Summary
Synaptotagmin-1 and neuronal SNARE proteins play key roles in evoked synchronous neurotransmitter release. However, it is unknown how they cooperate to trigger synaptic vesicle fusion. Here we report atomic-resolution crystal structures of Ca2+- and Mg2+-bound complexes between synaptotagmin-1 and the neuronal SNARE complex, one of which was determined with diffraction data from an X-ray free electron laser, leading to an atomic-resolution structure with accurate rotamer assignments for many sidechains. The structures revealed several interfaces, including a large, specific, Ca2+-independent, and conserved interface. Tests of this interface by mutagenesis suggest that it is essential for Ca2+-triggered neurotransmitter release in neuronal synapses and for Ca2+-triggered vesicle fusion in a reconstituted system. We propose that this interface forms prior to Ca2+-triggering, and moves en bloc as Ca2+ influx promotes the interactions between synaptotagmin-1 and the plasma membrane, and consequently remodels the membrane to promote fusion, possibly in conjunction with other interfaces.
Introduction
Membrane fusion is essential for many physiological processes in eukaryotic cells, including protein and membrane trafficking, hormone secretion, and neurotransmitter release1,2. Evolutionarily conserved SNARE (Soluble N-ethylmaleimide sensitive factor Attachment protein REceptor) proteins play a key role in these processes. Specific combinations of SNARE proteins are located on opposite membranes. Upon zippering into a highly stable four-helix bundle—the SNARE complex, they provide the energy for membrane fusion3,4. However, other factors are essential for regulation of membrane fusion. In particular, several key proteins are required for neurotransmitter release in addition to neuronal SNAREs5, but it is unknown at the atomic level of detail how these factors cooperate with SNAREs to promote synaptic transmission. One key factor is the Ca2+ sensor synaptotagmin, which consists of a short N-terminal luminal segment, a single transmembrane α-helix, an unstructured linker, and two Ca2+-binding C2 domains, termed C2A and C2B respectively, or C2AB together6. There are 16 isoforms of mammalian synaptotagmins that are localized to synaptic and secretory vesicles or the plasma membrane. Among these isoforms, synaptotagmin-1 (Syt1) is a Ca2+ sensor for evoked synchronous neurotransmitter release7. Synaptotagmin-2 and -9 are also involved in evoked synchronous neurotransmitter release for different subsets of neurons8. In contrast, synapotagmin-7 plays a role in “slower” asynchronous release9,10; moreover, these and other synaptotagmins as well act in other types of exocytosis5. In addition to its role in evoked synchronous release, Syt1 also clamps the frequency of miniature spontaneous events11–13.
Syt1 binds in a Ca2+-dependent manner to anionic membranes; during binding, anionic phospholipids and synaptotagmin C2 domains together coordinate calcium14–17. The membrane-synaptotagmin interaction has functional significance since the Ca2+ affinity of Syt1 for binding to anionic membranes and the Ca2+ sensitivity of neurotransmitter release are tightly correlated16,18. The Syt1 C2AB fragment can induce vesicle clustering19 and preferentially binds to curved membranes20,21. Moreover, C2 domains may penetrate the membrane upon Ca2+-binding22,23.
Syt1 also interacts with the neuronal SNARE complex based on immunoprecipitation and pull-downs24–27, single molecule fluorescence resonance energy transfer (smFRET)28, and nuclear magnetic resonance29,30 experiments. A gain-of-function mutation in the Ca2+-binding region of the C2A domain suggested that the Syt1-SNARE interaction may be functionally important26, but the molecular basis and the significance of the interaction between Syt1 and the SNARE complex remain unknown.
Several crystal structures of Syt1 C2A, C2B domains, and C2AB fragments are available31–33, as well as the structure of the neuronal SNARE complex3. However, the atomic-resolution structure of the complex between Syt1 and the neuronal SNARE complex (referred to as Syt1-SNARE complex) has been elusive. Single molecule methods allowed the study of the Syt1-SNARE complex under dilute conditions with spatially isolated neuronal SNARE complexes reconstituted in a supported bilayer28. The observed smFRET histograms28 suggested several possible interfaces between Syt1 and the SNARE complex. Other dynamic or approximate models of the C2AB-SNARE complex were obtained by nuclear magnetic resonance (NMR)29,30, but cannot be readily compared with the previous smFRET studies28 and the results presented here because of differences in conditions, particular covalent attachment of lanthanide labels30, and lack of atomic resolution.
Here we report atomic-resolution crystal structures of a Syt1-SNARE complex in two different crystal forms and in the presence of either Ca2+ or Mg2+. We found several interfaces, including a large structurally and evolutionarily conserved interface that is Ca2+-independent. Structure-based mutations of this interface disrupt evoked neurotransmitter release in primary neurons and Ca2+-triggered fusion in a reconstituted system.
Structure of the Syt1-SNARE complex
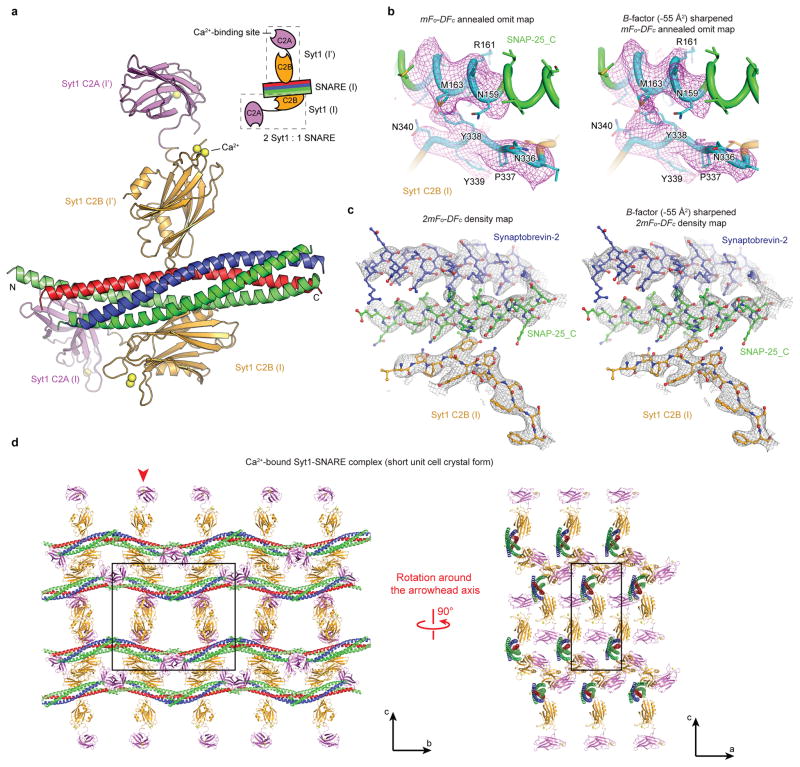
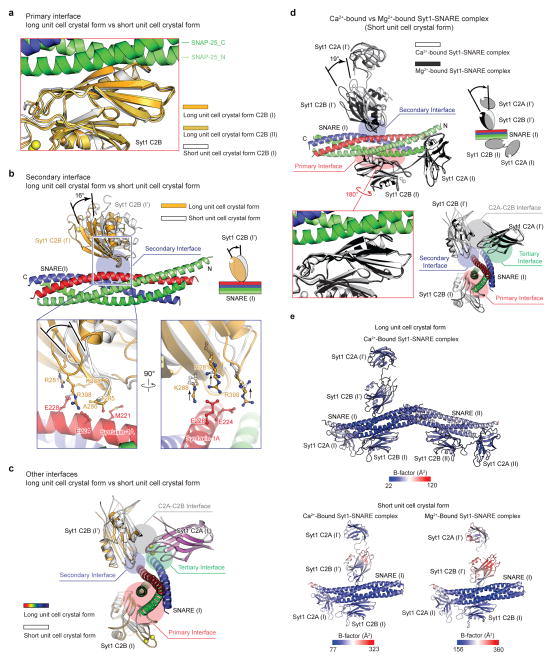
We designed and tested several chimeric constructs involving the Syt1 C2AB fragment (amino acid range 141–421) and the neuronal SNARE complex (Extended Data Figs. 1a, b, Methods). We crystallized the Ca2+- and Mg2+-bound Syt1-SNARE complexes (Extended Data Figs. 1d, e), and determined their structures (Fig. 1, Extended Data Table 1). The crystallization conditions were at near physiological pH and ionic strength (Methods and Extended Data Fig. 1e). We observed two crystal forms for the Ca2+-bound Syt1-SNARE complex, referred to as “short unit cell” and “long unit cell” crystal forms hereafter (Extended Data Figs. 2 and 3).
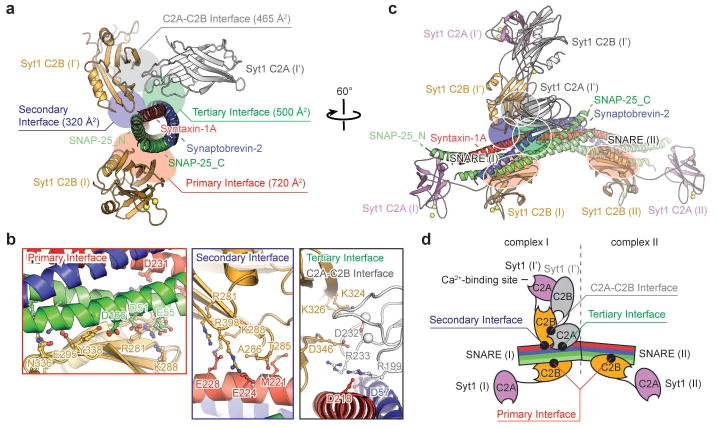
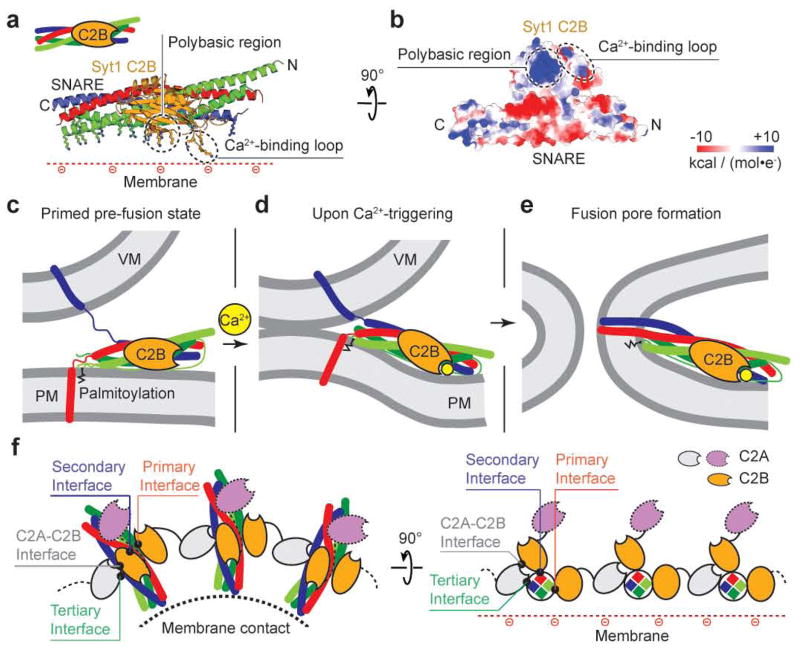
Figure 1. Crystal structure of the Syt1-SNARE complex.
a, Structure of the Ca2+-bound Syt1-SNARE complex (only showing complex I) in the long unit cell crystal form (Extended Data Table 1, Extended Data Fig. 2). Two Syt1 C2B domains (designated as I and I′) and one Syt1 C2A domain (related by crystallographic symmetry and coloured in white) form a total of three interfaces (primary, secondary, and tertiary) with the neuronal SNARE complex (synaptobrevin-2, syntaxin-1A, and the two SNARE domains of SNAP-25A: SNAP-25_N and SNAP-25_C). A fourth interface (C2A-C2B interface) is located between Syt1 C2B (I′) and Syt1 C2A (I′). Interface areas are provided in parenthesis. b, Close-up views of the four interfaces with labels for sidechains of interacting residues. The left sub-panel shows a superposition of both primary interfaces that occur in the long unit cell crystal form (root-mean-square-difference, RMSD = 0.34 Å, including Cα atoms of the SNARE complex and the Syt1 C2B domain forming the interface). The middle sub-panel shows the secondary interface. The right sub-panel shows both the tertiary interface and the C2A-C2B interface. c, Rotated view of panel a, but showing the entire asymmetric unit and the symmetry-related Syt1 C2AB fragment. Three Syt1 C2AB fragments (designated as I, I′ and II) bind to two SNARE complexes in the asymmetric unit. SNARE (II) only interacts with the C2B domain of one Syt1 C2AB fragment, Syt1 (II), via the same primary interface as observed in complex I. d, A schema corresponding to the structure shown in panel c.
The X-ray free electron laser (XFEL) of the Linac Coherent Light Source (LCLS) at SLAC National Accelerator Laboratory yielded substantially higher-quality diffraction data than the Advanced Photon Source (APS) NE-CAT microfocus synchrotron beamline at Argonne National Laboratory from similar crystals of the long unit cell crystal form (Extended Data Figs. 2a, b). The electron density maps obtained from the XFEL diffraction data were notably superior to those of the synchrotron datasets (Methods). In particular, the electron density maps calculated from the XFEL data set were of sufficient quality to obtain accurate rotamers for most sidechains, including those at the interfaces between molecules (Extended Data Figs. 2d–f). This is one of the first new crystal structures determined using XFEL diffraction data. Moreover, in contrast to the tens of thousands to millions of crystals typically used in XFEL-based crystallography experiments34, we obtained a reasonably complete data set from a few hundred images using 148 crystals (Methods).
Three distinct Syt1-SNARE interfaces
The crystal structures of the Syt1-SNARE complex reveal three interfaces between the SNARE complex and the Syt1 C2A and C2B domains (referred to as “primary”, “secondary”, and “tertiary”), as well as an interface between Syt1 C2 domains (referred to as “C2A-C2B interface”, Figs. 1a, b, Supplementary Videos 1 and 2). There are two essentially identical instances of the primary interface formed between Syt1 C2B domains and the SNARE complex in the long unit cell crystal form (Figs. 1c, d). The secondary interface involves another Syt1 C2B domain and the SNARE complex, while the tertiary interface involves a Syt1 C2A domain and the SNARE complex (Fig. 1b). These two C2 domains also form the C2A-C2B interface. All three interfaces between Syt1 and the SNARE complex fall within the range of smFRET efficiency histograms obtained previously28 (Extended Data Fig. 4). These interfaces may suggest how multiple Syt1 and SNARE complexes may simultaneously interact in the neuron (see below).
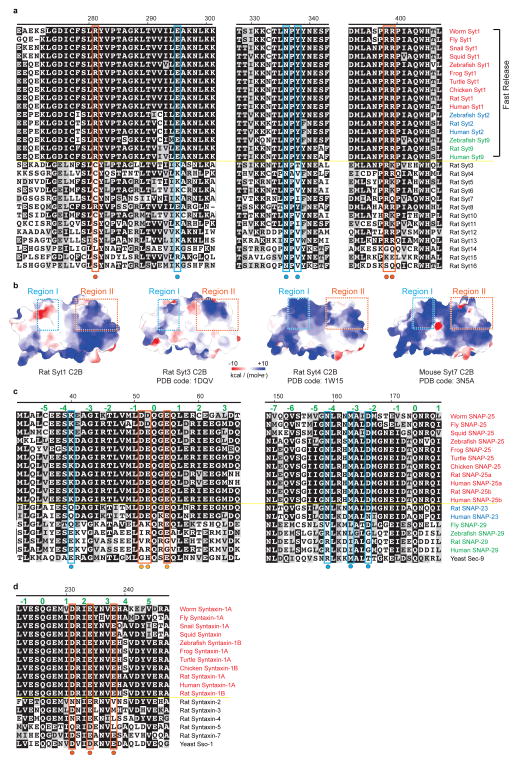
The largest, primary interface between the Syt1 C2B domain and the SNARE complex is very similar in both complexes (Fig. 1b) and in both crystal forms (Extended Data Fig. 5a), suggesting that it is not affected by crystal packing. The primary interface is also very similar in the Ca2+-bound as well as in the Mg2+-bound crystal structures (Extended Data Fig. 5d), implying a Ca2+-independent interface. The residues involved in the primary interface have relatively low temperature factors, among the lowest in these structures (Extended Data Fig. 5e), and the electron densities of the sidechains that form this interface are well-defined (Figs. 2c, d), suggesting a specific interaction. In a recent study, lanthanide labels were covalently attached to SNAP-25 residues 41 and 166 for pseudocontact chemical shift NMR measurements in the presence of 125 thiocyanate30; these covalent labels would likely disrupt the primary interface (Fig. 2c), so it is not possible to compare our crystal structures with this NMR study. The interacting residues of the primary interface are conserved across different species for synaptotagmins (Syt1, Syt2, Syt9, ref. 8), SNAP-25, and syntaxin-1A homologs involved in fast synchronous release (Extended Data Fig. 6). In contrast, the interacting residues show variation among other synaptotagmin, SNAP-25, and syntaxin homologs that are not known to be involved in fast synchronous release.
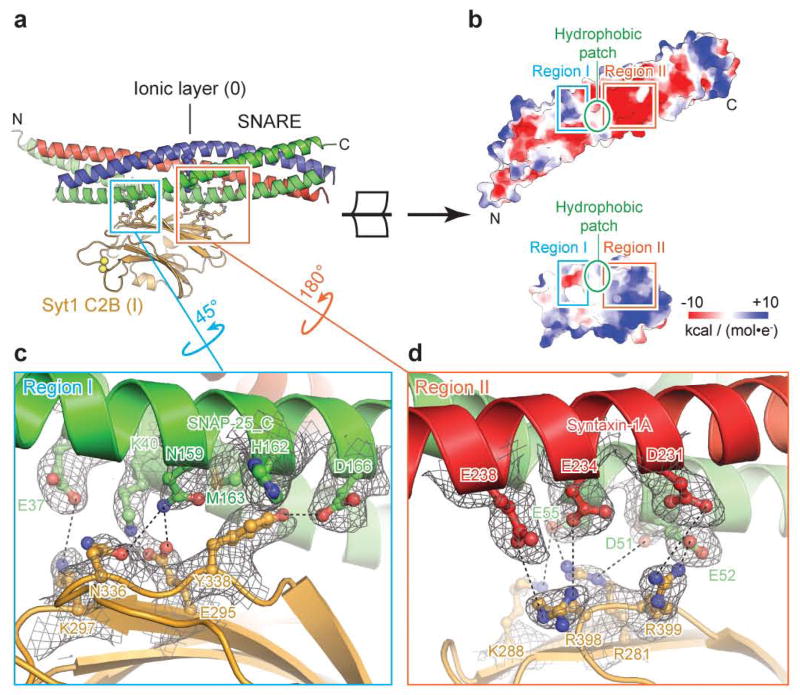
Figure 2. Primary interface between the Syt1 C2B domain and the neuronal SNARE complex.
a, Overview of the primary interface (complex I in the long unit cell crystal form) along with interacting residues (sticks and balls). b, Open-book view of the electrostatic potential map of the primary interface. The two polar regions I, II are connected by a hydrophobic patch (SNAP-25 I44, L47, V48 and Syt1 V292, L294, A402). c, d, Close-up views of regions I and II. Interacting residues are labeled, along with dashed lines that indicate hydrogen bonds or salt bridges. 2mFo-DFc electron density maps of the interacting residues are superimposed (grey mesh; contour level = 1.5 σ).
The C2B domain forming the secondary interface is slightly rotated with respect to the SNARE complex between the two crystal forms and between the Ca2+- and Mg2+-bound crystal structures (Extended Data Figs. 5b, d), although the interactions at the secondary interface itself are similar. The C2A domain that forms the tertiary interface is in the same orientation in both crystal forms and in the Ca2+- and Mg2+-bound crystal structures (Extended Data Figs. 5c, d) and the interacting side chains are in similar positions. The interactions that form the tertiary interface are primarily ionic, involving residues Syt1 R199 and R233, syntaxin-1A D218, and synaptobrevin-2 D57 (Fig. 1b) with well resolved electron density.
The conserved primary interface
We divide the primary interface into two regions (Fig. 2) dominated by polar interactions: region I comprises SNAP-25 E37, K40, N159, M163, D166 and Syt1 E295, K297, N336 and Y338; region II comprises SNAP-25 D51, E52, E55, syntaxin-1A D231, E234, E238, and Syt1 R281, K288, R398, and R399. The SNARE complex is positively charged and Syt1 is negatively charged in region I, whereas the opposite is observed for region II. In contrast, other synaptotagmin isoforms not involved in fast synchronous release exhibit sequence variations and differences in the electrostatic potential maps in both regions (Extended Data Figs. 6a, b), suggesting that the primary interface may be an interaction that is specific for fast synchronous release.
The primary interface is critical
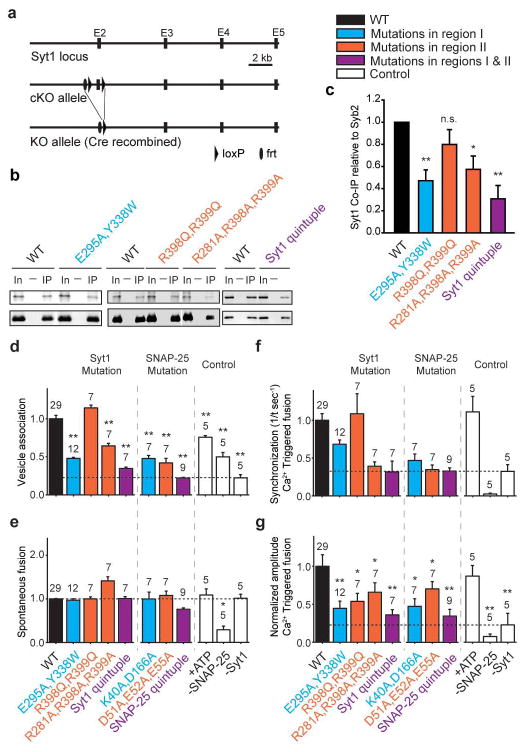
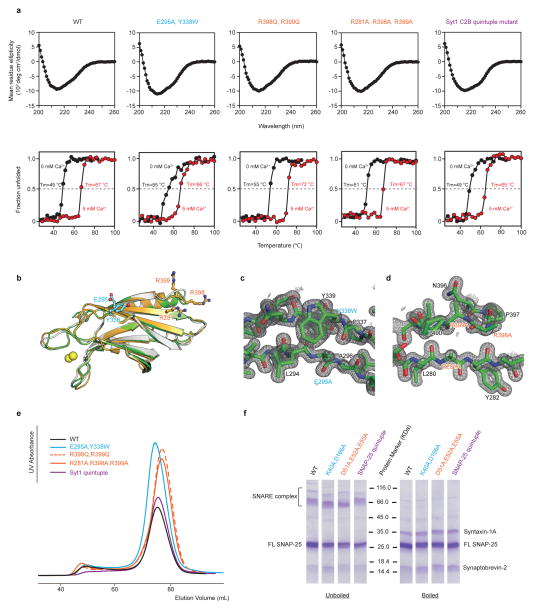
We designed mutations of the critical interacting residues of the primary interface based on the crystal structure and verified that all mutants result in properly folded Syt1 and SNARE complex (Methods and Extended Data Fig. 7). To test the interactions in neurons, we performed co-immunoprecipitation of syntaxin-1A in cultured Syt1 conditional knockout (cKO) neurons infected with viruses expressing WT Syt1 or Syt1 containing the designed C2B mutants (Fig. 3a and Methods). The region II Syt1 mutant (R398Q, R399Q) was reported previously35,36, and is used here for comparison. Syntaxin-1A was immunoprecipitated from lysates of these neurons, and the presence of co-immunoprecipitated proteins was assayed with monoclonal antibodies against Syt1 and synaptobrevin-2. Mutation of either regions I and II of the primary interface reduced Syt1 binding to syntaxin-1A/SNARE complex by ~50%, and simultaneous mutation of both regions (“Syt1 quintuple”) reduced binding to ~33% as compared to a Syt1 WT construct introduced in the same manner (Figs. 3b, c). These results suggest that both regions are required for efficient Syt1-SNARE binding.
Figure 3. Mutations of the primary interface affect binding and Ca2+-triggered single vesicle-vesicle fusion.
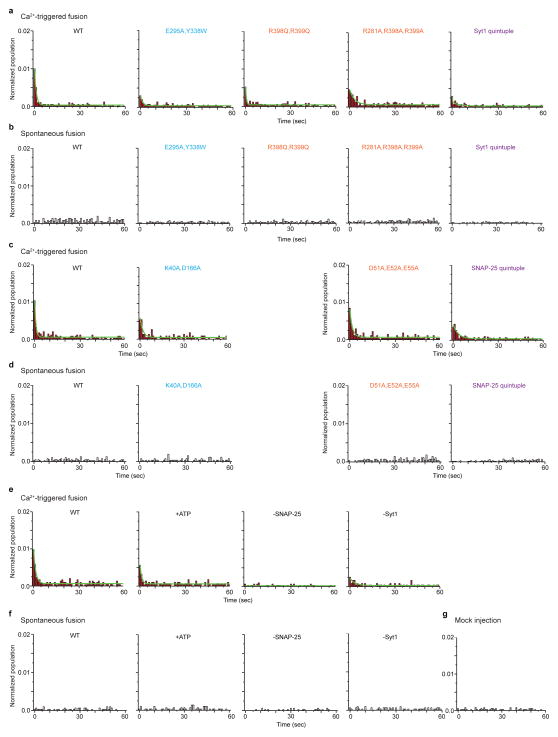
“Syt1 quintuple” refers to the Syt1 mutant (R281A, E295A, Y338W, R398A, R399A). “SNAP-25 quintuple” refers to the SNAP-25 mutant (K40A, D51A, E52A, E55A, D166A). The colour code is specified in the figure. a, Schematic diagram of the Syt1 cKO mice. The Syt1 exon 2 which contains the transmembrane domain is floxed. Cre-recombinase removes exon 2, ablating all cytoplasmic Syt1 sequences. b, Co-immunoprecipitation of either Syt1 (top row) or synaptobrevin-2 (bottom row) with a syntaxin-1A antibody in Syt1 cKO cultured neurons rescued with the indicated Syt1 mutant constructs. c, Quantification of co-immunoprecipitation of Syt1 normalized to synaptobrevin-2. Results are scaled to Syt1 WT levels. All data are means ± SEMs; statistical significance was analyzed by the Student’s t-test comparing the mutants with WT Syt1; **, P<0.01, n=4 for Syt1 E295A, Y338W and “Syt1 quintuple”; n.s., no significant difference, n=3 for Syt1 R398Q, R399Q; *, P<0.05, n=4 for Syt1 R281A, R398A, R399A. d–g, Bar graphs showing the effects of Syt1 and SNAP-25 mutants in fusion of single vesicles with reconstituted neuronal SNAREs, Syt1, and complexin-1 (Methods and ref. 38). d, Number of associated SV vesicles after incubation of SV-vesicles with surface-immobilized PM-vesicles for a 1 min period. e, Number of spontaneous fusion events over the subsequent 1-min observation period normalized by the number of associated SV vesicles. f, Synchronization, that is, decay rates (1/τ), of the histograms of fusion events upon 500 μM Ca2+-injection. Error bars are error estimates computed from the covariance matrix upon fitting the corresponding cumulative histograms with a single exponential decay function using a Levenberg-Maquardt technique. g, Amplitude of the first 1-sec time bin upon Ca2+-injection. Each value in this panel was normalized by the respective number of fusion events after Ca2+ injection. d, e, g, All data are means ± SEMs; the number of independent repeat experiments are depicted above the bars and in Extended Data Table 2; statistical significance was assessed by the Student’s t-test comparing all other conditions with WT (**, P<0.005; *, P<0.05). The cumulative fusion histograms are shown in Extended Data Fig. 8. Controls in panels d–g are in the presence of 3 mM ATP and in the absence of SNAP-25 and Syt1. As expected, Ca2+-triggered fusion required the presence of both SNAP-25 and Syt1; fusion is not affected by the presence of ATP.
To further investigate the function of the primary interface between the neuronal SNARE complex and Syt1, we used a single-vesicle content-mixing assay37,38 and tested the effect of both Syt1 and SNAP-25 mutants on association, spontaneous fusion, and Ca2+-triggered fusion of single vesicles with reconstituted full-length neuronal SNAREs, Syt1, and complexin-1 (Figs. 3d–g, Extended Data Fig. 8, Extended Data Table 2). The Syt1 mutants disrupting the interface between the SNARE complex and Syt1 as seen by co-immunoprecipitation (Figs. 3b, c) also reduced vesicle association to similar degrees for the four different mutants (left group of mutants in Fig. 3d). Our reconstituted assay also allowed testing of the interacting SNAP-25 residues (middle group of mutants in Fig. 3d), showing significant reduction of vesicle association as well. Thus, as expected, mutations on both the Syt1 and SNARE sides of the primary interface reduce the interaction between them. Likewise, both groups of mutants reduce the amplitude and decrease the synchronization of Ca2+-triggered fusion, but they do not affect spontaneous fusion (Figs. 3e–g). In particular, the “Syt1 quintuple” mutant significantly reduced Ca2+-triggered synchronization to the control level without Syt1.
To further characterize the interaction between Syt1 and the SNARE complex, we investigated the effect of ATP, which has an ionic shielding effect on certain cellular processes39,40. We observed no effect on both spontaneous and Ca2+-triggered fusion, and only a mild effect on vesicle association (Figs. 3d–g, right group of mutants), suggesting that the functionally important interactions between Syt1 and the SNARE complex are not affected by ionic shielding.
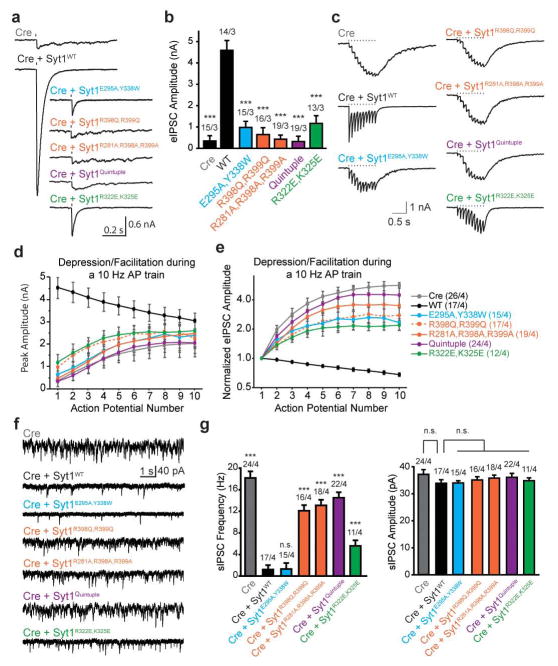
Evoked release requires primary interface
We assayed release electrophysiologically in Syt1 cKO neurons in which endogenous Syt1 was replaced with mutant Syt1 (Fig. 3a and Methods). As expected7, removal of Syt1 abolished synchronous release monitored by recording evoked inhibitory postsynaptic currents (eIPSCs), a phenotype that can be rescued by re-introduction of WT but not mutant Syt1 cDNA (Figs. 4a, b). Interestingly, the region I mutants retained some rescue ability, in contrast to region II mutants that were nearly non-functional (Figs. 4a, b), including the R398Q, R399Q mutant as previously reported36. The triple mutant (R281A, R398A, R399A), and the quintuple mutant that combines mutations in both regions showed even more severe phenotypes. In contrast, a mutant that affects the polybasic region of Syt1 (R322E, K325E) exhibited a milder phenotype as reported previously30. During high frequency stimulation, WT cultured hippocampal neurons displayed depression while Syt1 cKO neurons showed asynchronous release with robust facilitation (Figs. 4c–e). All Syt1 mutants underwent facilitation and the severity of the phenotype for each mutant correlated well with the results for single evoked release. Another known consequence of Syt1 removal is the unclamping of spontaneous release. Interestingly, the E295A, Y338W mutant could rescue this phenotype while the region II mutants (or combinations thereof) could not (Figs. 4f, g). There were no differences in spontaneous miniature IPSC amplitudes between WT and mutant rescues (Fig. 4g).
Figure 4. Mutations of the primary interface impair Syt1 function in Ca2+-triggered release.
Recording of inhibitory postsynaptic currents (IPSCs) from cultured Syt1 cKO hippocampal neurons infected with lentiviruses expressing Cre-recombinase and Syt1 mutants of the primary interface and a mutant of the polybasic region (R322E, K325E). All recordings were performed in the presence of CNQX (20 μM) and AP-5 (50 μM) using a high Cl− internal solution. a and b, Sample traces of evoked IPSCs from single action potentials (a) and quantification of peak amplitudes (b). Tick mark indicates stimulus delivery. All data are means ± SEMs; number of cells/independent cultures analyzed are depicted above the bars; statistical significance was assessed by one-way analysis of variance comparing all other conditions with WT rescue group (***, P<0.001). c–e, Syt1 mutants display facilitation, instead of depression, during high-frequency stimulation. c, Sample traces of 10 Hz trains; d and e quantification of absolute (d) and normalized (e) IPSC amplitudes during the train; numbers of cells/independent cultures analyzed are depicted in parentheses in the labels for each of the traces. f and g, Syt1 mutants are unable to clamp the frequency of spontaneous IPSCs. f, Sample sIPSC traces; g, quantification of event frequency (left) and amplitude (right). All data are means ± SEMs; the number of cells/independent cultures analyzed are depicted above the bars. Statistical significance was assessed by one-way analysis of variance comparing all other conditions with the WT rescue group (***, P<0.001; n.s., no significant difference).
Together, these observations support the notion that the primary Syt1-SNARE interface is critical for the role of Syt1 as a Ca2+ sensor of evoked release, but that this Syt1-SNARE interface is not required for spontaneous release in neurons, consistent with the differential effect of Syt1 mutations on spontaneous and evoked release11–13. We note that spontaneous release observed in our reconstituted system38 reflects the fusion probability at exactly zero Ca2+ concentration (that is, in absence of a Ca2+-sensor for mini-release) whereas spontaneous release in neurons is driven by resting Ca2+-concentrations and is largely blocked by removing all calcium41. This circumstance may explain the different behavior for the E295A, Y338W mutant in neuronal cultures and in the reconstituted system. Moreover, there is little effect on synchronization of the R398Q, R399Q mutant of Syt1 in the reconstituted system, which may indicate the existence of additional interactions involving these residues with other factors not present in our minimal reconstituted system.
Model of Syt1 mediated Ca2+ triggering
Several mechanisms and models have been proposed on how Syt1 and the neuronal SNARE complex cooperate29,42,43. These models, however, were devised based on biochemical data without atomic-resolution structural information. Our crystal structures of the Syt1-SNARE complex now suggest a parsimonious mechanism that involves a concerted action of the unit consisting of the Syt1 C2B domain and the SNARE complex (Fig. 5a). We first focus on this unit alone and then later place it in the context of full-length Syt1 and other interactions. The combined surface of the Syt1 C2B-SNARE unit forms a flat face with an extensive pattern of positive charges (Fig. 5b) that includes the polybasic region of Syt1 as well as basic residues at the membrane-proximal side of the neuronal SNARE complex. The effect of the Syt1 R322E, K325E mutant (Fig. 4) as well as reported observations29,30,35,44,45 suggests a functional role for the polybasic region.
Figure 5. Model of the role of the primary Syt1-SNARE interface.
a–b, Unit that is formed by the primary interface between Syt1 C2B and the SNARE complex. a, Cartoon representation with positively charged sidechains shown as sticks; b, electrostatic potential map looking towards the positively charged face of the Syt1 C2B-SNARE unit. c–e, Proposed function of the Syt1 C2B-SNARE unit. c, Initial state prior to Ca2+ triggering. The juxtamembrane linkers of synaptobrevin-2 and of syntaxin-1A were modeled as random coils37. d, Intermediate state after Ca2+-triggering when the membranes are close enough to promote stalk formation48. e, Fusion pore formation. Zigzag lines: palmitoylated cysteine residues of the SNAP-25 linker region. VM, vesicle membrane, PM, plasma membrane. f, Other interfaces found in the crystal structure, along with the primary interface, could form a connected network of SNARE complexes that surrounds the point of contact between membranes. The left panel is a top down view onto the point of contact between membranes, the right panel is a rotated projection view.
Prior to Ca2+ influx, we assume a membrane juxtaposed, hemifusion-free state (Fig. 5c), the likely starting state for fast Ca2+ triggered fusion37, and a partially folded trans-SNARE complex46. We assume that the Syt1 C2B-SNARE unit can assemble in this state since the primary interface involves residues within the folded region of the trans-SNARE complex. The palmitoylated cysteine residues of SNAP-25 would be able to interact with the plasma membrane, and the membrane-proximal side of syntaxin is closely juxtaposed with the membrane, as inferred by the requirement of tight membrane-coupling of syntaxin for evoked release47. Upon Ca2+ binding to the Syt1 C2B domain, the Ca2+ binding loops partially insert into the membrane23 with a preference for membrane curvature20,21, possibly supported by ionic interactions via the polybasic region and with continuing maintenance of the Syt1-SNARE interface. We propose that the Syt1 C2B-SNARE unit moves en bloc as an entity upon Ca2+-triggering. The simultaneous membrane interactions of the Ca2+ binding loops, of the polybasic region, and of membrane-proximal region of the SNARE complex would therefore require a deformation of the plasma membrane. This morphological change of the plasma membrane juxtaposes the membranes closer than the critical distance (0.9 nm) to promote stalk formation48 (Fig. 5d), and subsequently leads to fusion pore opening (Fig. 5e).
It is likely that Syt1 engages in multiple functionally relevant interactions with neuronal SNARE complexes, including the other interfaces found in our crystal structures. All of these interactions may be anchored by the primary interface, which is the most stable and extensive contact site. We speculate that multiple SNARE complexes could interact via Syt1 interactions employing some or all of the interfaces (primary, secondary, tertiary, C2A-C2B) that are observed in our crystal structures (Fig. 5f). In addition to the primary and secondary interfaces formed by C2B domains (gold), the tertiary interface involves a C2A domain (gray). This interface could be involved in displacing complexin from its interaction with the core of the SNARE complex since complexin binds in the groove between syntaxin-1A and synaptobrevin-249; partial displacement of complexin may be important for neurotransmitter release50. Interestingly, the gain-of-function mutation D232N in the Ca2+-binding region of the C2A domain26 is close to the tertiary interface and it is also part of the C2A-C2B interface (Fig. 1b). In the proposed network of interactions there are C2A domains (purple) that have no interactions with SNARE complexes; we envision that they could interact with a membrane. This entire Syt1-SNARE assembly would thus be poised to confer cooperativity upon Ca2+-triggering on a sub-millisecond timescale.
Methods
Strategy for crystallization of the Syt1-SNARE complex
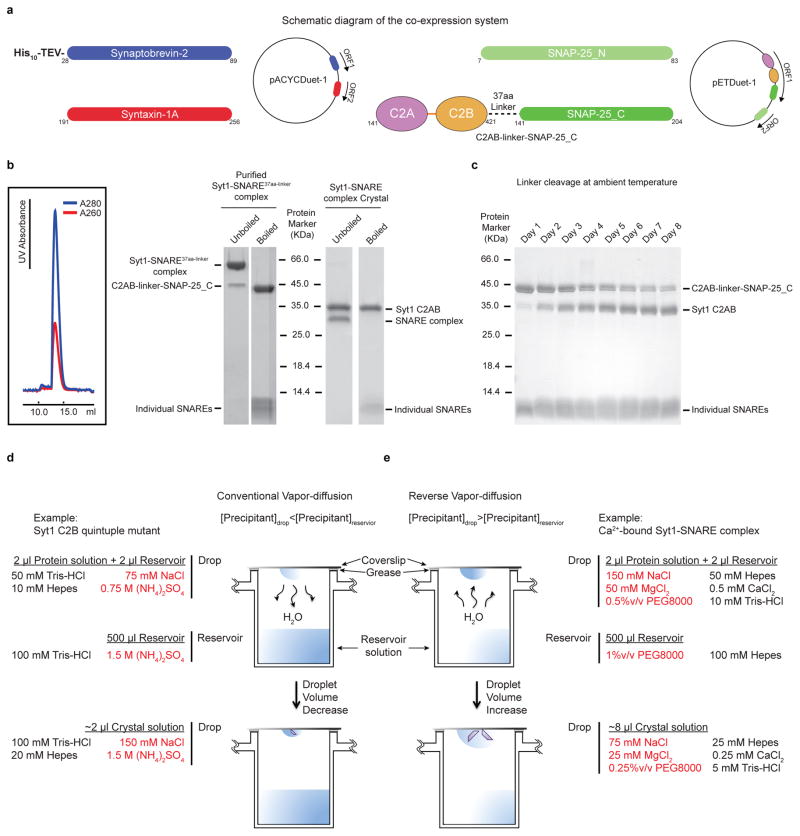
Successful crystallization of the Syt1-SNARE complex involved extensive testing of multiple designs of covalently linked chimeras between a Syt1 C2AB fragment and different SNARE domain fragments of the neuronal SNARE complex (both SNARE domains of SNAP-25, denoted SNAP-25_N and SNAP-25_C, as well as the SNARE domain of syntaxin-1A). Three different linker lengths for the chimeras were tested (16, 23, and 37 amino acids) derived from the linker sequence of the human Oct-1 transcription factor51. This particular linker sequence had been used previously to crystallize the Arf-arf GAP complex52. We also tested different truncations of the neuronal SNARE complex3,53. The resulting constructs were screened for protein expression, and homogeneity by ion exchange and size exclusion chromatography. The best constructs resulted in a Syt1-SNARE complex that eluted in a mono-disperse peak in the final size-exclusion chromatography step, and that remained stable in SDS-PAGE without boiling, a hallmark for neuronal SNARE complex formation (Extended Data Fig. 1b). Best results were obtained when the SNARE fragments were truncated as previously described53. The best candidates were used for crystallization trials (see below).
Cloning, expression and purification of the Syt1-SNARE complex
The SNAP-25 isoform used throughout this study is commonly referred to as isoform 2 or SNAP-25A. The C2AB fragment of rat synaptotagmin-1 (amino acid range 141–421) was fused to the amino terminus of the C-terminal SNARE domain of rat SNAP-25 (SNAP-25_C, amino acid range 141–204) via a 37 amino acid linker (sequence: NLSSDSSLSSPSALNSLSSPSALNSTASNSPGIEGLS) derived from the human Oct-1 transcription factor51 (Extended Data Fig. 1a) (referred to as C2AB-linker-SNAP-25_C).
The C2AB-linker-SNAP-25_C chimera, the rat SNAP-25_N fragment (amino acid range 7–83), the rat syntaxin-1A fragment (amino acid range 191–256), and the His-tagged rat synaptobrevin-2 fragment (amino acid range 28–89) were cloned into the Duet expression system (Novagen) following previous work with the neuronal SNARE complex54 (Extended Data Fig. 1a). These four protein constructs were co-expressed in E. coli, leading to complex formation in the host (referred to as Syt1-SNARE37aa-linker complex). Specifically, E. coli BL21(DE3) cells were grown overnight at 30 °C using auto-inducing LB medium55. After harvesting the cells by centrifugation, the pellet was resuspended in lysis buffer (50 mM Tris-HCl, pH8.0, 300 mM NaCl, 20 mM imidazole, 1 mM CaCl2, 0.5mM TCEP and EDTA-free protease inhibitor cocktail (Roche)) and lysed by three passes through the Emulsiflex C5 homogenizer (Avestin) at 15, 000 psi. After centrifugation, the cleared lysate was bound to a 4 ml bed volume of Ni-NTA beads (Qiagen) equilibrated in the lysis buffer. Beads were harvested by centrifugation and poured into a column, washed with the lysis buffer, and subsequently washed with the lysis buffer supplemented with additional 40 mM imidazole. The Syt1-SNARE37aa-linker complex was eluted with the lysis buffer supplemented with additional 350 mM imidazole.
Depending on purification of Ca2+-free or Ca2+-bound complex, EDTA or CaCl2 was included at specified steps. The fresh eluent of the Ni-NTA-affinity purified Syt1-SNARE37aa-linker complex was pooled and dialysed against dialysis buffer I (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5 mM TCEP; with 1 mM CaCl2 or 10 mM EDTA) for 3 to 4 h at 4 °C. The dialysate was supplemented with TEV protease, and further dialyzed in dialysis buffer II (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 0.5 mM TCEP; with 1 mM CaCl2 or none) overnight at 4 °C. After removal of uncleaved sample, the His-tag-cleaved complex was subjected to anion exchange chromatography (buffer A: 50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 0.5 mM TCEP, buffer B: 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.5 mM TCEP; both buffer A and B were supplemented with 1 mM CaCl2 or 2 mM EDTA) using a linear gradient of NaCl starting at 50 mM and ending at 250 mM. The peak fractions were pooled, concentrated, and loaded onto a Superdex 200 10/300 GL column (GE Healthcare) that was pre-equilibrated with SEC buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 0.5 mM TCEP) for Ca2+-free complex, and SEC buffer supplemented with 1 mM CaCl2 for Ca2+-bound complex (Extended Data Fig. 1b). The peak fractions containing Syt1-SNARE37aa-linker complex were pooled and concentrated to a final concentration of ~20 mg ml−1 for crystallization.
Cloning, expression and purification of WT and mutant Syt1 C2B domains
All WT and mutant synaptotagmin-1 C2B domains were prepared by using a standard PCR-based protocol. All PCR products were subcloned into the pGEX-6P-1 (GE Healthcare) and expressed as GST-tagged fusion proteins in E. coli BL21(DE3) cells at 30 °C overnight using auto-inducing LB medium55. After harvesting the cells by centrifugation, the sample was resuspended in lysis buffer containing 50 mM HEPES-Na, pH 7.5, 300 mM NaCl, 2 mM DTT and EDTA-free protease inhibitor cocktail (Roche), and then subjected to sonication and centrifugation. The supernatant was incubated with glutathione-sepharose beads (GE Healthcare). The resin was extensively washed with 50 ml of wash buffer I containing 50 mM HEPES-Na, pH 7.5, 300 mM NaCl, and 1 mM DDT, followed by 50 ml of wash buffer II containing 50 mM HEPES-Na, pH 7.5, 300 mM NaCl, 1 mM DTT, and 50 mM CaCl2 (ref. 19). The GST tag was cleaved overnight at 4 °C with PreScission protease (GE Healthcare) in cleavage buffer containing 50 mM HEPES-Na, pH 7.5, 300 mM NaCl, 1mM DTT, 2 mM EDTA. The cleaved proteins were purified by gel filtration on Superdex 75 (GE Healthcare) that was pre-equilibrated with SEC buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5 mM TCEP. The peak fractions were pooled and concentrated to a final concentration of ~30 mg ml−1 for circular dichroism analysis and crystallization.
Crystallization of the Ca2+-bound Syt1-SNARE complex
Crystallization trials performed by mixing the neuronal SNARE complex and Syt1 C2AB fragments were unsuccessful, probably due to the weak affinity between the two components and the aggregation propensity at higher concentrations, especially in the presence of Ca2+. To overcome these problems, crystallization trials were performed with chimeras between Syt1 C2AB and SNAP-25_C, as described above. The best three candidates consisted of three different linker lengths. Initially, thin crystalline plates that diffracted to about 30 Å were obtained from a construct connected by the 23 amino acid linker. Screening of volatile crystallization additives (such as, methanol, ethanol, 1,2-butanediol) led to improved diffraction to about 10 Å resolution, with sharp Bragg peaks. The complex with a longer 37 amino acid linker (Syt1-SNARE37aa-linker) was more soluble in SEC buffer compared to the one with the shorter 23 amino acid linker. Clusters of needle-shaped crystals were obtained for this construct at first. Using a reverse vapor-diffusion method led to thicker crystal plates (Extended Data Fig. 1e), and eventually produced Bragg peaks past 4 Å resolution. There were consistently two crystal forms in the same drop with identical morphology, referred to as “short unit cell” crystal form and “long unit cell” crystal form, respectively. The difference between the short and long unit cell crystal forms of the Ca2+-bound Syt1-SNARE complex can be approximately described by a doubling of the number of complexes, except that one of the interacting Syt1 C2AB fragments is absent (Fig. 1c, Extended Data Fig. 2g, Extended Data Fig. 3d).
Prior to setting up crystal trays, purified Syt1-SNARE37aa-linker complex (at a concentration of ~20 mg ml−1) was diluted to a final concentration of ~8 mg ml−1 supplemented with CaCl2 and MgCl2. The final buffer contained 20 mM Tris-HCl (pH 8.0), 300 mM NaCl, 100 mM MgCl2, 1 mM CaCl2, and 0.5 mM TCEP. The reservoir contained 100 mM HEPES-Na (pH 7.5) and 1% PEG 8000. Equal amount of protein and reservoir (2 μl) were mixed and incubated at 20 °C. The crystals appeared after 1 to 4 months, and were flash-frozen in a cryo-protecting solution containing the same constituents as the crystallization condition supplemented with 35% (v/v) sucrose.
Although Syt1 was initially covalently linked to the C-terminal half of SNAP-25 (SNAP-25_C), the linker was slowly cleaved at ambient temperature (Extended Data Fig. 1c). Moreover, crystal growth required 1 to 4 months and contained entirely cleaved complex (Extended Data Fig. 1b), enabling formation of the 2:1 and 3:2 stoichiometries in the asymmetric units of the short and long unit cell crystal forms, respectively. Thus, the crystal structures are likely not affected by the initial presence of the linker. We simply refer to the resulting crystal structure as that of the “Syt1-SNARE complex”.
Crystallization of the Mg2+-bound Syt1-SNARE complex
Crystals of Mg2+-bound Syt1-SNARE complex were grown using the reverse hanging-drop vapor-diffusion method. Purified Syt1-SNARE37aa-linker (at a concentration of ~20 mg ml−1) that was prepared without Ca2+ and in the presence of EDTA (as described above) was diluted to a final concentration of ~4 mg ml−1 and supplemented with 100 mM MgCl2. The final buffer contained 20 mM Tris-HCl (pH 8.0), 300 mM NaCl, 100 mM MgCl2, and 0.5 mM TCEP. The reservoir contained 100 mM HEPES-Na (pH 7.5) and 2.5% PEG3350. Similar to Ca2+-bound complex, the linker was also cleaved during crystallization. Mg2+-bound Syt1-SNARE complex crystals appeared after 2 months and were flash-frozen in a cryo-protecting solution containing the same constituents as the crystallization condition supplemented with 20% (v/v) glycerol.
Crystallization of the quintuple mutant of the Syt1 C2B domain
Crystals of the quintuple mutant (R281A, E295A, Y338W, R398A, R399A) of the Syt1 C2B domain were grown by the hanging-drop vapor diffusion method at 20 °C by mixing 2 μl protein solution (at a concentration of ~10 mg ml−1) in a buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.5 mM TCEP) with equal volume of reservoir solution containing 100 mM Tris-HCl (pH 8.5), 1.5 M ammonium sulfate.
XFEL data collection and processing
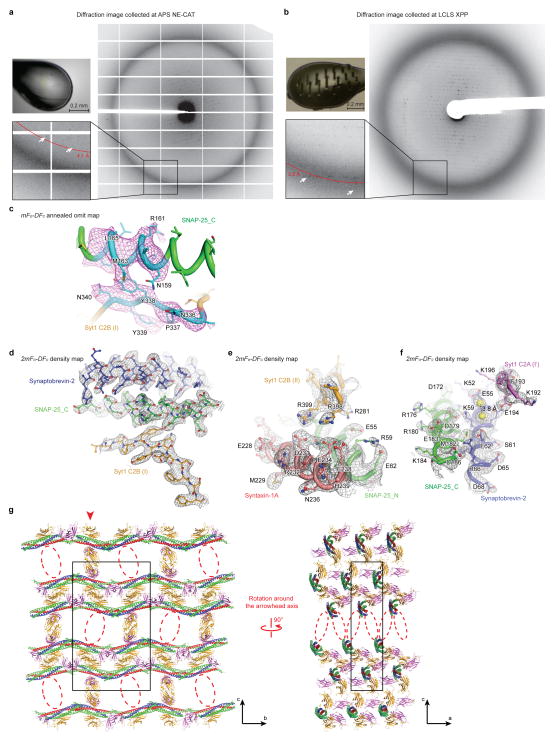
The diffraction data of crystals of the long unit cell crystal form of the Ca2+-bound Syt1-SNARE complex (Extended Data Table 1) were collected at the X-ray Pump Probe (XPP) endstation using the LCLS XFEL at the SLAC National Accelerator Laboratory at Stanford University. The XFEL beam was focused to 30 μm at a nominal energy of 9 keV and a pulse duration of 40 fs in self-amplified stimulated emission (SASE) mode56,57. Each 40 fs XFEL pulse at the XPP endstation at LCLS delivers 1012 photons, exceeding a dose of 30 MGy. The dose is so large that the diffraction volume is vaporized after exposure to a single XFEL pulse (Extended Data Fig. 2b). Nonetheless, the 40 fs laser pulse enables the so-called “diffraction before destruction” data collection58,59, whereby the diffraction pattern is recorded before the sample is destroyed.
The diffraction images were obtained from 148 frozen and cryo-protected crystals mounted in conventional cryo-loops at 100 K using a goniometer-based fixed-target sample delivery station60. XFEL diffraction tests of hydrated crystals at room temperature did not show any marked improvement in comparison to cryo-preserved crystals, at least in terms of visible limiting resolution of the observed diffraction pattern. Furthermore, the entire crystal was damaged when exposed at room temperature, requiring many more crystals and much more time to collect a complete diffraction dataset. As beam time and sample volume were limited, we decided to collect diffraction data at cryogenic temperature, since it was possible to obtain multiple diffraction images from the same frozen crystal.
In most cases, 2 – 20 diffraction images (depending on crystal size and quality) were obtained from a single crystal. Since the diffracting volume was destroyed by the XFEL beam, a different volume had to be exposed for every shot (Extended Data Fig. 2b). The crystal was translated at least 100 microns between exposures. During data collection we observed that improved diffraction quality could be achieved by placing consecutive shots far apart (i.e., across the length of the crystal from one another), then later “filling” between the first and second shots in the same “shuttling” manner. Finally, care was taken to collect exposures at a variety of spindle (ϕ) angles in order to maximize the completeness of the final dataset.
Of the 148 crystals used in the experiment, 113 crystals produced 578 images that could be processed; the other crystals did not diffract or showed multiple lattices. The long unit cell form occurred much more frequently (about 80%). Images indexed in the short unit cell crystal form were identified using a hierarchical clustering method61 and were omitted from further processing since it was not possible to obtain a complete dataset in this crystal form. Other images were rejected during data processing as described below. The 309 diffraction images (from 72 crystals) in the final selection for the long unit cell crystal form were indexed and integrated with the cctbx.xfel suite of data-processing software62–64, with the diffraction data processing parameters optimized by a grid search procedure (Lyubimov, et al., in preparation). The integrated diffraction data were subsequently scaled, merged and post-refined with the PRIME software65.
We optimized the data integration process using a combination of mosaic quality analysis64, highest number of bright reflections yielded by integration, and overlay of observed reflections on the actual integrated areas to determine if nearly all visible reflections were integrated, and no unobserved reflections were predicted and integrated. We inspected overlays of diffraction images and predicted reflection positions in order to fine-tune the computational approaches and optimize the data processing of the XFEL data. The iterative scaling and post-refinement approach utilized by PRIME allowed the construction of a complete diffraction dataset from the relatively small number of diffraction images.
Synchrotron beamline data collection
All other diffraction datasets (Extended Data Table 1) were collected using beamline 24ID-C of the Advanced Photon Source (APS) at Argonne National Laboratory (Argonne, IL). Diffraction data of the best crystals of both the Ca2+-bound short unit cell crystal form and the Mg2+-bound short unit cell crystal form of the Syt1-SNARE complex were indexed and integrated using the XDS software66, and scaled and merged using the SCALA program in CCP4 package67. Diffraction data of the quintuple mutant of the Syt1 C2B domain were indexed, integrated, scaled, and merged using the XDS software66.
Structure determination
The phases for all crystal structures of Syt1-SNARE complex were determined by molecular replacement with Phaser68 using the rat SNARE complex (PDB code 1N7S), the rat Syt1 C2A domain (PDB code 3F04), and the rat Syt1 C2B domain (PDB code 1UOW) as search models. The structures were iteratively rebuilt and refined using the programs Coot69, CNS1.370,71, and phenix.refine72 (Extended Data Table 1). Both the Ca2+- and Mg2+-bound Syt1-SNARE complex in the short unit cell crystal form were refined with phenix.refine72 using non-crystallographic symmetry (NCS) restraints, secondary structure restraints, and grouped B-factor refinement. The long unit cell crystal form of the Ca2+-bound Syt1-SNARE complex was initially refined using CNS 1.370,71, with DEN restraints73, restrained grouped B-factors and NCS restraints, then further refined with phenix.refine72 using non-crystallographic symmetry (NCS) restraints, secondary structure restraints, and individual B-factor refinement. mFo-DFc annealed omit maps (Extended Data Fig. 2c and Extended Data Fig. 3b) were calculated using phenix.refine72 using omit refinement consisting of three cycles of simulated annealing and grouped/individual B-factor refinement (grouped B-factor refinement for the Ca2+-bound Syt1-SNARE complex in the short unit cell crystal form, individual B-factor refinement for the Ca2+-bound Syt1-SNARE complex in the long unit cell form) with non-crystallographic symmetry (NCS) restraints and secondary structure restraints; residues 335 to 340 in Syt1 and 159–166 in SNAP-25 were omitted (that is, residues in region I of the primary interface between Syt1 and the SNARE complex), atoms within a 4 Å cushion around these omitted residues were kept fixed. The R values of the refined structures (Extended Data Table 1) are well within the range that is typical at the corresponding resolutions74. Ramachandran analysis with MolProbity75 indicated that 98% of the residues are in the favored regions and none are in disallowed regions for both the Ca2+- and Mg2+-bound Syt1-SNARE complexes in the short unit cell crystal form, and that 97% of the residues are in the favored regions and none are in disallowed regions for the Ca2+-bound Syt1-SNARE complex in the long unit cell crystal form. The quality indicators for the crystal structures of the Syt1-SNARE complex are well within acceptable ranges indicated by the “polygon” plot produced by phenix.refine72 and by the validation report of the deposited structures and diffraction data.
The phases for crystal structure of the quintuple mutant of the Syt1 C2B domain was determined by molecular replacement with Phaser68 using the rat Syt1 C2B domain (PDB code 1UOW) as the search model. The structure was iteratively built and refined using the program Coot69, and phenix.refine72 (Extended Data Table 1). The final model consists of four Syt1 C2B molecules in the asymmetric unit. Ramachandran analysis with MolProbity75 indicated that 98% of the residues are in the favored regions and none are in disallowed regions.
The LCLS XFEL produced superior electron density maps
The diffraction data obtained at the LCLS XFEL extended to substantially higher resolution than data collected at the APS NE-CAT microfocus synchrotron beamline from similar crystals of the long cell form (Extended Data Figs. 2a, b). It is notable that only 1 out of 85 screened crystals in the long unit cell form diffracted to 4.1 Å at APS, while 61 out of ~ 72 long unit cell crystals diffracted to at least 3.5 Å at LCLS. In fact, only a lack of available XFEL beamtime prevented us from collecting a complete diffraction dataset beyond 3.5 Å. We note that similar improvements in limiting resolution of XFEL versus synchrotron diffraction images have been observed for GPCRs60. Interestingly, while the short unit cell crystal form produced a limiting resolution of 3.6 Å at APS NE-CAT, the density maps are substantially less well defined for sidechains compared to the LCLS data set (compare Extended Data Figs. 2c–f and Figs. 3b, c).
Taken together, the LCLS diffraction data set proved notably superior to the particular synchrotron-derived datasets that we collected in terms of limiting resolution and quality of the electron density maps, and was thus essential for more accurately determining side chain positions. Moreover, in the LCLS XFEL crystal structure there is clear electron density for 19 Ca2+ bound to the Ca2+ binding sites of the Syt1 C2 domains and to a few additional sites on the surfaces of Syt1 and SNARE molecules (Extended Data Table 1). In contrast, electron density could be identified for only 7 Ca2+ in the short unit cell crystal form collected at the APS synchrotron. Taking into account the smaller number of molecules in the asymmetric unit of the short unit cell crystal form (2 Syt1 and 1 SNARE complex compared 3 Syt1 and 2 SNARE complexes), there are still fewer Ca2+ sites that were observed in maps derived from the data set collected at APS, suggesting that Ca2+-binding sites have been affected by radiation damage.
At present, it is difficult to assess the relative quality of XFEL diffraction data studied here with conventional rotation diffraction data measured at a synchrotron. We suspect that the standard diffraction data statistics (such as the merging R values) of rotation data are better due to the ability to directly measure full reflections (at least by summation of partials) without modeling partiality, which is still a relatively crude process even with the latest post-refinement approaches. In our opinion, these apparently poorer statistics obtained from the current state of the art XFEL diffraction data processing methods are more than offset by the improved quality of the resulting electron density maps for diffraction data of crystals collected at the XFEL (Figs. 2c, d, Extended Data Figs. 2c–f). These maps, superior to those of the short unit cell data crystal structure collected at the APS NE-CAT microfocus synchrotron beamline at comparable limiting resolution (Extended Data Figs. 3b, c), enabled us to better investigate the binding interfaces between Syt1 and the SNARE complex, and was thus essential for the structural portion of our study.
Validation and structure analysis
MolProbity75 was used for evaluating the geometry and quality of the models (Extended Data Table 1). The electrostatic potential maps were calculated and displayed using the UCSF Chimera package76 (Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311)). All other structure figures were prepared with PyMol (DeLano, 2002, The PyMOL Molecular Graphics System, http://www.pymol.org, Schrödinger, LLC). Interface areas were calculated by PISA77; note that the commonly used “buried surface area” is twice the “interface area”.
Design of mutations to disrupt the primary interface
We mutated combinations of region I interacting residues (Syt1 E295A, Y338W and SNAP-25 K40A, D166A), and region II interacting residues (Syt1 R281A, R398A, R399A and SNAP-25 D51A, E52A, E55A). We verified that the Syt1 mutants are properly folded by measuring circular dichroism (CD) spectra and thermal denaturation curves (Extended Data Fig. 7a), crystal structure determination of the Syt1 C2B quintuple mutant (R281A, E295A, Y338W, R398A, R399A) (Extended Data Figs. 7b–d, Extended Data Table 1), and size exclusion chromatography of Syt1 (Extended Data Fig. 7e). The crystal structure of the quintuple mutant is very similar to that of WT C2B (PDB code 2YOA) (main chain RMSD 0.43 Å, Extended Data Fig. 7b). We also established that the mutated SNARE complexes form properly (Extended Data Fig. 7f).
A subset of the interacting residues of region II were suggested to be functionally important in previous studies: Syt1 R398, R399 (refs. 35,36,78), and SNAP-25 D51, E52, E55 (refs. 27,79,80). However, the role of these residues was unclear in the absence of structural information, which now reveals that they are part of a larger interface between Syt1 and the SNARE complex. Moreover, interactions in region I (Fig. 2a) have not been implicated in any previous studies.
CD spectroscopy of WT and mutants of Syt1 C2B
CD measurements were conducted with CD spectrometer (Model 202-01, Aviv Biomedical, Inc.) equipped with a temperature controller. Data were collected with 10 μM samples of WT and mutant Syt1 C2B proteins in 20 mM Tris (pH 8.0), 150 mM NaCl buffer (without or with 5 mM CaCl2) over a wavelength range of 195 nm to 260 nm, with 1 nm increments, in a 1 mm path length cell at 25 °C. Temperature denaturation experiments were performed at a wavelength of 216 nm by increasing the temperature from 25 °C to 100 °C in 3 °C temperature increments, a two minute temperature equilibration time, and a 3 sec averaging time. The fraction of unfolded protein at each temperature was calculated by using the formula (Iobs-If)/(Iu-If), where Iobs is the observed mean residue ellipticity, and Iu and If are the mean residue ellipticities of the unfolded and folded states, respectively. Iu and If were estimated by extrapolation of the linear regions of the extremes of the denaturation curves.
Sequence alignment
The alignment was performed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and the figures were prepared with Boxshade3.21 (http://www.ch.embnet.org/software/BOX_form.html). List of UniProt or GeneBank accession numbers of Syt1 homologues: Caenorhabditis elegans (worm Syt1, P34693); Drosophila melanogaster (fly Syt1, P21521); Lymnaea stagnalis (snail Syt1, AAO83847.1); Doryteuthis pealeii (squid Syt1, BAA09866.1); Danio rerio (zebrafish Syt1, XP_005164929.1; zebrafish Syt2, XP_009294914.1; zebrafish Syt9, AAI52175.1); Xenopus tropicalis (frog Syt1, XP_002935685.2); Chelonia mydas (turtle Syt1, XP_007057315.1); Gallus gallus (chicken Syt1, P47191); Rattus norvegicus (rat Syt1, P21707; rat Syt2, P29101; rat Syt9, P47861; rat Syt3, P40748; rat Syt4, P50232; rat Syt5, Q925C0; rat Syt6, Q62746; rat Syt7, Q62747; rat Syt8, Q925B4; rat Syt10, O08625; rat Syt11, O08835; rat Syt12, P97610; rat Syt13, Q925B5; rat Syt14, M0R7W7; Rat Syt15, P59926; Rat Syt16, D3ZB68); Homo sapiens (human Syt1, P21579; human Syt2, Q8N9I0; human Syt9, O00445). List of UniProt or GeneBank accession numbers of SNAP-25 and syntaxin homologues: Caenorhabditis elegans (worm SNAP-25, NP_505641.2; worm syntaxin-1A, O16000); Drosophila melanogaster (fly SNAP-25, P36975; fly SNAP-29, NP_523831.1; fly syntaxin-1A, Q24547); Lymnaea stagnalis (snail syntaxin-1A, AAO83845.1); Doryteuthis pealeii (squid SNAP-25, AAM18191.1; squid syntaxin; CAA74913.1); Danio rerio (zebrafish SNAP-25, NP_001020729.1; zebrafish SNAP-29, NP_001243185.1; zebrafish syntaxin-1B, Q9I9P6); Xenopus laevis (frog SNAP-25, XP_005287463.1); Xenopus tropicalis (frog syntaxin-1A, NP_001072191.1); Chrysemys picta bellii (turtle SNAP-25, XP_007057315.1; turtle syntaxin-1A, XP_005294403.1); Gallus gallus (chicken SNAP-25, P60878; chicken syntaxin-1B, F5HN09); Rattus norvegicus (rat SNAP25a, P60881-2; rat SNAP-25b, P60881-1; rat SNAP-23, O70377; rat SNAP-29, Q9Z2P6; rat syntaxin-1A, P32851; rat syntaxin-1B, P61266; rat syntaxin-2, P50279-2; rat syntaxin-3, Q08849; rat syntaxin-4, Q08850; rat syntaxin-5, Q08851-2; rat syntaxin-7, O70257); Homo sapiens (human SNAP-25a, P60880-2; human SNAP-25b, P60880-1; human SNAP-23, O00161; human SNAP-29, O95721; human syntaxin-1A, Q16623-1); Saccharomyces cerevisiae (yeast sec-9, P40357; yeast sso-1, P32867).
Protein expression and purification for single vesicle-vesicle experiments
Full length cysteine-free rat synaptobrevin-2, rat syntaxin-1A, rat SNAP-25A (with all endogenous cysteines changed to serines), rat Syt1 (with all endogenous cysteines changed to alanine, except the cysteine residue at position 277), and WT rat complexin-1 were expressed and purified as previously described38. All mutants of Syt1 and SNAP-25 were generated using the Quick Change Site-Directed Mutagenesis Kit (Agilent) and expressed and purified using the same protocol.
Protein reconstitution for single vesicle-vesicle fusion experiments
Neuronal SNAREs and Syt1 represent a minimal system for Ca2+-triggered membrane fusion. Recent evidence for this notion came from a reconstituted single vesicle-vesicle assay that discriminates among vesicle association, lipid mixing, and content mixing37,38,81. Moreover, addition of complexin-1 greatly enhanced Ca2+-triggered amplitude and synchronization, and suppressed spontaneous release in this system38. A variety of complexin-1 truncations and mutations qualitatively reproduced effects observed in neuronal cultures for both spontaneous and Ca2+-triggered release38, lending credence to this reconstituted system to investigate mechanistic questions.
We used the same membrane compositions and protein densities as in our previous studies37,82. Likewise, the reconstitution protocol was similar82 with several changes as described in ref. 38. Briefly, one class of vesicles was reconstituted with both Syt1, or its mutants, and synaptobrevin-2 (referred to as “synaptic vesicle” - SV - vesicles), while another class of vesicles was reconstituted with syntaxin-1A and SNAP-25 or its mutants (referred to as “plasma membrane” - PM - vesicles), using the previously described lipid compositions. The protein-to-lipid ratios used were 1:200 for synaptobrevin-2 and syntaxin-1A, and 1:1000 for Syt1 and its mutants. A 3- to 5-fold excess of SNAP-25 and its mutants (with respect to syntaxin-1A) and 3.5 mol% PIP2 were added to the protein-lipid mixture for PM vesicles only. Dried lipid films were dissolved in 110 mM β-octyl glucoside (beta-OG) buffer containing purified proteins. Detergent-free buffer (20 mM HEPES-Na, pH 7.4, 90 mM NaCl, 0.1 % 2-mercaptoethanol) was then added to the protein-lipid mixture until the beta-OG concentration reached the critical micelle concentration 24.4 mM. The vesicles were then subjected to size exclusion chromatography using a Sepharose CL-4B column, packed under near constant pressure by gravity with a peristaltic pump (GE Healthcare) in a 5 ml column with a 2 ml bed volume, that was equilibrated with buffer V (20 mM HEPES-Na, pH 7.4, 90 mM NaCl, 20 μM EGTA, 0.1 % 2-mercaptoethanol) followed by dialysis into 2 L of detergent-free buffer V supplemented with 5 g of Bio-beads SM2 and 0.8 g/L Chelex 100 resin (Bio-Rad, Life Science Research). After 4 h, the buffer was changed with 2 L of fresh buffer V containing Bio-beads and Chelex, and dialysis continued for 12 h. During the preparation of SV vesicles, 50 mM sulforhodamine B (Invitrogen) was present in all solutions prior to the size exclusion chromatography step. As described previously81, the presence and purity of reconstituted proteins was confirmed by SDS-PAGE of the vesicle preparations, and the directionality of the membrane proteins (facing outward) was assessed by chymotrypsin digestion followed by SDS-PAGE. The size distributions of the SV and PM vesicles were analyzed by cryo-EM, as described previously37.
Single vesicle-vesicle content-mixing assay
We used the single vesicle-vesicle assay described in ref. 38. Briefly, SV vesicles were labeled with a soluble fluorescent content dye (sulforhodamine B) at a moderately self-quenching concentration; for simplicity in this work we did not include a lipid-dye since we were exclusively interested in the exchange of content, the correlation for neurotransmitter release. The PM vesicles were immobilized on a surface that was passivated with polyethylene glycol (PEG) and functionalized via streptavidin-biotin linkages. SV vesicles were then added in the presence of 2 μM complexin-1. We directly started monitoring the arrival of SV vesicles to surface-immobilized PM vesicles during the first minute acquisition period. A stepwise increase in fluorescence emission of a spot in the field of view indicated the formation of a SV-PM vesicle pair during the vesicle association period.
Unbound SV vesicles were then removed through extensive washing with vesicle-free buffer, while continuing real-time observation of the fluorescence intensity; consequently we did not observe any additional SV-PM vesicle associations after the washing step. While continuing the observation for another one-minute period, a second step-wise increase of fluorescence intensity appeared for some fraction of the associated SV vesicles, which indicated Ca2+-independent, that is, spontaneous fusion events (referred to has the spontaneous fusion period). Next, we injected 500 μM Ca2+ solution, and continued monitoring for another minute, referred to as the Ca2+-triggered fusion period. For associated SV vesicles that did not undergo spontaneous fusion during the second period, a step-wise increase in fluorescence intensity during the third period indicated a Ca2+ triggered fusion event. To determine the temporal arrival of Ca2+ in the evanescent field of our TIR microscope setup, soluble Cy5 dye was added with the Ca2+ buffer to monitor the emergence of fluorescence intensity. Thus, our improved single vesicle-vesicle assay enables one to monitor the association of SV vesicles, spontaneous, and Ca2+ triggered fusion events during the same data acquisition. Further details can be found in ref. 38.
Previous reconstitutions often employed lipid mixing, rather than content mixing, between vesicles with reconstituted neuronal SNAREs21,83. For example, increased ensemble lipid mixing was observed upon addition of the soluble Syt1 C2AB fragment and Ca2+ (ref. 21). However, in retrospect, this result was probably caused by an effect on vesicle association since multivalent binding of the soluble Syt1 C2AB fragment can induce vesicle clustering19. Moreover, subsequent work revealed major differences between using the soluble C2AB fragment and reconstituted, full-length Syt184,85. More importantly, assays based on lipid mixing alone can produce misleading results since lipid mixing can occur without content mixing86. Our single vesicle-vesicle assay thus uses full-length Syt1 and it monitors content mixing, a correlate for neurotransmitter release37,38,81.
Syt1 cKO mice
Syt1 cKO mice were generated by the European Conditional Mouse Mutagenesis Program (EUCOMM) and are available from the European Mouse Mutant Archive (EMMA) (EM:06829). Exon 2 of Syt1 containing the transmembrane domain is floxed and its removal results in a frame shift that produces a truncated protein. The Syt1 targeted mice were first crossed to FLPe mice in order to remove a gene trap cassette surrounded by frt sites. This cross yielded the cKO mice as schematically shown in Fig. 3a. Exposure to Cre-recombinase results in the total absence of synchronous release which is typical for Syt1 KO neurons (Fig. 4a, b).
Neuronal cultures
Neuronal cultures were produced from WT and Syt1 cKO mice as previously described87. Hippocampi were dissected from P0 pups, dissociated by papain digestion, and plated on Matrigel-coated glass coverslips. Neurons were cultured in vitro in MEM supplemented with B27 (Gibco), glucose, transferrin, fetal bovine serum, and Ara-C (Sigma) and were analyzed after 14–16 days.
Lentivirus production
For rescue experiments, we used a lentiviral construct carrying a synapsin promoter, an optional rat Syt1 cDNA, internal ribosome entry site (IRES), and a GFP-Cre-recombinase fusion sequence. The control plasmid (TB592) contained no rescuing cDNA, with rescuing plasmids carrying the following cDNAs: TB761 (WT), TB762 (R398, 399Q), TB765 (E295A, Y338W), TB767 (R281A, R398A and R399A), TB777 (R281A, E295A, Y338W, R398A and R399A). To make viruses, human embryonic kidney 293T cells were co-transfected with the lentiviral vector and three packaging plasmids. Supernatant containing the viruses was collected 48 h after transfection and was used to infect hippocampal neuronal cultures at DIV4. Cultures were used for biochemical or physiological analyses at DIV14-16.
Electrophysiological recordings in cultured neurons
Recordings were performed essentially as previously described10. The whole-cell pipette solution contained 40 mM CsCl, 90 mM K-gluconate, 1.8 mM NaCl, 1.7 mM MgCl2, 3.5 mM KCl, 0.05 mM EGTA, 10 mM HEPES, 2 mM Mg-ATP, 0.4 mM Na-GTP, 10 mM phosphocreatine, and 10 mM QX-314 (pH 7.4, adjusted with CsOH). The bath solution contained 140 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM HEPES, 10 mM glucose (pH 7.4, adjusted with NaOH). Evoked synaptic responses were triggered by a bipolar electrode. GABA-R-mediated IPSCs were pharmacologically isolated with CNQX (20 μM) and AP-5 (50 μM) in the bath solution and recorded at a −70 mV holding potential. Since the intracellular solution contains high internal Cl− levels, IPSCs evoke large inward currents.
Immunoprecipitation and quantitative immunoblotting
Cultured Syt1 cKO neurons infected with viruses expressing the desired mutants were solubilized in PBS (with 1 mM CaCl2, 0.2% Triton X-100, pH 7.4) supplemented EDTA-free protease inhibitor cocktail (Roche) for 1 h. The lysate was cleared by centrifugation at 16,000 g for 10 min at 4 °C and immunoprecipitation was performed by incubating with polyclonal antibodies to syntaxin-1 (438B) or preimune sera for 1 h at 4 °C, followed by incubation with 15 μl of a 50% slurry of protein-A Sepharose beads (GE Healthcare) for 2 h at 4 °C. Beads were washed 5 times with 1 ml extraction buffer, bound proteins were eluted with 2× SDS sample buffer containing 100 mM DTT and boiled for 20 min at 100 °C.
Co-precipitated proteins were separated by SDS-PAGE followed by detection with monoclonal antibodies against rat Syt1 (604.4, Synaptic Systems - this antibody does not detect mouse Syt1) and synaptobrevin-2 (cl. 69.1, Synaptic Systems). To allow for quantitative detection, dye-conjugated secondary antibodies were used (IRDye 800CW Donkey anti-Mouse IgG, Li-cor), membranes were scanned in an Odyssey scanner (Li-cor), and quantification was performed using Image Studio software (Li-cor). All experiments included a Syt1 WT group in addition to the desired mutants. The ratio of the Syt1/synaptobrevin-2 signal was determined and normalized with the Syt1 WT condition being set equal to 1.
Extended Data
Extended Data Figure 1. Purification and crystallization of the Syt1-SNARE complex.
a, Diagram of the Duet co-expression vectors (Novagen) that express the fragments of the neuronal SNARE complex and the C2AB-linker-SNAP-25_C chimera used for purification and crystallization of the Syt1-SNARE37aa-linker complex. The rat syntaxin-1A and His-tagged rat synaptobrevin-2 fragments were cloned into the vector pACYCDuet-1; the C2AB-linker-SNAP-25_C chimera and the SNAP-25_N fragment were cloned into the vector pETDuet-1 with amino acid ranges labeled. Dashed lines represent the 37 amino acid linker (Methods). b, The purified Syt1-SNARE37aa-linker complex eluted as a single peak during size-exclusion chromatography (profile on the left). Left gel, Coomassie blue-stained SDS-PAGE gel of the purified Syt1-SNARE37aa-linker complex (unboiled and boiled). Right gel, Coomassie blue-stained SDS-PAGE gel of dissolved crystals of the Syt1-SNARE complex that were grown over a period of two months starting from purified Syt1-SNARE37aa-linker (unboiled and boiled). Although Syt1 was initially covalently linked to SNAP-25_C, the linker was cleaved during crystallization. The comparison between boiled and un-boiled lanes is a hallmark showing that neuronal SNARE complex is fully formed. c, Boiled Coomassie blue-stained SDS-PAGE gel of the purified Syt1-SNARE complex in solution at ambient temperature at the specified time after purification. Cleavage is apparent on day one and progresses slowly over several days. d, Schema showing the commonly used vapor-diffusion technique: the drop contains a lower concentration of the precipitant than the reservoir. The crystallization of the quintuple mutant of Syt1 C2B is used as an example. e, Schema showing a reverse vapor-diffusion method that was used for crystallization of the Ca2+-bound Syt1-SNARE37aa-linker complex: the drop contains a higher concentration of the precipitant than the reservoir.
Extended Data Figure 2. Diffraction images, electron density maps, and crystal packing of the Syt1-SNARE complex in the long unit cell crystal form.
a, Only one out of 85 screened crystals in the long unit cell crystal form diffracted to 4.1 Å resolution at the APS NE-CAT microfocus synchrotron beamline (a total of 105 crystals were screened with 20 that indexed in the short unit cell crystal form). b, 61 out of ~72 crystals in the long unit cell crystal form diffracted to at least 3.5 Å resolution at the LCLS XFEL (a total of 148 crystals were diffracted, out of those 113 crystals produced 578 images that could be processed; 35 crystals did not diffract or showed multiple lattices). These exposures were taken along the crystal c axis. The left upper pictures in panels a and b show images of loop-mounted crystals after X-ray exposure. c, mFo-DFc annealed omit map (Methods) of the Ca2+-bound Syt1-SNARE complex in the long unit cell crystal form using diffraction data collected at the LCLS XFEL; omitted residues within region I of the primary interface (residues 335 to 340 in Syt1 and 159 to 166 in SNAP-25) are colored cyan. The contour level is 2.3 σ. d–f, representative 2mFo-DFc electron density maps of the Ca2+-bound Syt1-SNARE complex in the long unit cell crystal form using diffraction data collected at the LCLS XFEL. The contour level is 1.5 σ. g, Views of the crystal lattice perpendicular to the bc (left) and to the ac (right) planes of the Ca2+-bound Syt1-SNARE complex in the long unit cell crystal form. The particular layer shown on the right corresponds to the red arrowhead in the left panel (only a slice corresponding to the layer is shown, creating the appearance of two disconnected groups of molecules – these groups are actually connected via interactions with the neighboring layers). The red dashed oval indicates the “missing” Syt1 C2AB fragment compared to the short unit cell crystal form (Extended Data Fig. 3d).
Extended Data Figure 3. Asymmetric unit, electron density maps and crystal packing of the Syt1-SNARE complex in the short unit cell crystal form.
a, Asymmetric unit of the Ca2+-bound Syt1-SNARE complex in the short unit cell crystal form at 3.6 Å resolution using diffraction data collected at the APS NE-CAT microfocus synchrotron beamline (Extended Data Table 1). The colour code is the same as in Fig. 1c. Two Syt1 C2AB fragments (distinguished by the designators I and I′) bind to the same SNARE complex in the asymmetric unit (see schema). b, mFo-DFc annealed omit map (Methods) of the Ca2+-bound Syt1-SNARE complex in the short unit cell crystal form collected at the APS NE-CAT microfocus synchrotron beamline; omitted residues within region I of the primary interface (residues 335 to 340 in Syt1 and 159 to 166 in SNAP-25) are colored cyan. The contour level is 2.3 σ. Left side: without B-factor sharpening, right side: with B-factor sharpening. c, Representative 2mFo-DFc electron density map of the Ca2+-bound Syt1-SNARE complex for the short unit cell crystal form using diffraction data collected at the APS NE-CAT microfocus synchrotron beamline. The contour level is 1.5 σ. Left side: without B-factor sharpening, right side: with B-factor sharpening. b and c: The sharpening B-factor (−55 Å2) was set to make the lowest atomic B-factor of the short unit cell crystal form comparable to that of the long unit cell crystal form. Even with B-factor sharpening, the electron density map of the long unit cell crystal form collected at the LCLS XFEL is superior to that of the short unit cell crystal form collected at the APS NE-CAT microfocus synchrotron beamline. d, Views of the crystal lattice perpendicular to the bc (left) and to the ac (right) planes of the Ca2+-bound Syt1-SNARE complex in the short unit cell crystal form. The particular layer shown on the right corresponds to the red arrowhead in the left panel. The unit cell is outlined by a black box.
Extended Data Figure 4. Single molecule FRET efficiency distributions of the Syt1-SNARE complex vs. FRET efficiency values calculated from the Syt1-SNARE interfaces observed in the crystal structure.
Shown are histograms of intermolecular single molecule FRET (smFRET) efficiency values that were measured between pairs of covalently attached organic labels on the Syt1 C2AB fragment and the SNARE complex28 (also shown as large spheres superimposed on the interfaces observed in the crystal structure). Arrowheads indicate FRET efficiencies calculated from the crystal structure of the Ca2+-bound Syt1-SNARE complex in the long unit cell crystal form (complex I) for the primary, secondary and tertiary interfaces, using the methods and approximations described in ref. 28 to simulate the positions of dye centers in order to calculate the FRET-efficiency values. Only the dye pair combinations between the nearest C2 domain (including the C2A-C2B linker) and the SNARE complex were calculated for the three interfaces. Note that due to the presence of transitions between different states the histogram reflect a combined effect of interaction interfaces. The label at position A61 would have disrupted the tertiary interfaces between the C2A domain and the SNARE complex, explaining the discrepancy for these labels (indicated by open triangles). In retrospect, the top smFRET-derived model28 and the primary interface observed in the crystal structure primarily differed in the orientation of the C2B domain. Moreover, the top smFRET derived model predicted the approximate location primary interface on the neuronal SNARE (see Fig. 4c in ref. 28).
Extended Data Figure 5. Comparison of the two crystal forms and the Ca2+- and Mg2+-bound crystal structures of the Syt1-SNARE complex.
a, Superposition of the primary interfaces of the Ca2+-bound Syt1-SNARE complex structure in the long unit cell crystal form (gold and bright-orange) and in the short unit cell crystal form (white). The primary interface is very similar in both crystal forms: the RMSD for the primary interface between both crystal forms is 0.38 Å (bright-orange) and 0.42 Å (white) for complex I and complex II, respectively (including Cα atoms of the SNARE complex and the Syt1 C2B (I) domain forming the interface). b, Superposition of complex I in the long unit cell crystal form with the asymmetric unit of the short unit cell crystal form, but only showing the secondary interface (light blue shaded disk) between Syt1 C2B (I′) and the SNARE complex (I). The bottom panels show close-up views of the secondary interface: left, interacting residues (sticks and balls); right, a 90° rotated view of the view shown in the left panel. The Syt1 C2B (I′) domain is rotated by 16 degrees between the two crystal forms and, as a consequence, the interactions between residues R281, K288, R398 of the Syt1 C2B (I′) domain and residues E224, E228 of syntaxin-1A are slightly changed by this rotation. Interestingly, residues Syt1 R281, K288 and R398 are involved in both the primary (Fig. 2) and secondary interfaces. c, Superposition of complex I in the long unit cell crystal form with the asymmetric unit of the short unit cell crystal form, showing all interfaces. d, Superposition of the Ca2+-bound (white) and Mg2+-bound (black) crystal structures of the Syt1-SNARE complex, both in the short unit cell crystal form. The lower left sub-panel shows a close-up view of the primary interface, indicating that it is very similar in both the Ca2+- and Mg2+-bound crystal structures. The Syt1 C2B domain that forms the secondary interface (light blue shaded disk) is rotated by 19 degrees between the Ca2+- and Mg2+-bound complexes. The lower right sub-panel is a rotated view of the complex, and also showing the tertiary interface (light green shaded disk), and the C2A-C2B interface that involve asymmetry-related Syt1 C2A domain (I′) (gray shaded disk). e, B-factor coloured cartoon representations of the asymmetric units of the Ca2+-bound long unit cell crystal form (upper), the Ca2+-bound short unit cell crystal form (lower left), and the Mg2+-bound short unit cell crystal form (lower right) of the Syt1-SNARE complex. Note that the primary interfaces have relatively low B-factors, similar to the majority of the structure, while parts of the C2A and C2B domains involved in the secondary and tertiary interfaces have higher B-factors, possibly indicating increased flexibility.
Extended Data Figure 6. Sequence alignments of Syt1, SNAP-25, and syntaxin-1A from different homologs.
a, Sequence alignment of Syt1 homologues, showing the sequences around the primary interface of the Syt1-SNARE complex. Note that rat Syt5 refers to UniProt ID Q925C0, zebrafish Syt9 refers to GeneBank accession number AAI52175, rat Syt9 refers to UniProt ID P47861, and human Syt9 refers to UniProt ID O00445. b, Electrostatic potential surfaces of the known crystal structures of synaptotagmin-1, synaptotagmin-3, synaptotagmin-4 and synaptotagmin-7; the dashed rectangles indicate the regions that correspond to the primary interface regions I and II of the Syt1-SNARE complex. c, Sequence alignment of different SNAP-25 homologs, showing the sequences around the primary interface of the Syt1-SNARE complex. d, Sequence alignment of different syntaxin homologs, showing a sequence range around the primary interface of the Syt1-SNARE complex. In all panels, the interacting residues of the primary interface are indicated by solid circles and coloured boxes for region I (cyan) and region II (red-orange).
Extended Data Figure 7. Syt1 and SNAP-25 mutants are well folded.
a, Upper panels, CD spectra of WT and mutant Syt1 C2B domains in the absence of Ca2+. Lower panels, thermal denaturation was monitored by molar ellipticity at a wavelength of 216 nm in the absence of Ca2+ (black) and in the presence of 5 mM Ca2+ (red). The specified melting temperatures were estimated as the mid-point of the melting curves (Methods). b, Superposition of the Syt1 C2B domains from the Ca2+-bound Syt1-SNARE complex in the short unit cell crystal form (gold), the crystal structure of the quintuple mutant (R281A, E295A, Y338W, R398A, R399A) of the Syt1 C2B domain (green), and the crystal structure of the isolated Syt1 C2B domain (white, PDB code 2YOA). c and d, Representative m2Fo-DFc electron density maps of the crystal structure of the quintuple mutant of the Syt1 C2B domain (Extended Data Table 1) contoured at 2.0 σ. The labels refer to the mutated residues. e, Overlay of SEC profiles of full-length Syt1 mutant proteins used in the single vesicle-vesicle fusion assay (Figs. 3d–g). f, Coomassie blue-stained SDS-PAGE with and without boiling of neuronal SNARE complexes formed by full-length SNAP-25 and its mutants, syntaxin-1A, and synaptobrevin-2, using the proteins that were used in the single vesicle-vesicle fusion assay (Methods).
Extended Data Figure 8. Probability of fusion vs. time upon 500 μM Ca2+-injection and spontaneous fusion for Syt1 and SNAP-25 mutants.
Shown are the data that were used to generate Figs. 3d–g. The number of independent experiments and analyzed events are provided in Extended Data Table 2. a–d, Cumulative histograms of probability of fusion vs. time for Syt1 mutants upon 500 μM Ca2+-injection (a) and spontaneous fusion (b), and SNAP-25 mutants upon 500 μM Ca2+-injection (c) and spontaneous fusion (d). Control experiments: e, Ca2+-triggered fusion; f, spontaneous fusion with 3 mM ATP, without SNAP-25 or Syt1; and g, mock injection without Ca2+.
Extended Data Table 1.
Crystallographic data and refinement statistics
| Ca2+-bound Syt1-SNARE complex (long unit cell crystal form) | Ca2+-bound Syt1-SNARE complex (short unit cell crystal form) | Mg2+-bound Syt1-SNARE complex (short unit cell crystal form) | Quintuple mutant of Syt1C2B | |
|---|---|---|---|---|
| Data collection | ||||
| Beamline | SLAC-LCLS | APS-NECAT | APS-NECAT | APS-NECAT |
| Space group | P212121 | P21212 | P21212 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 69.6, 171.1, 291.9 | 69.1, 171.6, 146.9 | 69.1, 171.8, 146.6 | 68.9, 107.6, 110.8 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 20.0–3.50 (3.62–3.50)* | 50.0–3.60 (3.73–3.60) | 85.9–4.10 (4.32–4.10) | 50.0–1.65 (1.69–1.65) |
| Rmerge(%) (rotation)† | -- | 6.7 (74.1) | 10.4 (64.6) | 6.40 (91.7) |
| Rmerge(%) (still)† | 39.7 (32.2) | -- | -- | -- |
| CC1/2 | 92.7 (35.5) | 99.9 (89.5) | 99.7 (64.6) | 99.9 (78.3) |
| I/σI | 7.7 (2.6) | 12.3 (1.9) | 8.8 (1.9) | 18 (2.3) |
| Completeness (%) | 87.6 (65.6) | 99.7 (98.7) | 95.9 (97.0) | 99.0 (98.5) |
| Multiplicity (rotation)† | -- | 14.5 (12.4) | 3.4 (3.5) | 6.7 (6.8) |
| Multiplicity (still)† | 5.0 (1.8) | -- | -- | -- |
| Refinement | ||||
| Resolution (Å) | 20.0–3.50 (3.62–3.50) | 50.0–3.60 (3.73–3.60) | 50.0–4.10 (4.25–4.10) | 50.0–1.65 (1.71–1.65) |
| No. reflections | 39174 (2884) | 20846 (2004) | 13519 (1320) | 98575 (9676) |
| Rwork/Rfree | 0.322/0.353 | 0.249/0.289 | 0.276/0.323 | 0.150/0.178 |
| No. atoms | ||||
| Protein | 10890 | 6506 | 6510 | 5033 |
| Ca2+ | 19 | 7 | 0 | 0 |
| Mg2+ | 0 | 0 | 4 | 0 |
| B-factors | ||||
| Protein | 49 | 158 | 194 | 23.1 |
| Ca2+ | 32 | 187 | - | - |
| Mg2+ | - | - | 202 | - |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.003 | 0.004 | 0.003 | 0.019 |
| Bond angles (°) | 0.758 | 0.862 | 0.714 | 1.763 |
Values in parentheses are for the highest resolution shell.
“(rotation)” refers to rotation diffraction data collected at the APS synchrotron and “(still)” refers to still diffraction data collected at the LCLS XFEL.
Extended Data Table 2.
Data summary table for the single vesicle-vesicle fusion experiments with Syt1 and SNAP-25 mutants
| No. of spontaneous fusion events | No. of Ca2+ triggered fusion events | Total no. of analyzed events (no. of associated vesicle pairs) | No. of independent experiments (N) | |
|---|---|---|---|---|
| Syt1 WT | 90 | 167 | 3764 | 5 |
| 100 | 262 | 4632 | 6 | |
| 49 | 152 | 3035 | 5 | |
| 64 | 173 | 3564 | 4 | |
| 159 | 256 | 6503 | 9 | |
| Syt1 mutants | ||||
| E295A, Y338W | 113 | 246 | 5872 | 12 |
| R398Q, R399Q | 166 | 217 | 6724 | 7 |
| R281A, R398A, R399A | 133 | 271 | 5134 | 7 |
| Syt1 quintuple | 49 | 143 | 5241 | 7 |
| SNAP-25 mutants | ||||
| K40A, D166A | 61 | 139 | 2549 | 7 |
| D51A, E52A, E55A | 81 | 150 | 2717 | 7 |
| SNAP-25 quintuple | 105 | 261 | 6741 | 9 |
| Control | ||||
| +ATP | 59 | 129 | 2830 | 5 |
| -SNAP-25 | 24 | 36 | 3040 | 5 |
| -Syt1 | 57 | 58 | 2379 | 5 |
| Mock | - | 25 | 1947 | 5 |
Supplementary Material
Acknowledgments
We thank Mark S. Padolina for help with protein purification, the Northeastern Collaborative Access Team (supported by NIH P41 GM103403) at Advanced Photon Source for X-ray data collection, the SSRL/LCLS scientists Elizabeth L. Baxter, Paul Ehrensberger, Thomas I. Eriksson, Yiping Feng, Michael Hollenbeck, Elena G. Kovaleva, Scott E. McPhillips, Silke Nelson, Jinhu Song, Yingssu Tsai, Vladimir Vinetsky, and Diling Zhu for their invaluable assistance with data collection at the LCLS XPP facility. Use of the Stanford Synchrotron Radiation Lightsource (SSRL) and Linac Coherent Light Source (LCLS), SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). Portions of this research were carried out at the Linac Coherent Light Source (LCLS) at the SLAC National Accelerator Laboratory. LCLS is an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. This research was supported in part by the National Institutes of Health (R37MH63105 to A.T.B.; MH086403 to T.C.S.; GM095887 and GM102520 to N.K.S and A.S.B); and by a HHMI Collaborative Innovation Award (HCIA) to A.T.B. and W.I.W.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Contributions Q.Z. designed, expressed, purified, and crystallized the Syt1-SNARE complexes and the Syt1 C2B quintuple mutant, performed CD experiments, and designed the mutants to disrupt the primary interface. Q.Z. and M.Z. collected all diffraction data, determined, and refined the crystal structures. Y.L. and J.D. performed the reconstituted single vesicle-vesicle experiments. T.B. performed the co-immunoprecipitation and electrophysiological experiments of neuronal cultures. A.Y.L., M.U., O.B.Z., N.K.S., A.S.B., W.I.W., and A.T.B. analyzed and processed the LCLS diffraction data. A.E.C. and S.M.S. designed the goniometer-based setup at LCLS-XPP and helped with data collection. R.M., M.C., and H.T.L helped with data collection at LCLS-XPP. R.A.P and Y.L. expressed and purified proteins for the single vesicle-vesicle experiments. U.B.C. helped with the comparison between the crystal structure and the smFRET data. Q.Z, M.Z, Y.L., T.B., T.C.S, and A.T.B. wrote the manuscript.
The coordinates of the atomic models and corresponding structure factors of the Syt1-SNARE complexes have been deposited in the Protein Data Bank (PDB) under the accession codes 5CCG, 5CCH, 5CCI, 5CCJ.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothman JE. The Principle of Membrane Fusion in the Cell (Nobel Lecture) Angew Chemie Int Ed. 2014:n/a–n/a. doi: 10.1002/anie.201402380. [DOI] [PubMed] [Google Scholar]
- 3.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 4.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 5.Südhof TC. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron. 2013;80:675–90. doi: 10.1016/j.neuron.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pang ZP, Südhof TC. Cell biology of Ca2+-triggered exocytosis. Curr Opin Cell Biol. 2010;22:496–505. doi: 10.1016/j.ceb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geppert M, et al. Synaptotagmin I: a major Ca2+ sensor for transmitter release at a central synapse. Cell. 1994;79:717–727. doi: 10.1016/0092-8674(94)90556-8. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Mashimo T, Südhof TC. Synaptotagmin-1, -2, and -9: Ca(2+) sensors for fast release that specify distinct presynaptic properties in subsets of neurons. Neuron. 2007;54:567–81. doi: 10.1016/j.neuron.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Wen H, et al. Distinct roles for two synaptotagmin isoforms in synchronous and asynchronous transmitter release at zebra fish neuromuscular junction. 2010 doi: 10.1073/pnas.1008598107/-/DCSupplemental. www.pnas.org/cgi/doi/10.1073/pnas.1008598107. [DOI] [PMC free article] [PubMed]
- 10.Bacaj T, et al. Synaptotagmin-1 and synaptotagmin-7 trigger synchronous and asynchronous phases of neurotransmitter release. Neuron. 2013;80:947–59. doi: 10.1016/j.neuron.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshihara M, Littleton JT. Synaptotagmin I functions as a calcium sensor to synchronize neurotransmitter release. Neuron. 2002;36:897–908. doi: 10.1016/s0896-6273(02)01065-6. [DOI] [PubMed] [Google Scholar]
- 12.Maximov A, Südhof TC. Autonomous function of synaptotagmin 1 in triggering synchronous release independent of asynchronous release. Neuron. 2005;48:547–54. doi: 10.1016/j.neuron.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Kochubey O, Schneggenburger R. Synaptotagmin Increases the Dynamic Range of Synapses by Driving Ca2+-Evoked Release and by Clamping a Near-Linear Remaining Ca2+ Sensor. Neuron. 2011;69:736–748. doi: 10.1016/j.neuron.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Davletov Ba, Sudhof TCa. Single C2 Domain From Synaptotagmin-I Is Sufficient for High-Affinity Ca2+/Phospholipid Binding. J Biol Chem. 1993;268:26386–26390. [PubMed] [Google Scholar]
- 15.Brose N, Petrenko aG, Südhof TC, Jahn R. Synaptotagmin: a calcium sensor on the synaptic vesicle surface. Science. 1992;256:1021–5. doi: 10.1126/science.1589771. [DOI] [PubMed] [Google Scholar]
- 16.Fernández-Chacón R, et al. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–9. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 17.Vrljic M, et al. Post-translational modifications and lipid binding profile of insect cell-expressed full-length mammalian synaptotagmin 1. Biochemistry. 2011;50:9998–10012. doi: 10.1021/bi200998y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee JS, et al. Augmenting neurotransmitter release by enhancing the apparent Ca2+ affinity of synaptotagmin 1. Proc Natl Acad Sci U S A. 2005;102:18664–9. doi: 10.1073/pnas.0509153102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Araç D, et al. Close membrane-membrane proximity induced by Ca(2+)-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–17. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- 20.Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell. 2009;138:709–21. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–8. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Rizo J, Südhof TC. Mechanism of phospholipid binding by the C2A-domain of synaptotagmin I. Biochemistry. 1998;37:12395–12403. doi: 10.1021/bi9807512. [DOI] [PubMed] [Google Scholar]
- 23.Rufener E, Frazier AA, Wieser CM, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin C2B domain determined by site-directed spin labeling. Biochemistry. 2005;44:18–28. doi: 10.1021/bi048370d. [DOI] [PubMed] [Google Scholar]
- 24.Bennett MK, Calakos N, Scheller RH. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Sci (New York, NY) 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- 25.Li C, et al. Ca(2+)-dependent and -independent activities of neural and non-neural synaptotagmins. Nature. 1995;375:594–599. doi: 10.1038/375594a0. [DOI] [PubMed] [Google Scholar]
- 26.Pang ZP, Shin OH, Meyer AC, Rosenmund C, Südhof TC. A gain-of-function mutation in synaptotagmin-1 reveals a critical role of Ca2+-dependent soluble N-ethylmaleimide-sensitive factor attachment protein receptor complex binding in synaptic exocytosis. J Neurosci. 2006;26:12556–65. doi: 10.1523/JNEUROSCI.3804-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rickman C, et al. Conserved prefusion protein assembly in regulated exocytosis. Mol Biol Cell. 2006;17:283–94. doi: 10.1091/mbc.E05-07-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi UB, et al. Single-molecule FRET-derived model of the synaptotagmin 1-SNARE fusion complex. Nat Struct Mol Biol. 2010;17:318–324. doi: 10.1038/nsmb.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai H, Shen N, Araç D, Rizo J. A quaternary SNARE-synaptotagmin-Ca2+-phospholipid complex in neurotransmitter release. J Mol Biol. 2007;367:848–63. doi: 10.1016/j.jmb.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brewer KD, et al. Dynamic binding mode of a Synaptotagmin-1–SNARE complex in solution. Nat Struct Mol Biol. 2015;22:555–564. doi: 10.1038/nsmb.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutton RB, Davletov Ba, Berghuis aM, Südhof TC, Sprang SR. Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez I, et al. Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–1069. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 33.Fuson KL, Montes M, Robert JJ, Sutton RB. Structure of human synaptotagmin 1 C2AB in the absence of Ca2+ reveals a novel domain association. Biochemistry. 2007;46:13041–8. doi: 10.1021/bi701651k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redecke L, et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science. 2013;339:227–30. doi: 10.1126/science.1229663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaffaney JD, Dunning FM, Wang Z, Hui E, Chapman ER. Synaptotagmin C2B domain regulates Ca2+-triggered fusion in vitro: critical residues revealed by scanning alanine mutagenesis. J Biol Chem. 2008;283:31763–75. doi: 10.1074/jbc.M803355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue M, Ma C, Craig TK, Rosenmund C, Rizo J. The Janus-faced nature of the C(2)B domain is fundamental for synaptotagmin-1 function. Nat Struct Mol Biol. 2008;15:1160–8. doi: 10.1038/nsmb.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diao J, et al. Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. Elife. 2012;1:e00109. doi: 10.7554/eLife.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai Y, et al. Complexin inhibits spontaneous release and synchronizes Ca 2+-triggered synaptic vesicle fusion by distinct mechanisms. Elife. 2014;3:1–14. doi: 10.7554/eLife.03756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suh BC, Hille B. Electrostatic interaction of internal Mg2+ with membrane PIP2 Seen with KCNQ K+ channels. J Gen Physiol. 2007;130:241–56. doi: 10.1085/jgp.200709821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park Y, et al. Controlling synaptotagmin activity by electrostatic screening. Nat Struct Mol Biol. 2012;19:991–7. doi: 10.1038/nsmb.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J, Pang ZP, Shin OH, Südhof TC. Synaptotagmin-1 functions as a Ca2+ sensor for spontaneous release. Nat Neurosci. 2009;12:759–66. doi: 10.1038/nn.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnakumar SS, et al. Conformational dynamics of calcium-triggered activation of fusion by synaptotagmin. Biophys J. 2013;105:2507–2516. doi: 10.1016/j.bpj.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, et al. Calcium sensitive ring-like oligomers formed by synaptotagmin. Proc Natl Acad Sci U S A. 2014;111:13966–71. doi: 10.1073/pnas.1415849111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masumoto T, et al. Ca2+-independent syntaxin binding to the C2B effector region of synaptotagmin. Mol Cell Neurosci. 2012;49:1–8. doi: 10.1016/j.mcn.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 45.Mackler J, Reist N. Mutations in the second C2 domain of synaptotagmin disrupt synaptic transmission at Drosophila neuromuscular junctions. J Comp Neurol. 2001;436:4–16. [PubMed] [Google Scholar]
- 46.Gao Y, et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–3. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou P, Bacaj T, Yang X, Pang ZP, Südhof TC. Lipid-Anchored SNAREs Lacking Transmembrane Regions Fully Support Membrane Fusion during Neurotransmitter Release. Neuron. 2013:1–14. doi: 10.1016/j.neuron.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aeffner S, Reusch T, Weinhausen B, Salditt T. Energetics of stalk intermediates in membrane fusion are controlled by lipid composition. Proc Natl Acad Sci U S A. 2012;109:E1609–18. doi: 10.1073/pnas.1119442109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, et al. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 50.Tang J, et al. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–87. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 51.Van Leeuwen HC, Strating MJ, Rensen M, De Laat W, Van Der Vliet PC. Linker length and composition influence the flexibility of Oct-1 DNA binding. EMBO J. 1997;16:2043–2053. doi: 10.1093/emboj/16.8.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ismail Sa, Vetter IR, Sot B, Wittinghofer A. The structure of an Arf-arfgap complex reveals a Ca2+ regulatory mechanism. Cell. 2010;141:812–821. doi: 10.1016/j.cell.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 53.Ernst Ja, Brunger AT. High resolution structure, stability, and synaptotagmin binding of a truncated neuronal SNARE complex. J Biol Chem. 2003;278:8630–8636. doi: 10.1074/jbc.M211889200. [DOI] [PubMed] [Google Scholar]
- 54.Cipriano DJ, et al. Processive ATP-driven disassembly of SNARE complexes by the N-ethylmaleimide sensitive factor molecular machine. J Biol Chem. 2013 doi: 10.1074/jbc.M113.476705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 56.Bonifacio R, Pellegrini C, Narducci LM. Collective instabilities and high-gain regime in a free electron laser. Opt Commun. 1984;50:373–378. [Google Scholar]
- 57.Kondratenko aM, Saldin EL. Generation of coherent radiation by a relativistic electron beam in an ondulator*. Part Accel. 1980;10:207–216. [Google Scholar]
- 58.Neutze R, Wouts R, van der Spoel D, Weckert E, Hajdu J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature. 2000;406:752–7. doi: 10.1038/35021099. [DOI] [PubMed] [Google Scholar]
- 59.Solem JC. Imaging biological specimens with high-intensity soft x rays. J Opt Soc Am B. 1986;3:1551. [Google Scholar]
- 60.Cohen AE, et al. Goniometer-based femtosecond crystallography with X-ray free electron lasers. Proc Natl Acad Sci. 2014;111:17122–17127. doi: 10.1073/pnas.1418733111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeldin OB, et al. Data Exploration Toolkit for serial diffraction experiments. Acta Crystallogr Sect D Biol Crystallogr. 2015;71:352–356. doi: 10.1107/S1399004714025875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hattne J, et al. Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers. Nat Methods. 2014;11:545–8. doi: 10.1038/nmeth.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sauter NK, Hattne J, Grosse-Kunstleve RW, Echols N. New Python-based methods for data processing. Acta Crystallogr D Biol Crystallogr. 2013;69:1274–82. doi: 10.1107/S0907444913000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sauter NK, et al. Improved crystal orientation and physical properties from single-shot XFEL stills. Acta Crystallogr D Biol Crystallogr. 2014;70:3299–309. doi: 10.1107/S1399004714024134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uervirojnangkoorn M, et al. Enabling X-ray free electron laser crystallography for challenging biological systems from a limited number of crystals. Elife. 2015;4:e05421. doi: 10.7554/eLife.05421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kabsch W. XDS. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr Sect D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 70.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 71.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 72.Adams PD, et al. PHENIX_: building new software for automated crystallographic structure determination. Acta Crystallogr Sect D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 73.Schröder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Urzhumtseva L, Afonine PV, Adams PD, Urzhumtsev A. Crystallographic model quality at a glance. Acta Crystallogr Sect D Biol Crystallogr. 2009;65:297–300. doi: 10.1107/S0907444908044296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pettersen EF, et al. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 77.Krissinel E, Henrick K. Inference of Macromolecular Assemblies from Crystalline State. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 78.Young SM, Neher E. Synaptotagmin Has an Essential Function in Synaptic Vesicle Positioning for Synchronous Release in Addition to Its Role as a Calcium Sensor. Neuron. 2009;63:482–496. doi: 10.1016/j.neuron.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 79.De Wit H, et al. Synaptotagmin-1 docks secretory vesicles to syntaxin-1/SNAP-25 acceptor complexes. Cell. 2009;138:935–46. doi: 10.1016/j.cell.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 80.Mohrmann R, et al. Synaptotagmin interaction with SNAP-25 governs vesicle docking, priming, and fusion triggering. J Neurosci. 2013;33:14417–30. doi: 10.1523/JNEUROSCI.1236-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kyoung M, et al. In vitro system capable of differentiating fast Ca2+-triggered content mixing from lipid exchange for mechanistic studies of neurotransmitter release. Proc Natl Acad Sci U S A. 2011;108:E304–13. doi: 10.1073/pnas.1107900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kyoung M, Zhang Y, Diao J, Chu S, Brunger AT. Studying calcium-triggered vesicle fusion in a single vesicle-vesicle content and lipid-mixing system. Nat Protoc. 2013;8:1–16. doi: 10.1038/nprot.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee HK, et al. Dynamic Ca2+-dependent stimulation of vesicle fusion by membrane-anchored synaptotagmin 1. Science. 2010;328:760–3. doi: 10.1126/science.1187722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lai Y, Lou X, Jho Y, Yoon TY, Shin YK. The synaptotagmin 1 linker may function as an electrostatic zipper that opens for docking but closes for fusion pore opening. Biochem J. 2013;456:25–33. doi: 10.1042/BJ20130949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Z, Liu H, Gu Y, Chapman ER. Reconstituted synaptotagmin I mediates vesicle docking, priming, and fusion. J Cell Biol. 2011;195:1159–70. doi: 10.1083/jcb.201104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zick M, Wickner WT. A distinct tethering step is vital for vacuole membrane fusion. Elife. 2014;3:e03251. doi: 10.7554/eLife.03251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maximov A, Pang ZP, Tervo DGR, Südhof TC. Monitoring synaptic transmission in primary neuronal cultures using local extracellular stimulation. J Neurosci Methods. 2007;161:75–87. doi: 10.1016/j.jneumeth.2006.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.