Abstract
Whole genome and exome sequencing tests are increasingly being ordered in clinical practice, creating a need for research exploring the return of results from these tests. A goal of the Clinical Sequencing and Exploratory Research (CSER) consortium is to gain experience with this process to develop best practice recommendations for offering exome and genome testing and returning results. Genetic counselors in the CSER consortium have an integral role in the return of results from these genomic sequencing tests and have gained valuable insight. We present seven emerging themes related to return of exome and genome sequencing results accompanied by case descriptions illustrating important lessons learned, counseling challenges specific to these tests and considerations for future research and practice.
Keywords: case studies, clinical genomics, exome sequencing counseling, genetic, genome sequencing, genomic medicine, incidental ersonalized findings, medicine, return of results
Background
The application of exome and genome sequencing tests in clinical medicine requires thorough exploration of the practical challenges, as well as the ethical issues and societal implications associated with the use of this technology. The research projects comprising the National Human Genome Research Institute (NHGRI) and National Cancer Institute (NCI), Clinical Sequencing and Exploratory Research (CSER) consortium began in late 2011 with the goal “to support both the methods development needed to integrate [genomic] sequencing into the clinic and the ethical, legal and psychosocial research required to responsibly apply personal genomic sequence data to medical care” [1]. One of the primary aims of the CSER program is to gain experience with and develop best practice recommendations for the return of results from exome and genome sequencing tests.
To date, these genomic sequencing tests have been primarily used for specific diagnostic purposes. One of the difficulties with the application of this technology to clinical diagnostic testing is choosing when exome and genome tests are the appropriate tool, and algorithms to aid in this decision have been proposed [2]. Challenges also arise with the generation of genomic sequence data that includes variants unrelated to the testing indication, or incidental findings (IFs). How best to manage the return of IFs from exome and genome tests is a topic of debate. Recommendations from the American College of Medical Genetics and Genomics (ACMG) addressed the return of IFs from clinical genomic sequencing tests [3], supporting the return of pathogenic variants in a specified list of genes associated with highly penetrant, medically actionable conditions. An amendment to these recommendations [4] includes a provision enabling patients the option to ‘opt-out’ of the return of IFs prior to testing. Statements specifically addressing return of genomic sequencing results to research participants have also been published [5] endorsing the need for research that explores returning a wide range of results as long as they meet an actionability threshold defined by the study and the participant has been consented for their return. The processes used by six CSER consortium sites to address their project’s questions of which IFs to disclose, and how, have been previously described and highlight the similarities and differences in approaches to resolving this issue [6]. Multiple factors appear to influence the views of researchers regarding what results to disclose including the evidence supporting the clinical significance of the variant being returned and concerns for participant well-being and researcher responsibility [7].
Literature exploring patient and/or participant preferences in the return of genomic results have shown that there is variability in the type of results individuals want to receive, and how they want to receive them [8,9]. Context (i.e., personal and family history) likely plays a role in the response to possibly learning results and their dissemination to other family members [10]. Cultural differences have also been observed with respect to interest in exome and genome sequencing, preferences for results and expectations [11,12].
The application of exome and genome sequencing tests in pediatric populations raises unique and challenging ethical concerns related to return of IFs and patient and/or participant preferences. Parental perceptions of the value of IFs for their children are likely an important factor when soliciting preferences for return of results [13]. Genetic counselors play a central role in engaging children and adolescents old enough to be a part of informed consent discussions and results disclosure sessions [14]. Guidelines taking into account issues of developing autonomy in pediatric participants, the duty to mitigate harm, family dynamics and other related concerns have been created [15].
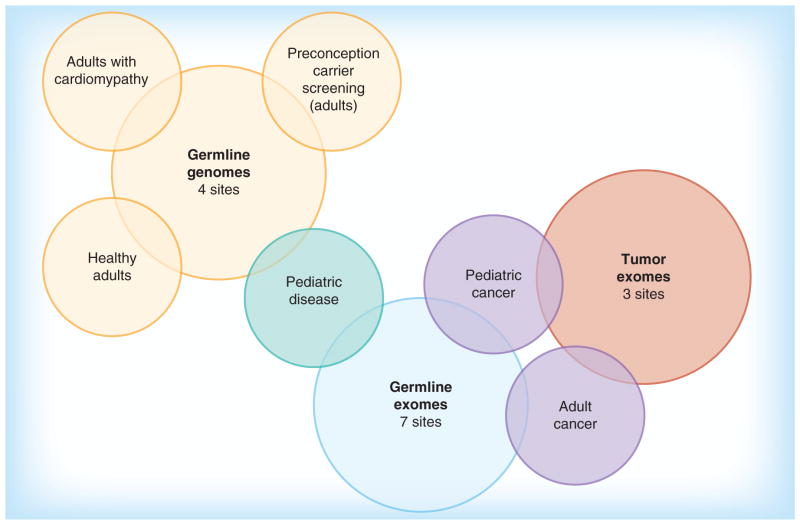
The cases described below highlight emerging themes genetic counselors in the CSER consortium have encountered in returning results from exome and genome sequencing to diverse populations. Children and adults are enrolled in these research studies with a variety of diagnostic indications, including developmental delay, congenital anomalies, cardiomyopathies and familial cancer. Return of results from sequencing of healthy individuals is also being explored. The number of sites performing germline exome and genome sequencing and tumor exome sequencing, as well as the phenotypes of participants, are presented in Figure 1. Types of results returned include diagnostic findings, IFs and carrier status. The lessons learned come from experiences with the return of both diagnostic results and IFs, and involve patient-participants and their family members. The term ‘patient-participant’ is used here to emphasize that, while these individuals are enrolled in a research study and are returned research results, their results may be incorporated into their clinical care and are generally returned in a clinical setting. The encounters described are meant to emphasize the particular challenge that arose with that specific patient-participant and family, and are not intended to be generalized to all cases of a similar nature.
Figure 1.
Collective experience with return of results from germline genome sequencing, germline exome sequencing and tumor exome sequencing in various clinical contexts .
Methods
Themes for lessons learned from the return of results from exome and genome sequencing tests were elicited from members of the CSER consortium Genetic Counseling Working Group. This Working Group was established in May 2013 with the mission to investigate current genetic counseling topics related to whole exome and genome sequencing and utilize experiences from the CSER consortium to inform best practices and launch related genetic counseling research. The Working Group has 39 members, the majority of whom are genetic counselors involved in a variety of roles within the CSER consortium research projects and have held past or current positions in clinical genetics practice. Themes were discussed and refined, and ultimately chosen by group consensus. Selection of themes was based on two factors: experience with illustrative cases and relevance to the topic of return of results. Case descriptions and lessons learned were summarized by the genetic counselor most closely affiliated with the case and were compiled. Genetic counselors contributing cases have extensive experience with return of results visits for research participants in their respective CSER consortium projects, as well as experience with return of clinical genetic test results. All cases involved patient-participants and families enrolled in Institutional Review Board (IRB)-approved protocols at their home institution. A method framework illustrating our approach is presented in Figure 2.
Figure 2.
Framework for identifying and refining themes and illustrative cases.
Results
A summary of the themes, lessons learned and challenges specific to the return of exome and genome sequencing results described in the cases below are presented in Table 1.
Table 1.
Summary of themes, lessons learned and challenges specific to the return of exome and genome sequencing results.
| Theme | Lesson(s) learned | Challenges specific to exome and genome sequencing |
|---|---|---|
| Managing expectations in pretest and post-test counseling, negative findings do not mean the condition is not genetic | Elicit perceived goals and expectations both during informed consent and after return of results to identify and address misconceptions | Belief that all pathogenic genetic variation can be identified and the clinical significance will be clear |
| Context matters: follow-up for recommendations from IFs in healthy and ill patient-participants | Both healthy and ill patient-participants who receive IFs may face challenges with adherence to screening/testing recommendations. Ill patient-participants may focus on the diagnostic results and over-interpret a negative result as ‘good news’ | Limited pretest discussion of the unanticipated condition(s) and implications of results. (Ill) Emphasizing importance of follow-up for medically actionable IFs in the context of more acute concerns. (Healthy) Lack of personal/family history may affect motivation and access to care |
| Considerations for returning large amounts of data | Discussing multiple findings may require two or more visits, separate report sections for the discussion of each finding and an accompanying counseling letter | Returning and discussing several additional findings unrelated to the primary diagnosis |
| Differing responses from families to the same result | Identify and explore unique thoughts and emotions of the patient-participant | Anticipating reactions when the range of possible results has not been discussed in detail prior to testing |
| Challenges with follow-up testing for family members of sequenced patient- participants | Targeted clinical follow-up testing may not be available to or feasible for relatives/partners | Follow-up testing for genes covered by genome/exome testing may not be available outside of a research study |
| Navigating the atypical presentation of well-known Mendelian condition | Challenging to identify resources, anticipate future medical implications and predict the manifestation of the condition in other family members | Providing genetic counseling for patients with atypical presentations will take more time and take place more often than with targeted gene sequencing |
| Deceased patients and communicating results with family members | Establish a plan during informed consent regarding whom, if anyone, to return results to posthumously including what type of results to share | Deciding what results to share from broad range of possible findings increased likelihood due to use of exome and genome sequencing tests in later stage cancer patients |
IF: Incidental finding.
Theme 1: Managing expectations in pretest & post-test counseling, negative findings do not mean the condition is not genetic
Testing scenario
A 5-year-old female with severe intellectual disability, hypotonia and a brain malformation was enrolled in a study which performs germline exome sequencing and returns primary diagnostic findings related to developmental delay, as well as IFs. Previous clinical genetic testing for this patient-participant, a microarray and mitochondrial disease testing, were nondiagnostic. During the informed consent counseling session the patient-participant’s mother articulated a misconception about the scope of exome sequencing when she asked, “so this test looks at everything?” At that time, the genetic counselor engaged the patient-participant’s mother in a detailed discussion regarding the types of genetic variation exome sequencing may or may not identify, and the possible results that could be returned, including their implications for her daughter’s future care.
Return of results
No diagnostic results or IFs were identified for return to the patient-participant. The family was given these negative results at an in-person clinic appointment with a medical geneticist and genetic counselor. The parents summarized their understanding of these results as “good news, because we do not think it is genetic.” At this point the limitations of exome sequencing were reiterated and the significance of a negative result was explained again. The strong possibility that their daughter could still have a genetic diagnosis that cannot be identified at this time was emphasized.
Lessons learned
This case illustrates the need to elicit perceived goals and expectations both during the informed consent conversation and after return of results so that misconceptions surrounding the limitations of exome or genome sequencing can be identified and addressed. While this issue arises in the interpretation of all negative genetic test results and particularly in prenatal testing, it is exacerbated by the false impression that, given their large scale approach, exome and genome sequencing tests can identify all types of pathogenic genetic variation, and that the clinical significance of all variants identified will be clear.
Theme 2: Context matters: follow-up for recommendations from IFs in healthy & ill patient-participants
Testing scenario (healthy patient-participant)
A 40-year-old reportedly healthy woman whose child has a congenital heart defect (CHD) was enrolled in a study which performs exome sequencing and returns IFs based on the preferences of the patient-participant. The patient-participant had the option of receiving all, none or certain types of IFs. She elected to receive all IF results.
Return of results
A pathogenic variant in the MYH7 gene associated with a risk for hypertrophic cardiomyopathy (HCM) was identified in the patient-participant. HCM is characterized by hypertrophy of the left ventricle of the heart and the symptoms vary from none to progressive heart failure or sudden cardiac death. The MYH7 pathogenic variant is not associated with structural heart disease and therefore was unrelated to the CHD diagnosed in her child, and was returned as an IF. Individuals with a genetic predisposition to develop HCM are recommended to have regular cardiac screening and their first-degree relatives should be tested for the pathogenic variant in the family. During results disclosure, the patient-participant reported no personal or family history of HCM and she had never had an echocardiogram. She expressed an appropriate degree of concern at the visit and an assessment of depression (Personal Heath Questionnaire-9) and anxiety (Beck Anxiety Inventory) symptoms showed no clinically significant change before and after results return. One week after the return of this result the patient-participant requested testing kits for all of her healthy children since her child with CHD would be evaluated for HCM at his next routine cardiology appointment. To date, this patient-participant has not returned the testing kits for her other children. In a study phone interview one-year after return of results, she indicated that she had shared her results with her physician and was planning to have cardiac screening, but was very busy with her son’s care. She had also shared her results and recommendations for genetic testing and cardiac screening with her parents and siblings soon after she received her results but they had not yet pursued evaluations.
Lessons learned
This case illustrates that healthy patient-participants who receive IFs from exome or genome sequencing that indicate the need for further screening/testing may have unique challenges with respect to following up on recommendations. It is possible that these IFs are less expected, particularly in the context of a negative family history, or that the healthy patient-participant is less connected to the health care system. In this circumstance, genetic counselors need to balance emphasizing the importance of follow-up screening/care in the absence of symptoms and preventing undue anxiety for conditions with reduced penetrance and variable expressivity. Assessing patient-participant understanding is important with all genetic testing; however, it should be emphasized in this scenario. While pretest counseling for IFs includes a broad discussion of many different types of conditions and the possibility that results could indicate a need for screening and/or prophylactic treatment, it is not the detailed discussion that occurs in a disease specific pretest counseling session. Additional research into the uptake of recommendations and psychological impact following disclosure of IFs to healthy individuals may help genetic counselors to develop additional counseling tools for these cases.
Testing scenario (ill patient-participant)
The international family of a 16 month old male diagnosed with a brain tumor was enrolled in a study which involves tumor and blood whole exome sequencing of childhood cancer patients and the return of results including: tumor-specific variants categorized by significance to clinical care, and pathogenic variants and variants of uncertain significance in cancer susceptibility genes and medically actionable IF genes. The parents of this patient-participant provided blood to be used for interpretation of variants identified by germline exome sequencing of the child.
Return of results
At disclosure, the patient-participant’s clinical oncologist and a study genetic counselor were present along with the father. Tumor exome sequencing did not identify changes that would currently impact the patient-participant’s clinical cancer care. Germline exome sequencing also did not identify pathogenic variants in cancer predisposition genes. However a medically actionable IF – a maternally inherited pathogenic variant in the SCN5A gene previously reported in individuals with Long QT syndrome (LQTS) – was identified. The oncologist began the session by explaining that the test did not identify pathogenic variants in the germline related to tumor predisposition syndromes. The father was then informed about the IF related to LQTS and the recommendation was made for a referral to cardiology for the patient-participant and the mother based on this result. Despite identification of the SCN5A pathogenic variant, the father’s response was relief. The father disclosed that his fear in attending the visit was based on his concern that the test would find a variant in the tumor sequence that would suggest chemo-resistance. He described these results as ‘good’ and stated that now the worry was behind him. The patient-participant was evaluated by cardiology. He was asymptomatic and described by the cardiologist as “genotype positive, phenotype negative” with a need for continued follow-up. Maternal follow-up status is unknown as the family has since returned to their home country.
Lessons learned
This case illustrates that sick patient-participants may need additional counseling regarding how a negative report does or does not affect their prognosis, and that follow-up for IFs can be challenging in this context. For patient-participants and their parents undergoing tumor sequencing, the potential impact of results on treatment or prognosis in the immediate future may be most important. It is imperative that efforts are made to ensure that a patient-participant does not overinterpret a negative report and recognizes that a negative tumor report does not necessarily provide ‘good’ news or ‘bad’ news. A unique challenge raised by exome or genome sequencing in this scenario is the need to emphasize the importance of follow-up for medically actionable IFs in the context of more acute concerns. More generally it is important to establish a research infrastructure to longitudinally follow genotype positive IF cases. In this case, the child’s “genotype positive, phenotype negative” status, demonstrates the evolving clinical characterization that will come along with genomic testing.
Theme 3: Considerations for returning large amounts of data
Testing scenario
A 17-year-old female with probable LQTS and a strong maternal family history of LQTS was enrolled in a study which performs germline exome sequencing for pediatric patient-participants with a variety of phenotypes and returns results related to the primary indication and IFs including carrier status for selected Mendelian disorders. Of note, the patient-participant’s father had a diagnosis of cardiomyopathy that was thought not to be hereditary. Her mother, with patient-participant assent, opted for her to receive pathogenic and likely pathogenic results related to cardiac disease, immediately actionable IF results and IFs related to actionable adult-onset disorders and carrier status. In the informed consent discussion, the patient-participant admitted to being fearful of the unknown and commented “You are not going to tell me anything bad, right?”
Return of results
Four variants of unknown significance related to cardiac diseases were identified: one in the SNTA1 gene (associated with LQTS), one in the MYH7 gene and one in the SGCD gene (both associated with cardio-myopathy) and one in the DSP gene (associated with arrythmogenic right ventricular dysplasia). An additional 5 IFs were returned including a pathogenic variant in the RYR1 gene (associated with malignant hyperthermia) and variants indicating that the participant is a carrier for four recessive conditions. The maternally inherited SNTA1 variant was strongly suspected to be the underlying genetic cause for the patient-participant’s cardiac symptoms and segregated with the condition in her maternal family. The MYH7 variant was paternally inherited and may be related to the patient-participant’s father’s history of cardio-myopathy. The return of results visit lasted 2 hours and the participant appeared to feel overwhelmed and anxious, though she did ask appropriate questions. A copy of the 11-page research report was given to the patient-participant and her parents at the beginning of the return of results session and at times they each skipped ahead to other sections/genes, which caused more confusion.
Lessons learned
This case illustrates challenges that arise with returning and discussing several additional findings unrelated to the primary diagnosis, a scenario unique to exome and genome sequencing tests. While panel gene tests ordered in standard clinical genetics practice for individuals with genetically heterogeneous conditions can lead to uncertainty in the interpretation of results when variants are identified in more than one gene, the complexity is compounded here by IFs that would not have been included in a panel. As this can overwhelm the patient-participant, physicians and genetic counselors may elect to return results from these tests in two or more visits. It may also be helpful to separate the results report into sections and distribute it to participants as the discussion of that section is completed. An accompanying counseling letter or a simplified results report for future reference may also facilitate understanding in scenarios where a large amount of complex information is discussed at one time. This will likely also facilitate communication regarding the results with other family members.
Theme 4: Differing responses from families to the same result
Testing scenario
Two unrelated 6-year-old females with developmental delay and microcephaly were both independently enrolled in a study that performs germline exome sequencing and returns primary diagnostic findings related to developmental delay, as well as IFs. Prior tests showed a normal female karyotype on one patient and both had microarray testing which was nondiagnostic.
Return of results
No diagnostic results or IFs were identified for either child. Both families were given these negative results in person at a clinic appointment with a medical geneticist and genetic counselor. After the results were explained, the two families responded with contrasting reactions. One family reported that they were glad to have not been given a diagnosis because they did not want a ‘label’ to be put on their daughter. The other family said they were disappointed and wished they had an answer.
Lessons learned
This case illustrates that, similar to receiving results from traditional genetic tests, families receiving the same result from exome or genome sequencing tests may have different responses to the information presented. Predicting the reaction of patient-participants and their families to results is more challenging in the context of exome or genome sequencing since the range of possible results has not been discussed in detail prior to testing. In this case, it is likely that these families also had different reactions to receiving the negative results from the clinical tests their daughters had prior to enrollment in the study. Similar to the clinical setting, those involved in returning results to research participants and families need to be mindful to not enter interactions with a preconceived notion of how the individual(s) will feel and how they will respond to the results disclosed. Instead it is important to take the time to elicit and explore their unique thoughts and emotions in the context of the personal and family history of the patient-participant and their family.
Theme 5: Challenges with follow-up testing for family members of sequenced patient-participants
Testing scenario
A 34-year-old healthy woman actively considering pregnancy was enrolled in a study which performs germline genome sequencing and returns carrier status results and medically actionable IFs from predefined gene-disease lists. Carrier results and IFs are disclosed at separate visits and family history is not elicited at the time of enrollment.
Return of results
This patient-participant was found to be heterozygous for a pathogenic variant in the SLC3A1 gene, identifying her as a carrier for cystinuria. Cystinuria is a renal condition associated with the development of stones in the kidney, ureter and bladder. When a carrier result is disclosed to a female patient-participant, the study’s protocol dictates recruitment of the male partner for genome sequencing. At the initial consent visit, this patient-participant indicated that her husband may not be interested in enrolling in the study, so study personnel considered that he may want to pursue clinical carrier testing for cystinuria after learning the patient-participant’s carrier status. It was subsequently discovered that clinical carrier testing for cystinuria is not available in a US laboratory. The only way for the patient-participant’s partner to have follow-up testing would be to enroll in a research study performing genomic sequencing that returns carrier status results. At her return of results visit which she attended without her husband, the patient-participant was not concerned about the risk of having a child with cystinuria since she did not perceive it to be a serious condition. She reiterated that she did not expect her husband to be interested in enrolling in the study and he did not.
Lessons learned
This case illustrates that targeted clinical follow-up testing may not be available to the relatives or partners of participants undergoing exome or genome sequencing since clinical single gene tests are not available for all of the genes sequenced by these tests. This may also be true for the relatives or partners of those who have clinical exome and genome sequencing tests. Furthermore, the cost of clinical testing for those who do not elect to enroll in research studies may not be covered by insurance and self-pay may be prohibitive for some. It may be prudent to consider the availability of targeted clinical follow-up testing when deciding what genes to sequence and what variants to return from exome and genome sequencing tests.
Theme 6: Navigating the atypical presentation of well-known Mendelian condition
Testing scenario
A 66-year-old female with a diagnosis of HCM was enrolled in a study which performs whole genome sequencing and returns diagnostic results and IF results of more general significance, including known pathogenic and likely pathogenic Mendelian variants, certain variants of uncertain significance in Mendelian genes, carrier status for Mendelian diseases and known pharmacogenomics associations for five commonly used medications [16]. Five years prior to her enrollment, this patient-participant underwent a targeted genetic testing panel for 11 genes associated with HCM and the results were negative.
Return of results
A pathogenic variant was found in PTPN11, a gene that was not included in this patient-participant’s previous genetic test. The particular PTPN11 variant reported is associated with LEOPARD syndrome. LEOPARD is an acronym for the cardinal features of the condition which are lentigines, ECG conduction abnormalities, abnormal genitalia, retardation of growth and sensorineural deafness [17]. HCM is also a feature of LEOPARD syndrome; however, the patient-participant did not have a history of any of the cardinal findings. Her physician returned this result and explained that the PTPN11 pathogenic variant is likely the cause of her HCM and that other family members, in particular her two daughters, could also be tested. The patient-participant responded enthusiastically to this molecular diagnosis and expressed that she would like to research the condition on her own.
Lessons learned
This case illustrates that exome and genome sequencing technologies will likely identify individuals with pathogenic variants who have nonclassic phenotypes. This challenging scenario will occur more frequently compared with a directed single gene or panel approach to diagnostic testing. Resources for most Mendelian disorders are targeted to individuals with a classic presentation making it challenging for those with nonclassic phenotypes to research and understand their conditions. Clinicians will need to help patient-participants navigate resources and figure out what information pertains to them. Also, with little to no data, clinicians will have to field participant questions regarding what to expect in the future and whether a condition will manifest more severely in other relatives who inherit the same pathogenic variant.
Theme 7: Deceased patient-participants & communicating results with family members
Testing scenario
A 43-year-old woman with a diagnosis of inflammatory breast cancer diagnosed at age 42 was enrolled in a study which performs tumor and germline exome sequencing and returns results related to driver mutations in the tumor for therapeutic decision making, as well as germline secondary findings (defined as variants in genes that are not expected but may explain the phenotype) and IFs. Her family history was notable for a brain tumor diagnosed in her sister at age 37 (anaplastic astrocytoma/glioblastoma) and multiple myeloma diagnosed in her mother at age 77.
Return of results
A TP53 pathogenic variant was identified in 14% of the patient-participant’s germline reads raising concern for germline mosaicism for the variant. The TP53 gene is associated with Li–Fraumeni syndrome, a cancer predisposition syndrome associated with an increased risk for several types of cancer including breast cancer, sarcomas, leukemia, lymphomas and adrenocortical cancer. Additional testing was recommended in a new blood sample as well as other tissue types. Unfortunately, the patient-participant had died by the time testing was completed. The consent form did not identify a process for return of results in this situation. After discussion of the case among study personnel, results were disclosed to the patient-participant’s husband given the significant clinical impact of the germline TP53 pathogenic variant for her family members. The patient-participant’s daughter underwent site specific analysis for the pathogenic variant and was negative. The patient-participant’s sister who had a history of a brain tumor presented separately to an outside institution for clinical genetics evaluation. Based on her personal and family history she met clinical criteria for TP53 germline testing and opted to pursue full TP53 sequencing and deletion/duplication analysis and no mutations were detected.
Lessons learned
This case illustrates the importance of establishing a plan with patient-participants during the informed consent conversation regarding with whom, if anyone, to share results if they are identified posthumously. Additionally, outlining what types of results to share with that alternative contact person is recommended since several different types of results can be identified by exome and genome sequencing tests. This issue is important with the current application of exome and genome sequencing tests for patients and participants in the cancer setting who are at the end stages of their disease. In this case the return of the patient-participant’s results to her family members may have given peace of mind to her daughter who tested negative and provided some guidance for her sister’s clinical genetic testing, even though she was eligible and elected to proceed with full sequencing of the TP53 gene instead of single site analysis. Future research exploring how best to consent patient-participants for return of results to family members in the event of their death, and how best to return results to those designated, is needed.
Discussion
This paper presents selected case descriptions that illustrate some of the types of challenges faced to date by CSER consortium genetic counselors returning exome and genome sequencing results and what has been learned from these encounters. The themes which emerged are not mutually exclusive, for example, cases that involve the return of negative results also require consideration of the context of these disclosures. Also, all possible challenges are not expected to be represented by one or more of the highlighted themes.
Challenges specific to exome & genome sequencing
These cases illustrate that the challenges faced with exome and genome sequencing differ from those faced with single gene and panel genetic tests. The importance of managing patient-participant expectations before and after return of results is heightened with exome and genome sequencing. Patient-participants are counseled that this technology produces enormous amounts of sequence data, thus it is especially important to discuss the limitations of testing, including challenges in interpreting the clinical significance of variants, and the meaning of negative results. For several CSER consortium projects, and increasingly in the clinical oncology genetics setting, germ-line genome or exome sequencing is being performed in tandem with tumor sequencing in individuals with a diagnosis of cancer. The potential impact of tumor sequencing results on treatment or prognosis in the immediate future may be the priority of these patient-participants and their families, and it is important that the patient-participant understands what a negative result does or does not mean. In this context, and that of ill patient-participants in general, the return of IFs raises the challenge of managing adherence to the recommend follow-up plan for those who are dealing with acute medical problems. Ill individuals also need to consider which, if any, results they would like to be returned to family members posthumously, thus adding to the complexity of the informed consent conversation.
Unique challenges also arise in counseling healthy individuals returned IFs from exome and genome sequencing tests. Returning results associated with conditions for which healthy patient-participants have no experience requires balancing the importance of adherence to follow-up recommendations with the uncertainty about penetrance and expressivity. As indications for exome and genome sequencing expand, variants will increasingly be identified in individuals with nonclassic phenotypes. Counseling resources to support patient-participants in this situation, and systems to track long term outcomes in these individuals and their families will be needed to address this.
Best practice considerations
Further exploration into the incorporation of exome and genome sequencing tests into clinical care will inform future genetic testing and return of results practices. The range and number of possible results returned from these tests is larger than that from single gene and gene panel testing. Additional visits to discuss the different types of results returned and the development of tools to facilitate patient-participant understanding (separate reports for different types of results, summary counseling letters or reports, etc.) should be considered [18]. Research suggests that the two-step process for returning results may be preferred by both medical professionals and parents/adolescents considering genomic testing [14]. As genetic testing practices evolve to include exome and genome sequencing, the possibility of returning results from these tests for which follow-up testing of relatives and/ or partners is not available or financially possible may also require attention. Further research will inform if the availability of targeted clinical follow-up testing for relatives and/or partners necessitates consideration when choosing genes to include on exome and genome sequencing tests for variant interpretation.
Ethical & practical issues
The integration of exome and genome sequencing into medical practice raises both ethical and practical challenges. Many ethical challenges are faced in the informed consent discussion for these tests. For example, finding an appropriate balance in the amount of information discussed so that patient-participants can make an informed choice without being overloaded is essential [19], and this balance is likely specific to the individual being consented. Soliciting informed patient-participant preferences for the types of results they wish to receive, when applicable, is also part of the informed consent process of several CSER consortium projects. Defining which types of results to offer for return requires balancing a patient-participant’s right to know their genomic results and the obligation of the medical professional to do no harm. For example, returning results related to conditions that are not medically actionable is not currently recommended in clinical practice; however, patient-participants and their families have shown interest in this type of information [14]. Finally, ethical challenges specifically related to the return of IFs have been raised including how best to approach the return of these results to family members of living and deceased individuals, and how to treat results generated from data and specimens submitted to biobanks [20,21].
One practical challenge demonstrated by the above cases is the time required to educate the patient-participant about the different types and implications of diagnostic and IF results that can be generated by exome and genome sequencing tests. Similarly, results disclosure may include discussion of multiple conditions with different inheritance patterns, a variety of personal and family implications and follow-up recommendations. Another practical challenge is the frequency in which exome and genome testing identifies pathogenic variants with reduced penetrance associated with mild or nonclassic phenotypes. As genomic testing expands our scientific knowledge of these variants will improve, but presently genetic counselors are challenged with educating patient-participants about these complex results as well as implementing a plan to communicate updates to variant interpretations. To avoid exhausting genetic counseling resources, alternative methods beyond the traditional model of genetics results disclosure will need to be explored.
Future directions
The cases presented in this article involve diverse patient-participant populations and clinical settings; however, the authors recognize that there is much that remains to be learned and explored in the area of return of results from exome and genome sequencing. An area of future research is the connection between the process and content of informed consent for these tests and the outcomes of subsequent return of results. Helping patient-participants to have realistic expectations about the types and implications of results that may be learned from genomic sequencing through the informed consent process is likely to have a positive impact on how they understand and act on results returned. Likewise, as we gain more experience in return of results from exome and genome sequencing we will be better able to provide a realistic framework for these expectations and goals. For example, refining the estimated frequency of IFs returned for different populations [22,23] will enable more accurate pretest discussions regarding how many and what type of results are expected to be identified. It is expected that the process of informed consent will evolve [24,25] and that best practices with regards to informed consent for genomic testing will mitigate some of the challenges faced with return of results. Another area for future research is an evaluation of returning results through alternative methods other than the traditional model of a genetics professional meeting face-to-face with a participant. This will require the development of models of care delivery so that the incorporation of wide scale genomic sequencing can be supported in a practical and ethical way [26].
Conclusion
The experience of the CSER consortium genetic counselors with return of results from exome and genome sequencing has led to a variety of lessons learned that will inform the future development of best practice recommendations. Many challenges faced in the return of these genomic sequencing results are different from those encountered in returning results from traditional genetic tests and this is largely due to the broad range of possible results and the need to educate patient-participants both pre- and post-testing. Addressing these challenges will inform the future practice of genomic testing. Further research is needed regarding the evolution of the informed consent process and its relationship with return of results, and how return of results will be affected and supported by the incorporation of different models of results disclosure. Genetic counselors are in the position to meaningfully contribute to this research and will play a central role in the dissemination of findings to nongenetics physicians, key stakeholders and, most importantly, patients and research participants considering exome and genome sequencing.
Future perspective
The adoption of exome and genome sequencing tests into routine clinical care is expected to increase in the next 5–10 years as the cost of this testing continues to decrease and interest in genetic testing grows. Clinical exome and genome sequencing tests are also likely to be ordered more frequently by nongenetic providers. This increase in utilization will generate a need to develop integrated tools to manage the volume of sequence data produced, including a centralized database to enable consistent and accurate variant interpretation by research and clinical laboratories. Systems to track long term patient care and economic outcomes will be needed to ensure that new genomic sequencing technologies are implemented cost effectively. Evidence based guidelines will also be required to advise the informed consent and return of results processes. While not highlighted by the cases presented in this report, challenges specific to exome versus genome sequencing warrant further discussion and exploration. Finally, disclosure practices for exome and genome sequencing results will likely need to expand beyond the traditional paradigm of face-to-face counseling to include alternative methods that support different patient-participant preferences and increase access to this technology. The role of the genetic counselor in this area is evolving, but is expected to include supporting nongenetics physicians, developing educational materials and providing support for return or results questions for nonmedically actionable variants.
Executive summary.
Background
The application of exome and genome sequencing in clinical medicine requires thorough investigation of the practical challenges, as well as the ethical issues and societal implications associated with the use of this technology.
The cases described highlight emerging themes genetic counselors in the Clinical Sequencing and Exploratory Research (CSER) consortium have encountered in returning results from exome and genome sequencing tests to diverse participant populations, including children and adults with a variety of diagnostic indications as well as healthy individuals.
Methods
Themes for lessons learned from the return of exome and genome sequencing results were elicited from members of the CSER consortium Genetic Counseling Working Group, summarized by the genetic counselor most closely affiliated with the case and were compiled.
Results
Seven themes with illustrative case descriptions are presented and include: 1) managing expectations in pretest and post-test counseling, negative findings do not mean the condition is not genetic; 2) context matters: follow-up for recommendations from incidental findings in healthy and ill patient-participants; 3) considerations for returning large amounts of data; 4) differing responses from families to the same result; 5) challenges with follow-up testing for family members of sequenced patient-participants; 6) navigating the atypical presentation of well-known Mendelian condition deceased patient-participants and 7) communicating results with family members.
Discussion
The application of genome and exome sequencing tests raises new challenges for return of results.
Further exploration into the incorporation of exome and genome sequencing in genetics and nongenetics clinics will inform future genetic testing and return of results practices.
Both practical and ethical challenges arise with the integration of exome and genome sequencing into medical practice.
There is much that remains to be learned and explored in the area of return of results from exome and genome sequencing including the connection between informed consent and return of results and exploring alternative models to return results.
Conclusion
The experience of the CSER consortium genetic counselors with return of results from genomic sequencing has led to a variety of lessons learned that will inform the future development of best practice recommendations.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work was funded by: UO1HG0006546, R21HG006596, R01HG006600, U01HG006485, U01HG006500, U01HG006492, UM1HG007301, UM1HG007292, UM1 HG006508, U01HG006487, U01HG006507, U01HG007307, R01HG006615, R21HG006613, R21HG006594, R01HG004500, R01HG006618, R01CA154517, R21HG006612. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest;
•• of considerable interest
- 1.National Human Genome Research Institute. Clinical Sequencing Exploratory Research (CSER) www.genome.gov.
- 2.Xue Y, Ankala A, Wilcox WR, Hegde MR. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med. 2014 doi: 10.1038/gim.2014.122. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 3••.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. Provides recommendations for reporting incidental findings from clinical genomic sequencing and describes a minimum list of actionable conditions/genes/variants that are recommended to be returned. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American College of Medical Genetics. ACMG Updates Recommendation on “Opt Out” for Genome Sequencing Return of Results. www.acmg.net.
- 5••.Jarvik GP, Amendola LM, Berg JS, et al. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet. 2014;94(6):818–826. doi: 10.1016/j.ajhg.2014.04.009. An important distinction between clinical research and medical care. They recommend that research results that meet an actionability threshold be offered for return to consenting participants; however, participants have a right to decline potentially medically actionable results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg JS, Amendola LM, Eng C, et al. Processes and preliminary outputs for identification of actionable genes as incidental findings in genomic sequence data in the Clinical Sequencing Exploratory Research Consortium. Genet Med. 2013;15(11):860–867. doi: 10.1038/gim.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klitzman R, Buquez B, Appelbaum PS, Fyer A, Chung WK. Processes and factors involved in decisions regarding return of incidental genomic findings in research. Genet Med. 2014;16(4):311–317. doi: 10.1038/gim.2013.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy Bollinger J, Bridges JF, Mohamed A, Kaufman D. Public preferences for the return of research results in genetic research: a conjoint analysis. Genet Med. 2014;16(12):932–939. doi: 10.1038/gim.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright MF, Lewis KL, Fisher TC, et al. Preferences for results delivery from exome sequencing/genome sequencing. Genet Med. 2014;16(6):442–447. doi: 10.1038/gim.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitch K, Joseph G, Guiltinan J, Kianmahd J, Youngblom J, Blanco A. Lynch syndrome patients’ views of and preferences for return of results following whole exome sequencing. J Genet Couns. 2014;23(4):539–551. doi: 10.1007/s10897-014-9687-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu JH, Crouch J, Jamal SM, Tabor HK, Bamshad MJ. Attitudes of African Americans toward return of results from exome and whole genome sequencing. Am J Med Genet. 2013;161A(5):1064–1072. doi: 10.1002/ajmg.a.35914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu JH, Crouch J, Jamal SM, Bamshad MJ, Tabor HK. Attitudes of non-African American focus group participants toward return of results from exome and whole genome sequencing. Am J Med Genet. 2014;164A(9):2153–2160. doi: 10.1002/ajmg.a.36610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sapp JC, Dong D, Stark C, et al. Parental attitudes, values, and beliefs toward the return of results from exome sequencing in children. Clin Genet. 2014;85(2):120–126. doi: 10.1111/cge.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levenseller BL, Soucier DJ, Miller VA, Harris D, Conway L, Bernhardt BA. Stakeholders’ opinions on the implementation of pediatric whole exome sequencing: implications for informed consent. J Genet Couns. 2014;23(4):552–565. doi: 10.1007/s10897-013-9626-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm IA, Savage SK, Green RC, et al. Guidelines for return of research results from pediatric genomic studies: deliberations of the Boston Children’s Hospital Gene Partnership Informed Cohort Oversight Board. Genet Med. 2014;16(7):547–552. doi: 10.1038/gim.2013.190. [DOI] [PubMed] [Google Scholar]
- 16.Vassy JL, Lautenbach DM, Mclaughlin HM, et al. The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials. 2014;15:85. doi: 10.1186/1745-6215-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NCBI Bookshelf. GeneReviews. LEOPARD Syndrome. www.ncbi.nlm.nih.
- 18.Zhao JQ, Haga SB. Promoting the participant-researcher partnership. Genet Med. 2014;16(3):228–230. doi: 10.1038/gim.2013.118. [DOI] [PubMed] [Google Scholar]
- 19.Rigter T, Henneman L, Kristoffersson U, et al. Reflecting on earlier experiences with unsolicited findings: points to consider for next-generation sequencing and informed consent in diagnostics. Hum Mut. 2013;34(10):1322–1328. doi: 10.1002/humu.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Wolf SM. Return of individual research results and incidental findings: facing the challenges of translational science. Ann Rev Genomics Hum Genet. 2013;14:557–577. doi: 10.1146/annurev-genom-091212-153506. Discusses ethical considerations in returning research results and incidental findings. Challenges include management of what information participants are owed by researchers and the necessity of bridging the link between research and clinical care for the return of results process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen NL, Karlson EW, Malspeis S, Lu B, Seidman CE, Lehmann LS. Biobank participants’ preferences for disclosure of genetic research results: perspectives from the Our Genes, Our Health, Our Community project. Mayo Clin Proc. 2014;89(6):738–746. doi: 10.1016/j.mayocp.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Dorschner MO, Amendola LM, Turner EH, et al. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet. 2013;93(4):631–640. doi: 10.1016/j.ajhg.2013.08.006. The frequency of actionable, pathogenic incidental findings varies across ethnic groups, and propose the need to develop a robust and diverse centralized resource to provide comprehensive variant classification information on pathogenicity for clinical utility in genomic medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston JJ, Rubinstein WS, Facio FM, et al. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet. 2012;91(1):97–108. doi: 10.1016/j.ajhg.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Appelbaum PS, Parens E, Waldman CR, et al. Models of consent to return of incidental findings in genomic research. Hastings Cent Rep. 2014;44(4):22–32. doi: 10.1002/hast.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Appelbaum PS, Waldman CR, Fyer A, et al. Informed consent for return of incidental findings in genomic research. Genet Med. 2014;16(5):367–373. doi: 10.1038/gim.2013.145. Suggests a critical need to establish new approaches to the informed consent process for returning incidental findings in genomic research due to constrained time limits for practitioners as well as concern around disclosing too much information to participants and overwhelming them, while also sharing enough information for individuals to make informed decisions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Yu JH, Jamal SM, Tabor HK, Bamshad MJ. Self-guided management of exome and whole-genome sequencing results: changing the results return model. Genet Med. 2013;15(9):684–690. doi: 10.1038/gim.2013.35. Proposes a self-guided management model for the return of results using information systems approach, so individuals have control over which sequencing results are returned based on their personal value system. They present a dynamic resource that should be “managed” rather than “returned” in an ever-evolving process adapting to rapid technological advances. [DOI] [PMC free article] [PubMed] [Google Scholar]