; P = 0.032, Fig. 3B). No difference in DFS and OS was found between TERC FISH-positive and negative patients. Similar results were observed for each of the genes adopting the ROC classification. Patients with high MYC GCN (≥2.83 per cell) showed a tendency for shorter DFS than those with low MYC GCN (median DFS, not reached vs. 33.3 mo; P = 0.06, Fig. 3C). Patients with high MYC GCN (≥2.83 per cell) had shorter OS than those with low MYC GCN (median OS, not reached vs. 42.9 mo; P = 0.02, Fig. 3D). No difference in DFS and OS was detected according to high TERC GCN (≥2.65per cell) or low TERC GCN (< 2.65 per cell).
TABLE 3.
Univariate Analysis of Disease-free Survival and Overall Survival for Variables Considered
| Disease-free Survival |
Overall Survival |
|||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P | HR | 95% CI | P |
| Sex (male/female) | 0.82 | 0.39-1.73 | 0.60 | 0.78 | 0.31-1.57 | 0.48 |
| Age (y) continuous | 1.01 | 0.98-1.05 | 0.25 | 1.03 | 1.00-1.06 | 0.042 |
| Smoking (never/ever) | 0.86 | 0.31-2.38 | 0.77 | 0.70 | 0.30-1.64 | 0.42 |
| Histology (squamous/other) | 1.27 | 0.76-2.13 | 0.35 | 1.05 | 0.65-1.68 | 0.85 |
| Grading (G3/G1-G2) | 1.53 | 0.92-2.55 | 0.10 | 1.5 | 0.93-2.40 | 0.095 |
| TNM stage (II-III/I) | 2.86 | 1.64-4.97 | < 0.0001 | 2.47 | 1.48-4.12 | 0.001 |
| MYC FISH (positive/negative)* | 1.84 | 1.05-3.24 | 0.032 | 1.74 | 1.05-2.90 | 0.032 |
| MYC GCN (≥2.83/<2.83 copies)† | 1.71 | 0.96-3.04 | 0.06 | 1.80 | 1.06-3.06 | 0.02 |
| TERC FISH (positive/negative)* | 1.49 | 0.84-2.63 | 0.16 | 1.53 | 0.91-256 | 0.10 |
| TERC GCN (≥2.65/<2.65 copies)† | 1.34 | 0.76-2.37 | 0.30 | 1.37 | 0.82-2.29 | 0.22 |
Using the University of Colorado Cancer Center (UCCC) criteria the tumors were considered FISH-positive when ≥4 gene copies displayed in ≥40% of tumor cells.
Receiver operating characteristic (ROC) classification.
CI indicates confidence interval; FISH, fluorescence in situ hybridization; GCN, gene copy number; HR, hazard ratio.
FIGURE 3.
Kaplan-Meier curves of disease-free survival (DFS) and overall survival (OS) according to MYC FISH-positive and MYC FISH- negative based on the University of Colorado Cancer Center (UCCC) criteria (A and B) and according to MYC gene copy number (GCN) ≥2.83 per cell and MYC GCN < 2.83 per cell based on the receiver operating characteristic (ROC) classification (C and D) in non–small cell lung cancer (NSCLC) patients. FISH indicates fluorescence in situ hybridization.
Two multivariate Cox regression models for DFS and OS were built according to UCCC criteria (model 1) and ROC classification (model 2). Both the multivariate Cox regression models included the variables (stage and age) that were found significant at the univariate analysis. Increased MYC FISH-positive according to the UCCC criteria or ROC classification remained significantly associated with worse DFS (HR, 1.92; 95% CI, 1.09-3.38; P = 0.02 and HR, 1.97; 95% CI, 0.97-3.54; P = 0.02, respectively) and OS (HR, 1.01; 95% CI, 1.07-2.97; P = 0.02 and HR, 2.05; 95% CI, 1.20-3.50; P = 0.008, respectively). The stage (II to III vs. I) was confirmed to be an independent poor prognostic factor for DFS (model 1: HR, 3.68; 95% CI, 2.01-6.73; P < 0.0001; model 2: HR, 3.52; 95% CI, 1.93-6.43; P < 0.0001) and OS (model 1: HR, 2.68; 95% CI, 1.56-4.61; P < 0.0001; model 2: HR, 2.88; 95% CI, 1.66-4.98; P < 0.0001) (Table 4).
TABLE 4.
Multivariate Analyses of Disease-free Survival and Overall Survival for Variables Considered
| Multivariate Analysis |
||||||
|---|---|---|---|---|---|---|
| Disease-free Survival |
Overall Survival |
|||||
| Variables | HR | 95% CI | P | HR | 95% CI | P |
| Model 1 | ||||||
| MYC FISH (positive/negative)* | 1.92 | 1.09-3.38 | 0.02 | 1.01 | 1.07-2.97 | 0.02 |
| TNM stage (II-III/I) | 3.68 | 2.01-6.73 | < 0.0001 | 2.68 | 1.56-4.61 | < 0.0001 |
| Age (y) continuous | 1.0 | 0.97-1.04 | 0.64 | 1.01 | 0.98-1.05 | 0.26 |
| Model 2 | ||||||
| MYC GCN (≥2.83/<2.83 copies)† | 1.97 | 0.97-3.54 | 0.02 | 2.05 | 1.20-3.50 | 0.008 |
| TNM stage (II-III/I) | 3.52 | 1.93-6.43 | < 0.0001 | 2.88 | 1.66-4.98 | < 0.0001 |
| Age (y) continuous | 1.01 | 0.97-1.04 | 0.49 | 1.02 | 0.98-1.05 | 0.18 |
Using the University of Colorado Cancer Center (UCCC) criteria the tumors were considered FISH-positive when ≥4 gene copies displayed in ≥40% of tumor cells
Receiver operating characteristic (ROC) classification.
CI indicates confidence interval; FISH, fluorescence in situ hybridization; GCN, gene copy number; HR, hazard ratio.
DISCUSSION
To our knowledge, this is the first study, investigating in the same time the prognostic role of both MYC and TERC GCN in early NSCLC patients. Our results showed that high MYC GCN, an event occurring in approximately half of the patients, was an independent prognostic factor in resected NSCLC. In our series, high MYC GCN (either by UCCC criteria or ROC classification) was significantly associated with worse OS. These results are essentially consistent suggesting that NSCLC tumors with increased MYC GCN harbor biologically aggressive phenotypes as previously reported by other authors.10,11 The study of Kubokura et al10 conducted on a small number (31 patients) of resected NSCLC patients showed that MYC amplification correlated with lymph node metastasis, suggesting a possible negative effect on survival. Recently, Iwakawa et al11 found in 65 cases of small-sized ADCs by GeneChip Human Mapping 10-K SNP array and in 162 stage I lung ADCs based on real-time genomic PCR that MYC amplification was a prognostic marker for patients with both small-sized and early-stage I lung ADCs. Two studies by Volm et al9,27 reported that tumors with MYC overexpression protein showed a significant increased of metastasis in early NSCLC patients.
In several studies, the frequency of MYC amplification in NSCLC has been variably reported to be from 5% to 88% in patients having NSCLC who were not treated with EGFR- TKIs.1,6,10 These studies have used FISH method but with different interpretation criteria, which may account for the wide range of rates detected. To address this question, we analyzed MYC gene amplification in NSCLC using FISH and 2 interpretation criteria: the UCCC criteria and the ROC classification. FISH is generally accepted as more accurate than RT- PCR for evaluating GCN because it allows in situ examination of tumor cells, with little or no contamination from non-neo- plastic cells. Nevertheless, the interpretation criteria of MYC GCN using FISH in NSCLC have not been well established. The UCCC criteria used in our manuscript at the state of the art is the standard scoring method applied in clinical studies regarding not only EGFR but also other target genes.24 In a recent study in advanced NSCLC, Cappuzzo et al,28 using ROC analysis, defined high MYC GCN as a mean of ≥2.8 copies per cell for the evaluation of MYC amplification to discriminate responsive from nonresponsive patients to EGFR- TKIs, and demonstrated that high MYC GCN was detected in 53.7% of patients without significant association with clinicopathologic features. In agreement with this study, we also found high MYC GCN in 53.1% (by ROC classification) of our population, without significant correlation between increased MYC GCN (either by UCCC criteria or ROC classification) and sex, smoking history, histology, grading, and stage. Kubokura et al10 reported a much higher incidence of MYC amplification in NSCLC (88%), probably because of the less stringent criteria used for defining gene amplification (MYC GA defined as MYC > centromere 8).
Beyond its prognostic value, MYC has been considered as a possible specific target of biological treatment as demonstrated in a preclinical mouse model of Ras-induced lung ADC.29 Interestingly, Cappuzzo et al28 showed that the contemporary amplification of MYC and eukaryotic translation initiation factor 3 subunit H (EIF3H) was associated with better outcome after gefitinib therapy; among EGFR-positive patients (FISH-positive and/or mutated), only individuals with increased MYC/EIF3H GCN had a significant tumor shrinkage. The results of this study showed for the first time that MYC and EIF3H were coamplified in NSCLC, and this biologic event could positively affect the response and survival of patients treated with EGFR-TKIs. However, these data originated from a retrospective analysis and must be validated with a prospective study. The mechanism responsible for the highest sensitivity to anti-EGFR agents in presence of MYC GCN gain is not clear; however, similar results have been detected in breast carcinomas.30
TERC has been suggested to be a novel putative target in 3q2631 for molecular therapy of NSCLC, as some strategies have been evaluated to target the proteins associated with telomerase such as small molecules, antisense RNA, and ribozymes32 but without a clear impact on the clinical practice up to now.
We found TERC FISH positivity in 36.2% of cases by the UCCC criteria and high TERC GCN in 51.3% of cases according to ROC classification, and no statistically significant difference in DFS and OS was observed between patients with positive and negative TERC GCN using both criteria. This observation confirmed that high TERC GCN is not a prognostic factor for NSCLC, but it showed phenotypic properties strongly associated with SCC histology using both criteria (UCCC, P = 0.0001; ROC, P < 0.0001). These data agree with earlier studies of NSCLC in which DNA copy number gains at 3q26 occurred more frequently in SCC than in ADC.18
Contrary to the observations of Yan et al23 that the amplification of the chromosomal arm 3q (in particular 3q26.2-q29) was significantly higher in smoker than in non-smoker SCC patients, we found no association between high TERC GCN and smoking history. However, this could be because of the low percentage of nonsmoker patients (7.9% cases) in our population.
Recently, Eid et al33 found that TERC amplification and grading were significantly correlated with cervix cancer. Furthermore, there was significant correlation between MYC amplification and grading. TERC and MYC genes amplification were correlated and showed an inverse correlation with patients’ age. The study highlighted the importance of using TERC and MYC copy number gain by FISH in cervix cancer or premalignant lesions as a sensitive technique for early diagnosis and poor prognostic assessment.
In conclusion, both criteria (UCCC criteria and ROC classification) used to evaluate MYC genomic status showed similar results regarding survival, even though ROC classification seems to be more appropriate in terms of predicting the prognosis of NSCLC patients. Moreover, our data suggest that NSCLC with high MYC GCN is able to develop a more aggressive behavior, possibly representing as a biomarker, as tumors with high MYC GCN showed a significantly worse survival. Furthermore, high TERC GCN showed to be as a typical feature of lung SCC. Finally, the limited number of patients and the short follow-up in our study require that these results be interpreted cautiously and be confirmed in large prospective trials.
ACKNOWLEDGMENT
The authors thank the technical assistance of the Cytogenetics Core of the University of Colorado Cancer Center for all FISH analyses.
Supported in part by grants from the Italian Association for Cancer Research (AIRC) to A.F., Umbria Association Against Cancer (AUCC) to L.C., and NCI grants P50 CA58187 and P30 CA046934 to M.V.G.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1.Forozan F, Karhu R, Kononen J, et al. Genome screening by comparative genomic hybridization. Trends Genet. 1997;13:405–409. doi: 10.1016/s0168-9525(97)01244-4. [DOI] [PubMed] [Google Scholar]
- 2.Westermark UK, Wilhelm M, Frenzel A, et al. The MYCN oncogene and differentiation in neuroblastoma. Semin Cancer Biol. 2011;21:256–266. doi: 10.1016/j.semcancer.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Paulson KG, Lemos BD, Feng B, et al. Array-CGH reveals recurrent genomic changes in Merkel cell carcinoma including amplification of L-Myc. J. Invest Dermatol. 2009;129:1547–1555. doi: 10.1038/jid.2008.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonson BE, Ihde BC, Makuch RW, et al. Myc family oncogene amplification in tumor cell lines established from small cell lung cancer patients and its relationship to clinical status and course. J Clin Invest. 1987;79:1629–1634. doi: 10.1172/JCI112999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coller HA, Grandori C, Tamayo P, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000;97:2360–2365. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lockwood WW, Chari R, Coe BP, et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27:4615–4624. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broers JL, Viallet J, Jensen SM, et al. Expression of c-myc in progenitor cells of the bronchopulmonary epithelium and in a large number of non-small cell lung cancers. Am J Respir Cell Mol Biol. 1993;9:33–43. doi: 10.1165/ajrcmb/9.1.33. [DOI] [PubMed] [Google Scholar]
- 8.Dehan E, Ben-Dor A, Liao W, et al. Chromosomal aberrations and gene expression profiles in non-small cell lung cancer. Lung Cancer. 2007;56:175–184. doi: 10.1016/j.lungcan.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Volm M, Drings P, Wodrich W, et al. Expression of oncoproteins in primary human non-small cell lung cancer and incidence of metastases. Clin Exp Metastasis. 1993;11:325–329. doi: 10.1007/BF00058052. [DOI] [PubMed] [Google Scholar]
- 10.Kubokura H, Tenjin T, Akiyama H, et al. Relations of the c-myc gene and chromosome 8 in non-small cell lung cancer: analysis by fluorescence in situ hybridization. Ann Thorac Cardiovasc Surg. 2001;7:197–203. [PubMed] [Google Scholar]
- 11.Iwakawa R, Kohno T, Kato M, et al. Myc amplification as a prognostic marker of early stage lung adenocarcinoma identified by whole genome copy number analysis. Clin Cancer Res. 2011;17:1481–1489. doi: 10.1158/1078-0432.CCR-10-2484. [DOI] [PubMed] [Google Scholar]
- 12.Tonon G, Wong KK, Maulik G, et al. High resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi YW, Choi JS, Zheng LT, et al. Comparative genomic hybridization array analysis and real time PCR reveals genomic alterations in squamous cell carcinomas of the lung. Lung Cancer. 2007;55:43–51. doi: 10.1016/j.lungcan.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Kendall J, Liu Q, Bakleh A, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng J, Funk WD, Wang SS, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 17.Fan YB, Ye L, Wang TY, et al. Correlation between morphology and human telomerase gene amplification in bronchial brushing cells for the diagnosis of lung cancer. Diagn Cytopahol. 2010;38:402–406. doi: 10.1002/dc.21235. [DOI] [PubMed] [Google Scholar]
- 18.Pei J, Balsara BR, Li W, et al. Genomic imbalances in human lung adenocarcinomas and squamous cell carcinomas. Genes Chromo-somes Cancer. 2001;31:282–287. doi: 10.1002/gcc.1145. [DOI] [PubMed] [Google Scholar]
- 19.Pelosi G, Del Curto B, Trubia M, et al. 3q26 amplification and polysomy of chromosome 3 in squamous cell lesions of the lung: a florescence in situ hybridization study. Clin Cancer Res. 2007;13:1995–2004. doi: 10.1158/1078-0432.CCR-06-2483. [DOI] [PubMed] [Google Scholar]
- 20.Foster NA, Banerjee AK, Xian J, et al. Somatic genetic changes accompanying lung tumor development. Genes Chromosomes Cancer. 2005;44:65–75. doi: 10.1002/gcc.20223. [DOI] [PubMed] [Google Scholar]
- 21.Heselmeyer K, Schrock E, du Manoir S, et al. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA. 1996;93:479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh B, Stoffel A, Gogineni S, et al. Amplification of the 3q26.3 locus is associated with progression to invasive cancer and is a negative prognostic factor in head and neck squamous cell carcinomas. Am J Pathol. 2002;161:365–371. doi: 10.1016/S0002-9440(10)64191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan WS, Song LY, Wei WD, et al. Chromosomal imbalance in primary lung squamous cell carcinoma and their relationship with smoking. Ai Zheng. 2005;24:47–52. [PubMed] [Google Scholar]
- 24.Varella-Garcia M. Stratification of non-small cell lung cancer patients for therapy with epidermal growth factor receptor inhibitors: the EGFR fluorescence in situ hybridization assay. Diagn Pathol. 2006;15:1–19. doi: 10.1186/1746-1596-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beasley MB, Brambilla E, Travis WD. The 2004 World Health Organization. Classification of lung tumors. Semin Roentgenol. 2005;40:90–97. doi: 10.1053/j.ro.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Cai T, Moskowitz CS. Semi-parametric estimation of the binomial ROC curve for a continuous diagnostic test. Biostatistics. 2004;5:573–586. doi: 10.1093/biostatistics/kxh009. [DOI] [PubMed] [Google Scholar]
- 27.Volm M, van Kaick G, Mattern J. Analysis of c-fos, c-jun, c-erbB1, c-erbB2 and c-myc in primary lung carcinomas and their lymph node metastases. Clin Exp Metastasis. 1994;12:329–334. doi: 10.1007/BF01753840. [DOI] [PubMed] [Google Scholar]
- 28.Cappuzzo F, Varella-Garcia M, Rossi E, et al. MYC and EIF3H coamplification significantly improve response and survival of non-small cell lung cancer patients (NSCLC) treated with gefitinib. J Thorac Oncol. 2009;4:472–478. doi: 10.1097/JTO.0b013e31819a5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soucek L, Whitfield J, Martins CP, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim C, Bryant J, Horne Z, et al. Trastuzumab sensitivity of breast cancer with co-amplification of HER2 and cMYC suggests proapoptotic function of dysregulated cMYC in vivo. Breast Cancer Res Treat. 2005;94:S6. (abstract 46) [Google Scholar]
- 31.Yokoi S, Yasui K, Iizasa T, et al. TERC identified as a probably target within the 3q26 amplicon that is detected frequently in non-small cell lung cancers. Clin Cancer Res. 2003;9:4705–4713. [PubMed] [Google Scholar]
- 32.Chen H, Li Y, Tollefsbol TO. Strategies targeting telomerase inhibition. Mol Biotechnol. 2009;41:194–199. doi: 10.1007/s12033-008-9117-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eid MM, Nossair HM, Ismael MT, et al. Clinical significance of hTERC and C-myc genes amplifications in a group of Egyptian patients with cancer cervix. Gulf J Oncolog. 2011;1:18–26. [PubMed] [Google Scholar]



