Abstract
The cuticle plays important roles in plant development, growth and defense against biotic and abiotic attacks. Crystallized epicuticular wax, the outermost layer of cuticle, is visible as white-bluish glaucousness. In crops like barley and wheat, glaucousness is trait of adaption to the dry and hot cultivation conditions, and hentriacontane-14,16-dione (β-diketone) and its hydroxy derivatives are the major and unique components of cuticular wax in the upper parts of adult plants. But their biosynthetic pathway and physiological role largely remain unknown. In the present research, we identified a novel wax mutant in wheat cultivar Bobwhite. The mutation is not allelic to the known wax production gene loci W1 and W2, and designated as W3 accordingly. Genetic analysis localized W3 on chromosome arm 2BS. The w3 mutation reduced 99% of β-diketones, which account for 63.3% of the total wax load of the wild-type. W3 is necessary for β-diketone synthesis, but has a different effect on β-diketone hydroxylation because the hydroxy-β-diketones to β-diketone ratio increased 11-fold in the w3 mutant. Loss of β-diketones caused failure to form glaucousness and significant increase of cuticle permeability in terms of water loss and chlorophyll efflux in the w3 mutant. Transcription of 23 cuticle genes from five functional groups was altered in the w3 mutant, 19 down-regulated and four up-regulated, suggesting a possibility that W3 encodes a transcription regulator coordinating expression of cuticle genes. Biosynthesis of β-diketones in wheat and their implications in glaucousness formation and drought and heat tolerance were discussed.
Key Message
W3 is essential for β-diketone biosynthesis but suppresses its hydroxylation. Loss-of-function mutation w3 significantly increased cuticle permeability in terms of water loss and chlorophyll efflux.
Introduction
During transition from water to land colonization, plants developed an array of mechanisms for adaptation to the desiccation environment. One of these mechanisms is deposition of cuticle, a hydrophobic coat, to cover the aerial organ surfaces. In addition to function in protecting water loss, cuticle, as the interface between the sessile plants and their environment, plays important roles in plant defense against heat, UV radiation, pathogen and insect attacks [1]. Cuticle consists of the fundamental framework of cutin and intracuticular wax inserted in it and epicuticular wax overlaid on them. Cutin is the cell wall-bounded ester polymer of hydroxy fatty acids [2–4], and waxes are the very long chain fatty acids (VLCFAs) and their derivatives including alcohols, aldehydes, alkanes, ketones and wax esters [5]. Variation in epicuticular wax composition causes changes in plant appearance: glaucous or nonglaucous. Glaucousness, the bluish-white look, is the visible form of densely arrayed wax crystals.
In wax biosynthesis, de novo C16 and C18 fatty acids, the precursors, are first elongated into VLCFAs (C20 to C34) by fatty acyl elongase (FAE) complex. The VLCFAs can be converted to primary alcohols and further esterified with the C16 fatty acids into wax esters via the acyl reduction pathway [6–8] and converted to alkanes, aldehydes, secondary alcohols and ketones through decarbonylation pathway [9–11]. All the wax species are synthesized in epidermal cells, transported extracellular and deposited to cuticle. Wax synthesis and export are under tight developmental and environmental regulations. Molecular identification of the genes coding for the wax biosynthetic enzymes, wax transporters and wax regulators in the model plant Arabidopsis provided genetic and molecular supports to the VLCFA and associated pathways (reviewed in [12–14].
The VLCFA and associated pathways are conserved in higher plants, and many of their components of these pathways have also been identified in the grass genomes. Maize wax genes GLOSSY1 (GL1) [15, 16], GL2 [17], GL4 [18] and GL8 [19, 20] are homologous to ECERIFERUM3 (CER3), CER2, CER6 and β-keto acyl-CoA reductase 1 (KCR1) of Arabidopsis, respectively. A T-DNA insertion mutant in a CER6 homolog reduced leaf wax crystal density in rice [21]. Maize AP2 transcription factor GL15 involves in the transition from juvenile to adult leaf identity including wax composition [22], and HD-ZIP IV family transcription factor OCL1 regulates cuticular wax biosynthesis by activation of a fatty acyl-CoA reductase (FAR) and a putative wax transporter [23, 24].
In addition to the VLCFA and associated pathways, a diverse of plants, including barley and wheat (Triticum L.), employ another parallel wax biosynthetic pathway for biosynthesis of hentriacontane-14,16-dione (also known as β-diketone) and its derivatives. In wheat, the VLCFA pathways are active in the vegetative stage, but the β-diketone pathway predominates in the reproductive stage [25]. It is believed that β-diketone is synthesized using C14 and C16 fatty acids as precursors [26]. Wax profiling of barley mutants suggests that cer-q, cer-c and cer-u mutants respectively define the initial condensing reaction, subsequent chain extensions and hydroxylation [27], but little is known about the genetic components of this pathway. In common wheat (T. aestivum L., genomes AABBDD), two wax inhibitors and two wax production genes underlie the glaucousness variations: Iw1 on chromosome arm 2BS [28–31], Iw2 on 2DS [30–33], W1 on 2BS [30, 34] and W2 on 2DS [30]. In durum wheat (T. turgidum subsp. durum, genomes AABB), a third wax inhibitor Iw3 is located in the distal region of chromosome arm 1BS [35, 36]. We previously characterized a set of wheat near isogenic lines (NILs) differing at W1, W2, Iw1 and Iw2 loci and durum NILs differing at the Iw3 locus, and found that loss of both functional alleles of the W genes or presence of either Iw gene causes depletion of β-diketones and nonglaucous phenotype. While elimination of β-diketones is compensated by increase of aldehydes and primary alcohols in the Iw1 and Iw2 NILs [37], Iw3 inhibits β-diketone, reduces primary alcohols but increase aldehyde and alkanes in the glume wax [36]. Although expression of a FAR1 homolog was maintained at high level in the Iw NILs, no match was found between expression of 72 wax genes and β-diketone distribution pattern [36, 37]. All this suggests that β-diketones are synthesized through a not-yet-identified pathway.
We recently identified a nonglaucous (NG) mutant in Bobwhite (BW) wheat. Compared to the wild type BW, the mutant lost 63% of total wax load and almost all of the β-diketones, and showed a significant increase of water loss. Genetic and molecular analyses indicated that the mutant gene is not allelic to either W1 or W2 and expression of numerous cuticle genes of five different pathways was significantly down regulated. Here we report the results and their implications in Triticeae cuticle biosynthesis and drought tolerance.
Results
Genetic Analysis of the Wax Mutant
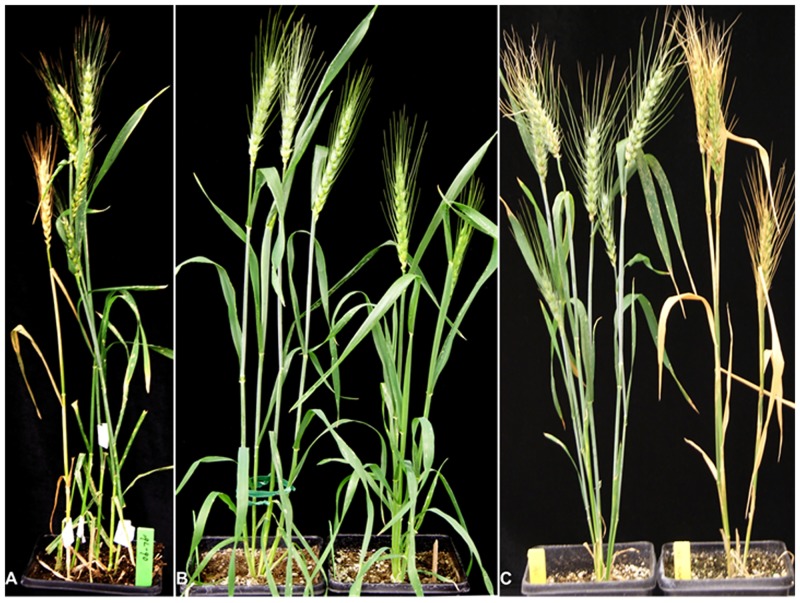
During intercrossing BW transgenic plants to combine the different RNAi transgenes, a NG plant was found in a small T1 population of the transgenic line #056 in the spring of 2010. It dried out prematurely (Fig 1A). We recovered two NG plants, NG1 and NG2, from the T2 populations derived from the glaucous T1 plants of the same pot. These NG plants had much lower level of glaucousness in leaves, sheaths, peduncle, and spikes as compared to the wild type BW (Fig 1B). The mutant plants died prematurely again in the late spring of 2011 although they were well watered (Fig 1C) probably due to the high temperature.
Fig 1. Phenotype of the nonglaucous mutant.
(A) The mutant was found in a small F2 population derived from a cross between two RNAi transgenic plants in BW background due to premature drying out. (B) Adult plants of BW (left) and the nonglaucous mutant (right) at anthesis. (C) Adult plants of BW (left) and the nonglaucous mutant (right) at grain-filling stage.
We started genetic analysis of this presumable NG mutant by crossing it with the wild type BW. The F1 plants were glaucous but their glaucousness level was intermediate between BW and the NG mutant (Fig 2A). A population of 361 F2 individuals segregated into 265 glaucous and 96 nonglaucous, which fits the 3 (glaucous) to 1 (nonglaucous) ratio (P = 0.38812; Table 1), indicating that a single-gene mutation underlies the nonglaucous phenotype. To test if the mutation was due to transgene insertion, we screened 38 nonglaucous F2 individuals derived from the cross between NG1 and BW for the transformation selection marker bar gene and RNAi transgene constructs [38] by targeting at the Ubq-bar and gus-nos junctions, respectively. Result showed that eight plants were negative for both the Ubq-bar and gus-nos junctions, and 7 additional plants were negative for the gus-nos junction. Furthermore, NG2 contained neither Ubq-bar nor gus-nos (S1 Fig). These data indicate that one bar locus was not linked with the RNAi locus in NG1 and that the nonglaucous mutation is independent of the transgene insertions.
Fig 2. Genetic analysis of w3 mutant.
(A) The F1 hybrid between BW and the nonglaucous mutant is intermediate between its parents in glaucousness intensity. (B) The F1 hybrid between the mutant and w1w2 double recessive line was glaucous. The scale bars indicate 1 cm.
Table 1. Phenotypes and segregations of glaucousness in hybrids and their progenies.
| Crosses | # of F1 | F1 Plant | F2 segregation | P values | P values | P values | P values |
|---|---|---|---|---|---|---|---|
| plants | phenotype | (gl : ng)* | (3 : 1) | (15 : 1) | (9 : 7) | (1 : 1) | |
| w3 x BW | 5 | glaucous | 106:38 | 0.70032 | 1.8 x 10−23 | 2.7 x 10−5 | 1.3 x 10−10 |
| w3 x CS-TDIC 2B | 5 | glaucous | 265:96 | 0.38812 | 5.9 x 10−56 | 7.9 x 10−11 | 15 x 10−18 |
| w1w2 x w3 | 5 | glaucous | 141:129 | 5.4 x 10−18 | 7.6 x 10−175 | 0.18217 | 0.46477 |
| w1w2 x BW | 5 | glaucous | 133:11 | 1.5 x 10−6 | 0.49112 | 2.4 x 10−24 | 1.2 x 10−81 |
| w3 x CS 2BS-1 | 6 | nonglaucous | 0:71 | 3.0 x 10−48 | 1.3 x 10−233 | 1.2 x 10−21 | 3.6 x 10−17 |
| w3 x CS 2BS-2 | 6 | nonglaucous | 0:71 | 3.0 x 10−48 | 1.3 x 10−233 | 1.2 x 10−21 | 3.6 x 10−17 |
| w3 x CS 2BS-3 | 6 | nonglaucous | 0:72 | 6.7 x 10−49 | 7.3 x 10−237 | 6.5 x 10−22 | 2.6 x 10−17 |
| w3 x CS 2BS-5 | 6 | nonglaucous | 0:68 | 2.8 x 10−46 | 8.1x 10−224 | 8.7 x 10−21 | 1.6 x 10−16 |
| w3 x CS 2BS-10 | 6 | nonglaucous | 0:72 | 6.7 x 10−49 | 7.3 x 10−237 | 6.5 x 10−22 | 2.6 x 10−17 |
| w3 x CS 2BS-14 | 6 | nonglaucous | 0:35 | 2.3 x 10−34 | 3.5 x 10−116 | 2.0 x 10−11 | 3.3 x 10−9 |
*gl, glaucous; ng, nonglaucous.
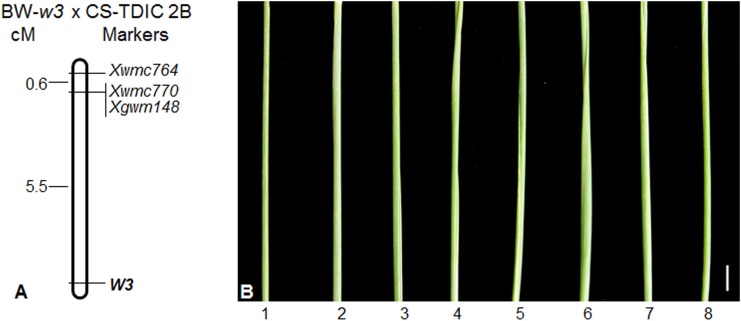
To test if the nonglaucous mutation is allelic to the known wax production genes in wheat, we crossed S-615 w1w2 near isogenic line (NIL), which is nonglaucous due to loss-of-function at the functionally redundant W1 and W2 loci [37], with BW and the NG2 mutant line. The F2 population derived from the cross between S615-w1w2 and BW segregated into 133 glaucous and 11 nonglaucous plants, well-fitting the 15 to 1 ratio (P = 0.49112; Table 1) and indicating that BW carries both W1 and W2 functional alleles. The F1 hybrid plants between S615-w1w2 and NG2 carry glaucousness at low intensity (Fig 2B), indicating that the nonglaucous mutation in BW is not allelic to either W1 or W2, but its wild type allele in S615-w1w2 complements to the functional alleles of the W1 and W2 loci carried by BW. According to the recommended rules for gene symbolization in wheat [39], we designated this new wax gene locus as W3 and the mutant as w3. A population of 270 F2 plants derived from the cross between the w3 mutant and the w1w2 NIL segregated into 141 glaucous and 129 nonglaucous, which fits a 1 to 1 (P = 0.46477) or 9 to 7 ratio (P = 0.18217; Table 1) and is more similar to the two-locus segregation ratios than a three-locus segregation. These results corroborate that W3 is not homologous to the known wax gene loci W1 and W2 and suggested that W3 is most probably linked with the W1 or W2 locus. To identify the chromosomal location of the W3 locus, we developed a mapping population from a cross between the w3 mutant and Chinese Spring (CS)-T. turgidum subsp. dicoccoides 2B substitution line (CS-TDIC 2B) and genotyped the 82 NG F2 individuals (168 gametes) using 2BS and 2DS SSR markers for detection of marker-W3 linkage. The result showed that W3 is linked with the 2BS SSR markers and 5.5 cM proximal to the marker loci Xwmc770 and Xgwm148 (Fig 3A).
Fig 3. Chromosomal localization of the W3 locus.
(A) Molecular mapping of the W3 locus on the chromosome arm 2BS. The markers are listed at the right side of the map, and genetic distances (cM) between the marker loci are indicated at the left side of the map. The W3 locus is indicated in bold. The top of the map is towards the telomere and the bottom is towards the centromere. (B) The peduncles of CS N2B-T2D (1), F1 hybrids of NG2 with N2B-T2D (2), with deletion line 2BS-1 (3), with 2BS-2 (4), with 2BS-3 (5), with 2BS-5 (6), with 2BS-10 (7) and with 2BS-14 (8). All the flag-leaf sheaths are nonglaucous. The scale bar indicate 1 cm.
We also crossed the w3 mutant with CS 2BS deletion lines (2BS-1, -2, -3, -5, -10 and -14). All these deletion lines and nullisomic 2B-tetrasomic 2D (N2B-T2D), in which lack of one pair of chromosome 2B is compensated by two pairs of chromosome 2D, are nonglaucous due to missing the W1 locus. The F1 hybrid plants between the w3 mutant and the 2BS deletion lines were nonglaucous similar to N2B-T2D, or carried nearly undetectable glaucousness (Fig 3B). No glaucous individuals were observed in their F2 populations (Table 1). This result corroborates that the W3 locus is located in the distal region of chromosome arm 2BS.
Furthermore, we genotyped BW and the w3 mutant using SSR markers located in the distal regions of the 42 wheat chromosome arms on wheat genetic maps. All 42 SSR markers were positive and detected monomorphism (S1 Table). This indicated BW and the w3 mutant shares the same genetic background.
Wax Morphology
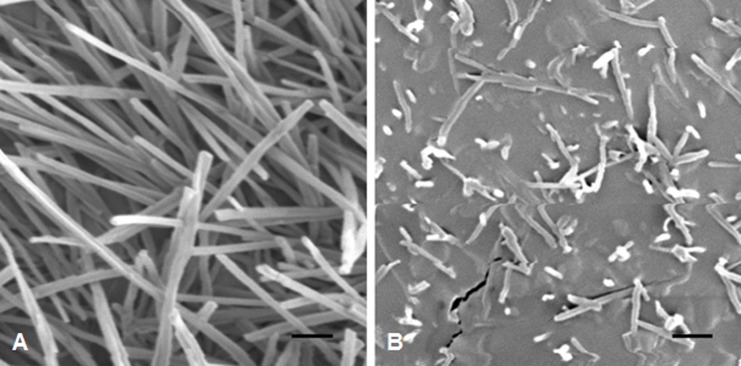
At stage F10.5.1, wheat plants are flowering, and glaucousness is fully developed. We inspected the cuticle surfaces of flag leaf sheaths of BW and w3 mutant under a scanning electron microscope (SEM) at this developmental stage. The electron micrographs showed clear differences between BW and w3 mutant (Fig 4). In BW, cuticle surface was covered with thick meshwork of long wax crystal tubes (Fig 4A). In contrast, short wax bodies were appressed to the leaf cuticle surface of w3 mutant at low density (Fig 4B). This result indicated that the W3 locus has a major effect on cuticle, epicuticular wax in particular, development. Considering that the great contrast in wax crystal morphology and density between BW and w3 mutant was found in the flag leaf sheath, we used this tissue for measuring cuticle permeability, profiling composition of the cuticular waxes and quantifying transcription of cuticle genes.
Fig 4. SEM micrographs of cuticle surfaces of flag leaf sheaths.
(A) BW and (B) w3 mutant. The scale bars indicated 1 μm.
Cuticle Permeability
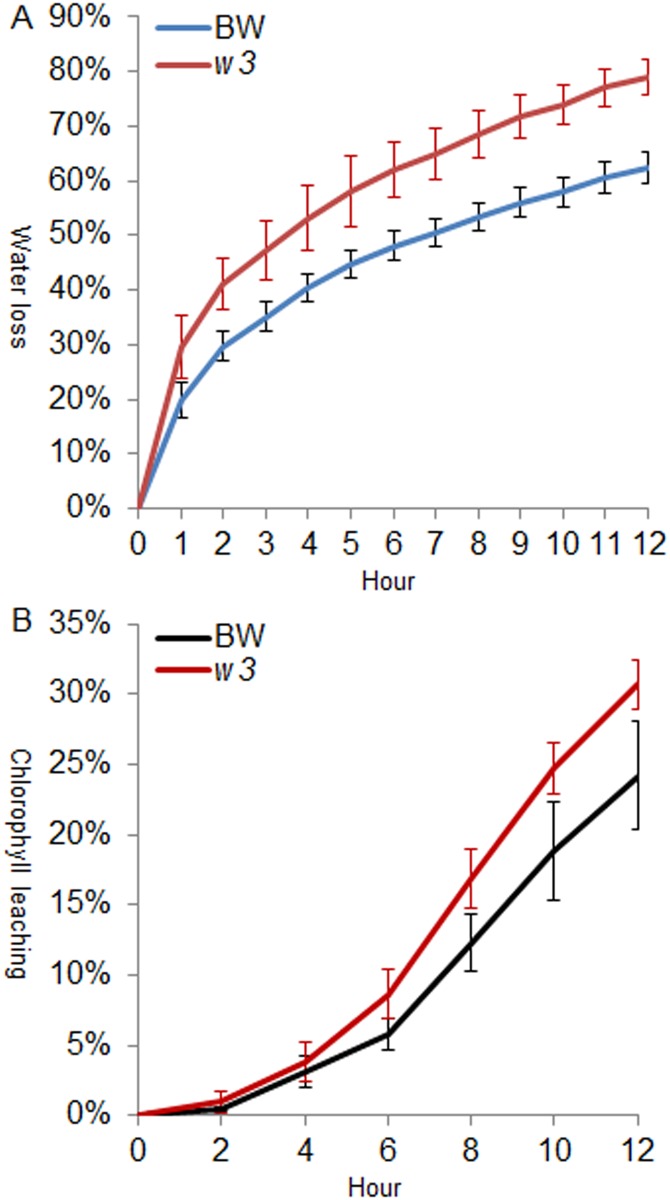
We measured the cuticle permeability in two experiments using the flag-leaf sheath: water loss in air and chlorophyll leaching in 80% ethanol. In the first experiment, significantly higher water loss rate was observed in the w3 mutant 1 h after detachment (P < 0.03303) and incremented throughout the time course (Fig 5A). We inspected stomata, and no significant difference was found in stomata density between BW and w3 mutant, ~60 stomata in one view field of 10x20 magnification under microscope. The stomata were closed within one hour after detachment. This suggests that the difference in water loss rate is attributed to cuticle permeability. In the second experiment, w3 mutant showed significantly higher chlorophyll efflux rate from the sixth to eighth hour of treatment (P < 0.02491; Fig 5B). Both lines were similar in total chlorophyll content after 48-hour extraction (P = 0.80435). This corroborates the conclusion from the water loss experiment that change of wax crystal morphology and density led to increase of cuticle permeability in w3 mutant.
Fig 5. Analysis of cuticle permeability of BW and w3 mutant.
Cuticle permeability was evaluated by air drying at room temperature (A) and by chlorophyll leaching in 80% ethanol (B). The numbers on the x-axes represent hours of treatment. Water loss or chlorophyll leaching at each time point is represented on the y-axes as percentages of the total water content or total chlorophyll content in the tissue. Measurements taken from six individuals were averaged.
Difference in water-loss rate was also found in spikes in relation to glaucousness. Six hours after detachment, top awns of the w3 mutant started to dry (Fig 6B). Twelve hours after detachment, spike of w3 mutant completely discolored and dried out; awns of w1w2 double recessive line were twisted and some glumes discolored, but BW and F1 between w3 mutant and w1w2 stayed in normal shape (Fig 6C). This indicates that W3 played an important role in preventing non-stomatal transpiration.
Fig 6. Spikes responses to dehydration.
Spike of BW (W1W1W2W2W3W3), w3 mutant (W1W1W2W2w3w3), w1w2 double recessive line (w1w1w2w2W3W3), and the F1 hybrid between w3 and w1w2 (W1w1W2w2W3w3) at 0 (A), 6 (B) and 12 h of dehydration (C). The designations are indicated on the top. The scale bars indicate 1 cm.
Wax Composition
We extracted wax from the flag-leaf sheath at stage F10.5.1 and comparatively profiled the wax composition of BW and the w3 mutant by GC-MS. Ninety-two wax molecules were identified, two thirds of which were trace and could not find a match in the National Institute of Standards and Technology database. These minors and unknowns account for ~10% total wax extract in BW, which had similar abundance in w3 mutant.
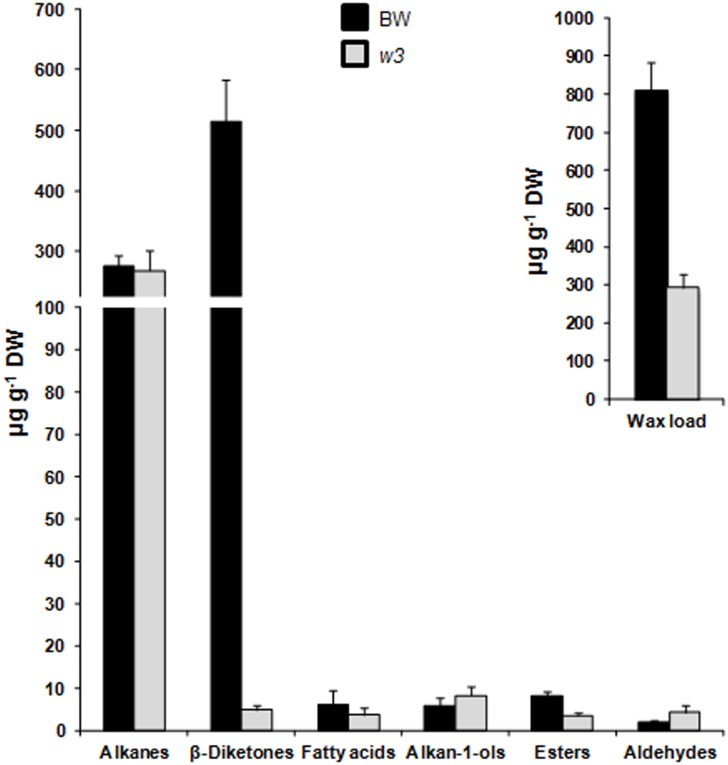
We analyzed the distribution patterns of major wax species in BW and w3 mutant. In BW, two major wax components, diketones and alkanes account for 63.3% and 34.0% of the total wax load, respectively. Remaining 2.7% are wax esters (1.0%), fatty acids (0.8%), primary alcohols (0.7%) and aldehydes (0.2%). Dramatic differences were found in total wax load and wax composition between BW and w3 mutant (Fig 7). Compared to BW, w3 mutant lost 64% of the total wax (P = 5 x 10−7; Fig 7). While alkanes were maintained unchanged (P = 0.68113; Fig 7), β-diketones reduced to 1% in w3 mutant (P = 1 x 10−8; Fig 7). As a result, alkanes account for over 90% of the total wax load of w3 mutant (Fig 7). Two types of diketones, β-diketone and hydroxy-β-diketones, were detected. Hydroxy-β-diketones account for 8.5% of the total β-diketones in BW. As a result, BW had a hydroxy-β-diketones to β-diketone ratio (OH-D/β-D) of 0.0925. In w3 mutant, β-diketone and hydroxy-β-diketones did not reduce proportionally: 181-fold reduction in β-diketone (P = 1 x 10−7), but 16-fold reduction in hydroxy-β-diketones (P = 0.00002; Fig 8). Two hydroxy-β-diketone isomers, 8- and 9-hydroxy hentriatane-14,16-dione, were detected. The 8-isomer reduced ~14-fold (P = 2 x 10−5) and the 9-isomer reduced ~22-fold (P = 0.00001) in w3 mutant. Therefore, the hydroxy-β-diketones content was roughly equal to that of β-diketone in w3 mutant with an OH-D/β-D of 1.014 (Fig 8), indicating a ~11-fold increase as compared to BW (P = 0.00695). This suggests a differential effect of the w3 mutation on biosynthesis of β-diketone and its hydroxylation.
Fig 7. Wax composition of BW and w3 mutant.
Total wax load and content of alkanes, β-diketones, primary alcohols (alkan-1-ols), aldehydes, wax esters, and fatty acids of the flag leaf sheaths were measured by GC-MS. The numbers on the y-axes indicate average content expressed as μg per g dried tissue (dry weight, DW). The error bars indicate standard deviation of the mean estimated from five biological replicates.
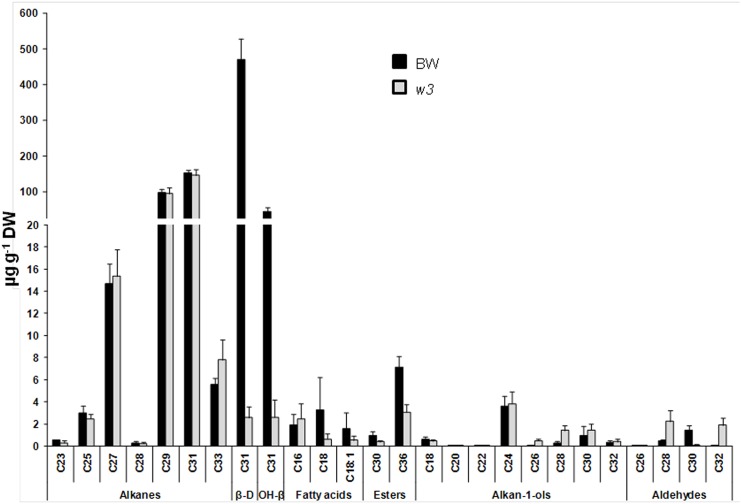
Fig 8. Variation of wax homologues between BW and w3 mutant.
Carbon atom numbers of alkanes, β-diketones, primary alcohols (alkan-1-ols), fatty acids, wax esters, and aldehydes are indicated on the x-axes. Their contents are indicated on y-axes as μg per g dried tissue (dry weight, DW). The error bars indicate standard deviation of the mean calculated from five biological replicates. β-D, β-diketone; and OH-β, hydroxy-β-diketones.
In addition to β-diketones, wax ester content, especially octadecanoic acid octadecyl ester (C36), was significantly decreased in w3 mutant (P = 0.00002; Figs 7 and 8). Contrary to diketones and wax esters, total aldehyde was significantly increased in w3 mutant (P = 0.01026; Fig 7). With regard to specific carbon length homologues of aldehydes, significant increase of C28 (P = 0.00309) and C32 (P = 0.00008) homologues and significant reduction of C26 (P = 0.00485) and C30 (P = 0.00008) homologues was observed in w3 mutant (Fig 8). Although no significant difference was found between BW and w3 mutant in the total primary alcohol and alkane content (Fig 7), changes were detected in some homologues of these wax species: C20 (P = 0.01215) alcohol was reduced in w3 mutant, but C22, C26 and C28 alcohols (P < 0.00036) and C33 alkane (P = 0.02579) were significantly increased in w3 mutant (Fig 8).
Transcription of Cuticle Genes
To gain insights into the W3-dependent genetic pathways, we profiled expression of 72 wheat wax candidate genes belonging to six categories [37] in the flag-leaf sheath of BW and w3-BW by qPCR. Of these 72 wax genes, five are responsible for cutin biosynthesis, 17 for fatty acyl elongation, 17 for VLCFA reduction and wax esterification, 21 for VLCFA decarbonylation, seven for wax transport and five for wax regulation (S2 Table). Result showed that expression of 19 genes was significantly down regulated and four up regulated (Fig 9). Of these 19 down-regulated genes, nine genes were down regulated more than twofold. These include five CER1 members, FAR5, KCR2, KCS-3 and LTP (Fig 9). Of the five down-regulated CER1 genes, CER1-1 showed greatest fold change, i.e. 3.4-fold down-regulation. Expression of four CER3 genes reduced only ~1.5-fold though they may also be involved in decarbonylation via interaction with CER1 gene products. By contrast, expression of CER4-3, CER4-12, MAH1-5 and WSD1 increased ~1.5-fold (Fig 9).
Fig 9. Transcriptional changes of wax genes in w3 mutant against BW.
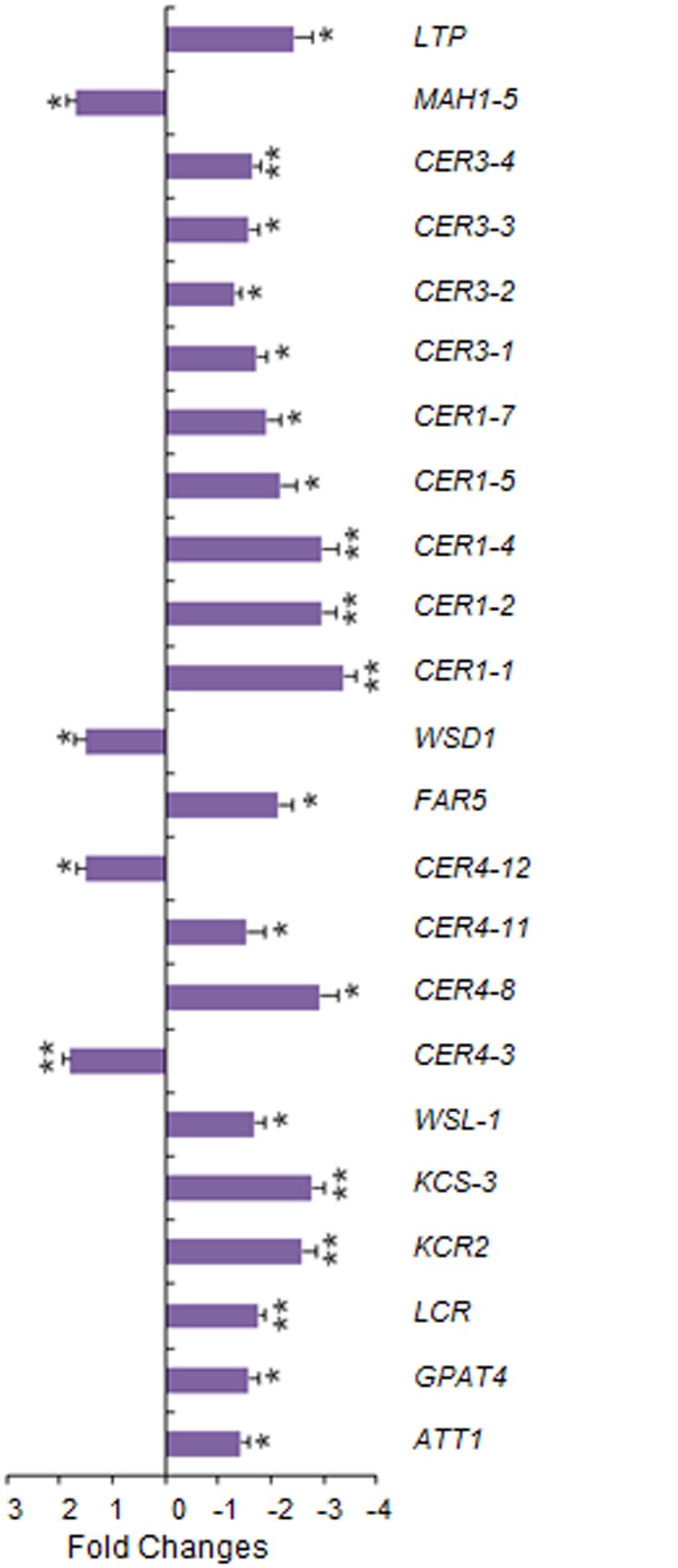
Genes with significant fold changes are depicted. The error bars represent standard deviation of the mean fold-change of mRNA levels calculated from four biological replicates. Asterisks indicate that the difference is significant at P< 0.05 (*) or at P < 0.01 (**). Expression data for all genes analyzed are listed in S2 Table.
Discussion
Glaucousness is one of the most eye-catching traits and has been used as a morphological marker in wheat genetic studies for ~80 years (reviewed in [30]). Its adaptive value in improving crop tolerance to drought and heat was recognized in 1980s [40, 41], and the climate change resumed interest in this trait more recently [36, 37, 42, 43]. Nevertheless, basic research of glaucousness in wheat lagged behind compared to other cereal crops, such as barley, a distant relative of wheat. One of the major reasons for this is lack of nonglaucous mutants in polyploid wheat. In this study, we identified a new β-diketone-deficit wax mutation w3 and investigated the function of β-diketone as a major wax component in drought tolerance. Molecular characterization of w3 mutant also shed light on diketone biosynthesis.
Wax Loci
In barley, a diploid species, 79 wax loci have been identified by 1,580 induced eceriferum mutants [44], of which 53 mutant loci were localized to the all 7 chromosomes [45]. Chromosome 2H, which is homoeologous to wheat chromosomes 2A, 2B and 2D, carries 11 wax loci, of which cer-c, cer-q, cer-u, cer-v and cer-zt are located on the short arm and cer-g, cer-n, cer-yb and gsh5 (cer-s) on the long arm. In contrast, only four wax loci, W1, W2, Iw1 and Iw2 have been identified in hexaploid wheat with W1 and Iw1 located on 2BS and W2 and Iw2 on 2DS [30]. In polyploid wheat, mutation rate are 5- to 10-fold higher than diploid species due to genetic redundancy [46, 47], but the mutant phenotypes are often masked by the wild type alleles of the homoeologous loci. The W1 and W2 genes are functionally redundant in forming glaucousness, but they differ in chromosomal locations [30] and expression profiles [37], suggesting a possibility that they are paralogous and their orthologs in the homoeologous chromosomes are silenced, deleted, neo- or sub-functionalized, which is an important feature of polyploid evolution [48]. In the present research, we showed W3 is located on chromosome arm 2BS and complements with W1 and W2 for β-diketone production. Different from W1 and W2, homoeologs of W3 in the A and D genomes of polyploid wheat lost their function in glaucousness formation. This makes W3 more tractable for genetic manipulation and molecular study of wheat wax.
In addition to BW, a single functional W3 locus is present in CS-TDIC 2B (Table 1), probably also in CS. But it was previously not detected in the CS aneuploids, including nullisomics, nulli-tetrasomics (NT) and deletion lines. This failure was because W3 is linked with W1. Deletion of either one causes the nonglaucous phenotype and prevents identification of another wax locus supposing that CS does not carry the W2 allele. We examined the 42 NT lines and found that only N2B-T2A and N2B-T2D were nonglaucous. We also examined all eight homozygous deletion lines available for chromosome arm 2BS (2BS-1, -2, -3, -5, -6, -10,-12 and -14) and found that all of them were nonglaucous. These results confirmed that W1 is located in the distal region of the chromosome arm [30], which prevents to identify additional wax loci on same chromosome arm although several wax loci located on homoeologous chromosome arm 2HS of barley [45]. No segregation in the F2 populations derived from the crosses between the w3 mutant and 2BS deletion lines suggests that both W1 and W3 are located in the distal bin. The w3 mutants may already exist in other wheat cultivars besides BW, but they are assumed to be the w1 mutant. Therefore, allelism test is essential for identifying new wax gene loci in addition to marker linkage mapping. We are mapping the W1 and W3 loci, and the closely linked molecular markers will provide more detailed information on the organization of these wax loci on 2BS.
The w3 mutation is independent of transgene insertions (S1 Fig), suggesting that it could have been induced by tissue culture process during transformation. It is well known that in vitro tissue culture generates genomic stress, reprograms cell development and causes genetic and epigenetic changes [49]. Some of these changes inherit as morphological or physiological mutants. Spontaneous mutation of other origin, however, cannot be excluded because we recently identified several other morphological mutants in BW, which did not go through tissue culture.
W3 Effect on Biosynthesis of β-Diketones
Like the wax genes W1 and W2 [37], W3 is essential for biosynthesis of β-diketones, but had little effect on biosynthesis of alkanes. This supports the proposal that β-diketones are synthesized through a genetic pathway different from VLCFA synthesis and associated alkane-forming decarbonylation pathways although both β-diketones and alkanes are odd-carbon waxes [26]. Two biosynthetic pathways have been proposed for β-diketone biosynthesis: condensation of two C16 β-ketoesters via a “biological Claisen reaction” [50] and a modified elongation reaction on a selectively protected β-keto acid precursor [51, 52]. Both proposals lack molecular evidence although the latter was based on biochemical analysis of the substrate and inhibitor specificity [26], wax metabolite profiling and genetic analysis of the barley cer-c, cer-q and cer-u mutants [51]. In the both proposed models, a decarboxylative reaction was included. The cer-q and cer-c mutations impair the keto-protected chain elongation and lead to C13 and C15 alkan-2-ol formation [51, 52]. These secondary alcohols, products of β-keto fatty acid decarbonylation, were not detected either in w1w2 [37] or the w3 mutant in this research, suggesting that these wheat wax production genes act in upstream of the pathway or β-diketone is synthesized through the “biological Claisen reaction” in wheat [50]. qPCR assays showed that transcription of three FAE genes, five CER1 and four CER3 homologs was significantly down-regulated in w3 mutant (Fig 9; S2 Table). In Arabidopsis, CER1 and CER3 are involved in VLCFA decarbonylation [11]. Because alkane content remained at the same level in w3 mutant as in the wild type, these decarbonylation genes were not involved in alkane synthesis, but probably participated in the β-diketone-forming decarboxylative reaction. Considering that decarbonylation is the last step of β-diketone synthesis, transcription of these CER1 and CER3 homologs is possibly regulated either directly by W3 or by a substrate-mediated feed-forward loop.
The hydroxy β-diketone isomers are derived from β-diketone by hydroxylation [27], hence their changes in w3 mutant are expected to be proportional. Contrary to this expectation, w3 mutant had a much higher HO-D/β-D ratio compared to BW. This result suggests that hydroxylation reaction enzymes in the BW background are less sensitive to depletion of the substrate, i.e. β-diketone, or W3 promotes synthesis of β-diketone but suppresses its hydroxylation. In agreement with the latter hypothesis, expression of MAH1-7, a homolog of MAH1 that is responsible for midchain alkane hydroxylation in Arabidopsis [10], increased 1.5-fold in the w3 mutant (Fig 9; S2 Table). In another research, we found that interaction between wax production genes W1 and W2 is required for biosynthesis of hydroxy-β-diketones and that up-regulation of MAH1-8 paralleled with β-diketone hydroxylation in the W1W2 double dominant genotype [37]. It would be interesting to see if W3 interacts with W1 and W2 by combining their loss-of-function mutations in the same genetic background and measuring their effect on the hydroxy β-diketone isomers.
In addition to FAE, decarbonylation and hydroxylase genes, expression of three cutin biosynthetic genes, FAR5 and the putative wax transporter LTP was also down regulated in the w3 mutant (Fig 9; S2 Table). It is hard to imagine how a single-gene mutation affects expression of genes functioning in five cuticle pathways. In the model plant Arabidopsis, mutation in wax regulator SHN of the AP2/EREBP family altered property of both wax and cutin and expression of genes in multiple pathways [53–55]. We measured expression of five known wax regulator genes, but none of them changed significantly (S2 Table). Accordingly, one possibility would be that W3 encodes a wax regulator that orchestrates transcription of the biosynthetic and transporter genes and those acting at the upstream of β-diketone pathway. This may explain the mutant effect on other wax species, such as wax esters, C33 alkane, C22, C26 and C28 alcohol, and aldehydes (Fig 8). An alternative and less likely scenario would be that W3 targets on the initial step of diketone biosynthesis and the expression of these 20 genes are regulated by substrate-mediated feed-forward loops or secondary responses as discussed earlier. In either case, our results suggest a possibility that β-diketone pathway in Triticeae uses some CER1 and CER3 homologs for decarbonylation and MAH1 homologs for hydroxylation.
β-Diketones and Glaucousness
Cuticular waxes from the upper parts of adult Triticeae plants can be divided into two types according to their major constituents: primary alcohol-rich and β-diketone-rich wax. The diploid wheat T. monococcum and T. urartu belong to the former and polyploid wheat T. turgidum and T. timopheevii to the latter type [56, 57]. Analyses of NG mutants of barley [27] and wheat [28, 36, 37, 58, 59] indicated that β-diketones play an important role in glaucousness development. Wax profiling of the wheat wax gene NILs identified three types of waxes from the flag leaf sheaths: β-diketone-rich wax from the glaucous NILs carrying W1w2, w1W2 or W1W2, alkane-rich wax from the nonglaucous NIL w1w2, and alcohol-rich wax from the nonglaucous NILs carrying the wax inhibitor Iw1 or Iw2 [37]. In BW, β-diketone and hydroxy-β-diketones account for 63% of the total wax load. In the w3 mutant, β-diketones reduced ~100-fold. At the same time, wax crystal tubes were almost invisible in the flag-leaf sheath (Fig 4B). This confirms the role of β-diketones in glaucousness determination.
Next to β-diketones, alkanes, mainly C29 and C31 homologues, constitute the second major wax component, accounting for 34% of the total wax in BW. Although only significant net change was found in C33 alkane (Fig 8), proportion of the whole alkanes was increased to 90% due to depletion of the β-diketones. Significant differences were also observed between BW and w3 mutant in aldehydes, primary alcohols, and wax esters, but they had very low abundance, less than 3% of total wax in BW, suggesting that they are less likely to contribute to the mutant phenotype.
β-Diketones and Abiotic Stress
Early physiological studies in cereal crops showed that glaucousness increased grain yield by >7% in barley [60] and wheat [40, 41]. More recent experiments performed in Mexico, where no rainfall was received during the growing cycle, showed glaucousness significantly contributed to grain yield even in irrigated condition [42].
Our recent research using the W1, W2, Iw1 and Iw2 NILs indicated that glaucousness significantly reduced cuticle permeability expressed as water loss and chlorophyll efflux. Difference in cuticle permeability was also found among the glaucous NILs, which differed in the hydroxy-β-diketones, and suggested a role of these hydroxy isoforms in drought tolerance [37]. In the present research, we showed that the w3 mutation impairs the biosynthesis of β-diketone and further deplete the hydroxy forms. At the same time, the w3 mutation significantly increased water loss and chlorophyll leaching (Fig 5). In the spike, w3 mutant showed even faster water loss than the w1w2 double recessive plant (Fig 6). One explanation is that the w3 mutant lost 99% of β-diketones (Fig 7), but they reduced 92% in the w1w2 NIL [37]. In addition to cuticular waxes, cutin proper plays an even greater role in protecting the non-controllable water loss as recently demonstrated in Arabidopsis [61] and barley [62]. In the w3 mutant, three of the five cutin genes assayed showed ~1.5-fold down-regulation (Fig 9; S2 Table). Transmission electron microscopic inspection of cutin organization in BW and w3 mutant may provide more insights into the effect of the w3 mutation on cutin foundation organization.
Another important function of glaucousness is to reflect extra irradiation [1]. In this respect, β-diketones greatly contribute to heat tolerance. The glaucousness can reduced the photosynthetic temperature by up to 0.7°C in the drought-stressed field [41]. We observed that well-watered w3 mutant plants died prematurely in the later spring seasons of 2010 and 2011 in a temperature poorly-controlled greenhouse room (Fig 1C). This did not occur in the fall and winter seasons, suggesting that loss of β-diketones in w3 mutant increased its susceptibility to the high temperature. Preliminary result from field experiment in 2014 showed that w3 mutation significantly reduced 1000-kernel weight (Wanlong Li and Karl Glover, unpublished data). Detailed physiological experiments need to be conducted using growth chambers with well-controlled temperatures and relative humidity to shed light on how the w3 mutation affects the thermal characteristics of adult wheat plants.
Materials and Methods
Plant Materials and Growing Conditions
Transgenic plant #056 was generated in BW by bombardment transformation of RNAi construct pVGS193 (Bi C, Trick HN, Gill BS and Li W, unpublished), which was developed using pANDA, a Gateway-based RNAi vector [38]. Other plant materials are listed in S3 Table with their accession numbers, wax genotypes, wax phenotypes and sources of seeds. The crosses were made by manual emasculation and pollination. All the plants were grown in 4”x4” pots containing Sunshine® potting mix #3 (Sun Gro Horticulture, Agawam, MA, USA) supplemented with Multicote®8 Controlled-Release Fertilizer (Haifa, Altamonte, FL, USA) in a greenhouse. Temperature in the greenhouse was set as 22° at day and 17° at night, and day length was 16 h.
Markers Genotyping and Linkage Mapping
Genomic DNA was isolated following the procedure described by [63]. Polymerase chain reaction (PCR) was conducted in 10 μl containing ~40 ng genomic DNA, 250 μM dNTPs, 200 nM primers, 0.5 unit of Taq polymerase, and 1× Green Go-Taq Reaction Buffer (Promega, Madison, WI, USA) and implemented on 9700 dual thermocyclers (ABI), which were programmed as 94°C for 5 min, then 40 cycles of 94°C 20 s, 55–60°C 30 s and 72°C 1 min 20 s, and finally 7 min at 72°C. The PCR products were separated by 1.5% agarose or 6% polyacrylamide gel electrophoresis.
We used MAPMAKER3.0 [64] for determining the order of marker and gene loci and the Kosambi mapping function for converting the recombination into genetic distance in terms of centi-Morgan (cM) [65]. An LOD score of 3.0 is applied to all marker loci.
Microscopy
For stoma counting and aperture observations, both sides of flag leaf sheaths were coated with 10% cellulose acetate dissolved in acetone using a paint brush. Air-dried for 5 min, the cellulose film was carefully peeled and the imprinted slides were observed under a light microscope at a magnification of 10x20. The fresh cuticle samples of flag leaf sheaths were sputtered with gold powder using the CrC-150 Sputtering System and inspected using a Hitachi S-3400N SEM (Hitachi, Tokyo, Japan).
Cuticle Trait Measurements
At Feekes’ stage 10.5.1 (F10.5.1) when wheat plants flower, flag leaf sheathes were detached and immersed in 30 ml 80% ethanol in a 50-ml falcon tube. The capped tube was agitated at 50 rpm on an orbital shaker. A 150-μl aliquot was removed at a two-hour time interval for measuring the absorbance at 648 nm and 664 nm on Synergy 2 Multi-Mode Microplate Reader (Biotek, Winooski, VT, USA). The sample was returned to the same tube after measurement. Six replicates of extraction were conducted each from an individual of the same line. Two measurements were performed for each extraction and their values were averaged for subsequent calculation. The sheaths were also subjected to air-drying at room temperature with 45% relative humidity. Their weight was measured at a one-hour time interval for 12 consecutive hours to evaluate the water loss rate using an AB54-S/FACT analytical balance (Mettler Toledo, Columbus, OH, USA) with an accuracy of ±0. 0001 g.
Wax Extraction and Metabolite Analysis
One flag leaf sheath was detached at stage F10.5.1 and immersed in 10 ml HPLC-grade chloroform containing 2 μg tetracosane as an internal reference, which is not present in plant wax, and gently agitated for 1 min, the tissue was rinsed in another tube containing 5 ml chloroform. Two extracts were merged and purified by filtration with Iso-Disc PTFE-13-2 filter (Sigma, St Louis, MO, USA). For each line, five biological replicates were extracted separately. Purified wax extract was dried under a nitrogen stream. Wax silylation, gas chromatography–mass spectrometry (GC-MS) profiling, and substance identification were performed at the W.M. Keck Metabolomics Research Laboratory of Iowa State University (Ames, IA, USA) on a fee-for-service basis. In brief, the dried wax extract was dissolved in 100 μl of BSTFA+TCMS (N,O-Bis(trimethylsilyl)trifluoroacetamide with 1% trimethylchlorosilane) and derivatized at 80°C for 30 minutes. The solution is dried under a stream of nitrogen and the residue is reconstituted in 50 μl chloroform and subjected to GC-MS analysis. GC-MS analysis was performed with an Agilent 6890 GC interfaced to a 5973 mass spectrometer. The HP-5ms column (30 m x 0.25 mm i.d. coated with a 0.25 μm film, Agilent Technologies) was used, and temperature gradient was programmed from 150 to 320°C at 5°C/min with helium flow rate at 1.0 ml/min. Operating parameters for MS were set to 70 eV of ionization voltage and 280°C of interface temperature.
Quantitative RT-PCR
Of the 72 quantitative RT-PCR (qPCR) primer pairs used, 64 were adopted from Zhang et al [37] and eight pairs from Kosma et al. [66] together with their gene name designations. At stage F9.0 when the flag leaf is fully emerged from the whorl and flag leaf sheath is rapidly elongating, the flag leaf sheath was collected and immediately frozen in liquid nitrogen and used for RNA isolation using Trizol reagent (Thermo Fisher Scientific Inc., Waltham, MA, USA) following the manufacturer’s instruction. Four biological replicates were used. After evaluating RNA integrity in agarose gel and quantification with Nanodrop ND-1000 (Thermo Scientific, Waltham, MA, USA), 1 μg total RNA was used for reverse transcription using QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA, USA). Approximately 5 ng cDNA was used as template for qPCR, which was performed on ABI 7900HT High-Throughput Real-Time Thermocycler (Thermo Fisher Scientific Inc.) using the iTaq™ SYBR® Green Supermix with ROX (Bio-Rad, Hercules, CA, USA). Two technical replicates were included for each biological replicate. Melt curve analysis was conducted to determine the amplified specificity of PCR products. TaRPII36 was used as the internal reference [37] and relative transcript abundance in the RNA samples was quantified using the 2-ΔΔCt method as described by [67].
Data Analysis and Statistics
Wax load, components, chlorophyll efflux, water loss, and wax gene transcription were measured from four or more biological replicates. The means, standard deviations and P values were estimated using Microsoft® Excel functions. Student’s t-tests were performed to evaluate statistical significance of the differences between BW and the NG mutant. Pearson's chi-squared test was calculated for the deviation of F2 segregation from the expected ratios. The cut-off for statistical significance was set to P-value ≤ 0.05.
Supporting Information
The upper band (arrow) was detected using a primer pair targeting on gus-nos junction (5’- CATGAAGATGCGGACTTACG-3’ and 5’- GCGCGCTATATTTTGTTTTC-3’). The lower band (arrow) was detected using a primer pair targeting on Ubq-bar junction (5’- GAAGTCCAGCTGCCAGAAAC-3’ and 5’- GCACCATCGTCAACCACTAC-3’). The designation of plant lines are indicated above the picture. BW, Bobwhite; NG1, nonglaucous mutant line 1; NG2, nonglaucous mutant line 2; CS, Chinese Spring. NG1, the female parent of the F2 population, carries the bar and RNAi transgene; NG2 is negative for either of them. BW and CS are the negative controls.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Drs. Bikram S. Gill (Kansas State University, Manhattan, KS, USA) and Koichiro Tsunewaki (Fukui Prefectural University, Fukui, Japan) for providing seeds.
Abbreviations
- BW
Bobwhite
- CS
Chinese Spring
- CS-TDIC 2B
Chinese Spring-T. turgidum subsp. dicoccoides 2B substitution line
- β-D
β-diketone
- CER
ECERIFERUM
- FAE
fatty acyl elongase
- FAR
fatty acyl-CoA reductase
- GL
GLOSSY
- KCR
β-keto acyl-CoA reductase
- KCS
β-keto acyl-CoA synthases
- LTP
lipid transfer protein
- MAH
midchain alkane hydroxylase
- NIL
near isogenic line
- NG
nonglaucous
- NT
nulli-tetrasomic
- OH-β
hydroxy β-diketone
- qPCR
quantitative real time PCR
- SEM
scanning electron microscope
- VLCFA
very long chain fatty acid
- WSD
Wax Ester Synthase/Acyl-Coenzyme A:Diacylglycerol Acyltransferase
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
This work was partly supported by the USDA/DOE Feedstock Genomics Program, South Dakota Agricultural Experimental Station (Brookings, SD) and South Dakota Wheat Commission (Pierre, SD).
References
- 1. Shepherd T, Wynne Griffiths D. The effects of stress on plant cuticular waxes. New Phytol. 2006;171(3):469–99. 10.1111/j.1469-8137.2006.01826.x . [DOI] [PubMed] [Google Scholar]
- 2. Kolattukudy PE. Biopolyester membranes of plants: cutin and suberin. Science. 1980;208(4447):990–1000. [DOI] [PubMed] [Google Scholar]
- 3. Nawrath C. The biopolymers cutin and suberin. Arabidopsis Book. 2002;1:e0021 10.1199/tab.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pollard M, Beisson F, Li Y, Ohlrogge JB. Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci. 2008;13(5):236–46. 10.1016/j.tplants.2008.03.003 [DOI] [PubMed] [Google Scholar]
- 5. Kolattukudy PE. Plant waxes. Lipids. 1970;5(2):259–75. [Google Scholar]
- 6. Vioque J, Kolattukudy PE. Resolution and purification of an aldehyde-generating and an alcohol-generating fatty acyl-coa reductase from pea leaves (Pisum sativum L.). Arch Biochem Biophys. 1997;340(1): 64–72. [DOI] [PubMed] [Google Scholar]
- 7. Rowland O, Zheng H, Hepworth SR, Lam P, Jetter R, Kunst L. CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 2006;142(3):866–77. 10.1104/pp.106.086785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li F, Wu X, Lam P, Bird D, Zheng H, Samuels L, et al. Identification of the wax ester synthase/acyl-coenzyme A: diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 2008;148(1):97–107. 10.1104/pp.108.123471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheesbrough TM, Kolattukudy PE. Alkane biosynthesis by decarbonylation of aldehydes catalyzed by a particulate preparation from Pisum sativum . Proc Natl Acad Sci U S A. 1984;81:6613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greer S, Wen M, Bird D, Wu X, Samuels L, Kunst L, et al. The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 2007;145(3):653–67. 10.1104/pp.107.107300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure JD, et al. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell. 2012;24(7):3106–18. 10.1105/tpc.112.099796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Samuels L, Kunst L, Jetter R. Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol. 2008;59:683–707. 10.1146/annurev.arplant.59.103006.093219 . [DOI] [PubMed] [Google Scholar]
- 13. Bernard A, Joubès J. Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog Lipid Res. 2013;52(1):110–29. 10.1016/j.plipres.2012.10.002 [DOI] [PubMed] [Google Scholar]
- 14. Lee SB, Suh MC. Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol Plant. 2013;6(2):246–9. 10.1093/mp/sss159 . [DOI] [PubMed] [Google Scholar]
- 15. Hansen JD, Pyee J, Xia Y, Wen TJ, Robertson DS, Kolattukudy PE, et al. The GLOSSY1 locus of maize and an epidermis-specific cDNA from Kleinia odora define a class of receptor-like proteins required for the normal accumulation of cuticular waxes. Plant Physiol. 1997;113:1091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sturaro M, Hartings H, Schmelzer E, Velasco R, Salamini F, Motto M. Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol. 2005;138(1):478–89. 10.1104/pp.104.058164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tacke E, Korfhage C, Michel D, Maddaloni M, Motto M, Lanzini S, et al. Transposon tagging of the maize Glossy2 locus with the transposable element En/Spm. Plant J. 1995;8(6):907–17. [DOI] [PubMed] [Google Scholar]
- 18. Liu S, Dietrich CR, Schnable PS. DLA-based strategies for cloning insertion mutants: cloning the gl4 locus of maize using Mu transposon tagged alleles. Genetics. 2009;183:1215–25. 10.1534/genetics.109.108936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dietrich CR, Perera MA, D Yandeau-Nelson M, Meeley RB, Nikolau BJ, Schnable PS. Characterization of two GL8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J 2005;42:844–61. [DOI] [PubMed] [Google Scholar]
- 20. Xu X, Dietrich CR, Delledonne M, Xia Y, Wen TJ, Robertson DS, Nikolau BJ, Schnable PS. Sequence analysis of the cloned glossy8 gene of maize suggests that it may code for a beta-ketoacyl reductase required for the biosynthesis of cuticular waxes. Plant Physiol. 1997;115(2):501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu D, Ranathunge K, Huang H, Pei Z, Franke R, Schreiber L, et al. Wax Crystal-Sparse Leaf1 encodes a beta-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta. 2008;228(4):675–85. 10.1007/s00425-008-0770-9 . [DOI] [PubMed] [Google Scholar]
- 22. Moose SP, Sisco PH. Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes & Development. 1996;10(23):3018–27. [DOI] [PubMed] [Google Scholar]
- 23. Javelle M, Vernoud V, Depege-Fargeix N, Arnould C, Oursel D, Domergue F, et al. Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor Outer Cell Layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol. 2010;154(1):273–86. 10.1104/pp.109.150540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Javelle M, Klein-Cosson C, Vernoud V, Boltz V, Maher C, Timmermans M, et al. Genome-wide characterization of the HD-ZIP IV transcription factor family in maize: preferential expression in the epidermis. Plant Physiol. 2011;157(2):790–803. 10.1104/pp.111.182147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tulloch AP. Composition of leaf surface waxes of Triticum species: Variation with age and tissue. Phytochemistry. 1973;12:2225–32. [Google Scholar]
- 26. Mikkelsen JD. The effects of inhibitors on the biosynthesis of the long chain lipids with even carbon numbers in barley spike epicuticular wax. Carlsberg Res Commun. 1978; 43(1):15–35. [Google Scholar]
- 27. von Wettstein-Knowles P. Genetic control of β-diketone and hydroxyl-β-diketone synthesis in epicuticular waxes of barley. Planta. 1972; 06:113–30. [DOI] [PubMed] [Google Scholar]
- 28. Adamski NM, Bush MS, Simmonds J, Turner AS, Mugford SG, Jones A, et al. The Inhibitor of wax 1 locus (Iw1) prevents formation of beta- and OH-beta-diketones in wheat cuticular waxes and maps to a sub-cM interval on chromosome arm 2BS. Plant J. 2013;74(6):989–1002. 10.1111/tpj.12185 . [DOI] [PubMed] [Google Scholar]
- 29. Simmonds JR, Fish LJ, Leverington-Waite MA, Wang Y, Howell P, Snape JW. Mapping of a gene (Vir) for a non-glaucous, viridescent phenotype in bread wheat derived from Triticum dicoccoides, and its association with yield variation. Euphytica. 2007;159(3):333–41. 10.1007/s10681-007-9514-3 [DOI] [Google Scholar]
- 30. Tsunewaki K, Ebana K. Production of near-isogenic lines of common wheat for glaucousness and genetic basis of this trait clarified by their use. Genes Genet Syst. 1999;74:33–41. [Google Scholar]
- 31. Wu H, Qin J, Han J, Zhao X, Ouyang S, Liang Y, et al. Comparative high-resolution mapping of the wax inhibitors Iw1 and Iw2 in hexaploid wheat. PLoS One. 2013;8(12):e84691 10.1371/journal.pone.0084691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nelson JC, Deynze AE, Sorrells ME, Autrique E, Lu YH, Merlino M, et al. Molecular mapping of wheat. Homoeologous group 2. Genome. 1995;38(3):516–24. [DOI] [PubMed] [Google Scholar]
- 33. Liu Q, Ni Z, Peng H, Song W, Liu Z, Sun Q. Molecular mapping of a dominant non-glaucousness gene from synthetic hexaploid wheat (Triticum aestivum L.). Euphytica. 2006;155(1–2):71–8. 10.1007/s10681-006-9302-5 [DOI] [Google Scholar]
- 34. Lu P, Qin J, Wang G, Wang L, Wang Z, Wu Q, et al. Comparative fine mapping of the Wax 1 (W1) locus in hexaploid wheat. Theor Appl Genet. 2015;128(8):1595–603. 10.1007/s00122-015-2534-9 [DOI] [PubMed] [Google Scholar]
- 35. Dubcovsky J, Echaide M, Giancola S, Rousset M, Luo MC, Joppa LR, et al. Seed-storage-protein loci in RFLP maps of diploid, tetraploid, and hexaploid wheat. Theor Appl Genet. 1997;95:1169–80. [Google Scholar]
- 36. Wang J, Li W, Wang W. Fine mapping and metabolic and physiological characterization of the glume glaucousness inhibitor locus Iw3 derived from wild wheat. Theor Appl Genet. 2014;127(4):831–41. 10.1007/s00122-014-2260-8 . [DOI] [PubMed] [Google Scholar]
- 37. Zhang Z, Wang W, Li W. Genetic interactions underlying the biosynthesis and inhibition of beta-diketones in wheat and their impact on glaucousness and cuticle permeability. PLoS One. 2013;8(1):e54129 10.1371/journal.pone.0054129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45(4): 490–5. [DOI] [PubMed] [Google Scholar]
- 39. McIntosh RA, Yamazaki Y, Dubcovsky J, Rogers J, Morris C, Somers DJ, et al. , editors. Catalogue of gene symbols for wheat 11th International Wheat Genetics Symposium; 2008; Brisbane Qld Australia: Sidney University Press. [Google Scholar]
- 40. Johnson DA, Richards RA, Turner NC. Yield, water relations, gas exchange, and surface reflectances of near-isogenic wheat lines differing in glaucousness. Crop Sci. 1983;23:318–25. [Google Scholar]
- 41. Richards RA, Rawson HM, Johnson DA. Glaucousness in wheat: Its development and effect on water-use efficiency, gas exchange and photosynthetic tissue temperature. Aust J Plant Physiol. 1986;13:465–73 [Google Scholar]
- 42. Monneveux P, Reynolds MP, González-Santoyo H, Peña RJ, Mayr L, Zapata F. Relationships between grain yield, flag leaf morphology, carbon isotope discrimination and ash content in irrigated wheat. Journal of Agronomy and Crop Science. 2004;190(6):395–401. 10.1111/j.1439-037X.2004.00116.x [DOI] [Google Scholar]
- 43. Bennett D, Izanloo A, Edwards J, Kuchel H, Chalmers K, Tester M, et al. Identification of novel quantitative trait loci for days to ear emergence and flag leaf glaucousness in a bread wheat (Triticum aestivum L.) population adapted to southern Australian conditions. Theor Appl Genet. 2012;124(4):697–711. 10.1007/s00122-011-1740-3 . [DOI] [PubMed] [Google Scholar]
- 44. Lundqvist U, Lundqvist A. Mutagen specificity in barley for 1580 eceriferum mutants localized to 79 loci. Hereditas. 1988;108:1–12. [Google Scholar]
- 45. Franckowiak JD, Lundqvist U. Descriptions of barley genetic stocks for 2011. Barley Genetics Newsletter 2011;41:54–202. [Google Scholar]
- 46. Slade AJ, Fuerstenberg SI, Loeffler D, Steine MN, Facciotti D. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat Biotechnol. 2005;23:75–81. [DOI] [PubMed] [Google Scholar]
- 47. Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, Berardi S, et al. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol 2009;9:115 10.1186/1471-2229-9-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8(2):135–41. 10.1016/j.pbi.2005.01.001 . [DOI] [PubMed] [Google Scholar]
- 49. Neelakandan AK, Wang K. Recent progress in the understanding of tissue culture-induced genome level changes in plants and potential applications. Plant Cell Rep. 2012;31(4):597–620. 10.1007/s00299-011-1202-z . [DOI] [PubMed] [Google Scholar]
- 50. Bianchi G. Plant waxes In: Hamilton RJ, editor. Waxes: chemistry, molecular biology and functions. Dundee, UK: The Oily Press; 1995. p. 175–222. [Google Scholar]
- 51. von Wettstein-Knowles P. Biosynthesis and genetics of waxes In: Hamilton RJ, editor. Waxes: chemistry, molecular biology and functions. Dundee, UK: The Oily Press; 1995. p. 91–129. [Google Scholar]
- 52. von Wettstein-Knowles P. Plant Waxes eLS: John Wiley & Sons, Ltd.; 2012. p. 1–11. [Google Scholar]
- 53. Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16(9):2463–80. 10.1105/tpc.104.022897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101(13):4706–11. 10.1073/pnas.0305574101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, et al. The transcription factor WIN1/SHN1 regulates Cutin biosynthesis in Arabidopsis thaliana. Plant Cell. 2007;19(4):1278–94. 10.1105/tpc.106.047076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tulloch AP, Hoffman LL. Leaf wax of Triticum aestivum . Phytochemistry. 1973;12:2217–23. [Google Scholar]
- 57. Tulloch AP, Baum BR, Hoffman LL. A survey of epicuticular waxes among genera of Triticeae. 2. Chemistry. Can J Bot. 1980;58:2602–15. [Google Scholar]
- 58. Barber HN, Netting AG. Chemical genetics of β-diketone formation in wheat. Phytochemistry. 1968; 7:2089–93. [Google Scholar]
- 59. Bianchi G, Figini ML. Epicuticular waxes of glaucous and nonglaucous durum wheat lines. J Agric Food Chem 1986;34(3):429–33. [Google Scholar]
- 60. Baenziger PS, Wesenberg DM, Sicher RC. The effects of genes controlling barley leaf and sheath waxes on agronomic performance in irrigated and dryland environments. Crop Sci. 1983; 23:116–20. [Google Scholar]
- 61. Lü S, Zhao H, Des Marais DL, Parsons EP, Wen X, Xu X, et al. Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 2012;159:930–44. 10.1104/pp.112.198697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen G, Komatsuda T, Ma JF, Nawrath C, Pourkheirandish M, Tagiri A, et al. An ATP-binding cassette subfamily G full transporter is essential for the retention of leaf water in both wild barley and rice. Proc Natl Acad Sci U S A. 2011;108(30):12354–9. 10.1073/pnas.1108444108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li W, Huang L, Gill BS. Recurrent deletions of puroindoline genes at the grain hardness locus in four independent lineages of polyploid wheat. Plant physiology. 2008;146:200–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1(2):174–81. [DOI] [PubMed] [Google Scholar]
- 65. Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–5. [Google Scholar]
- 66. Kosma DK, Nemacheck JA, Jenks MA, Williams CE. Changes in properties of wheat leaf cuticle during interactions with Hessian fly. Plant J. 2010;63(1):31–43. 10.1111/j.1365-313X.2010.04229.x . [DOI] [PubMed] [Google Scholar]
- 67. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The upper band (arrow) was detected using a primer pair targeting on gus-nos junction (5’- CATGAAGATGCGGACTTACG-3’ and 5’- GCGCGCTATATTTTGTTTTC-3’). The lower band (arrow) was detected using a primer pair targeting on Ubq-bar junction (5’- GAAGTCCAGCTGCCAGAAAC-3’ and 5’- GCACCATCGTCAACCACTAC-3’). The designation of plant lines are indicated above the picture. BW, Bobwhite; NG1, nonglaucous mutant line 1; NG2, nonglaucous mutant line 2; CS, Chinese Spring. NG1, the female parent of the F2 population, carries the bar and RNAi transgene; NG2 is negative for either of them. BW and CS are the negative controls.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.