Abstract
The goal of the 1000 Genomes Consortium is to characterize human genome structural variation (SV), including forms of copy number variations such as deletions, duplications, and insertions. Mobile element insertions, particularly Alu elements, are major contributors to genomic SV among humans. During the pilot phase of the project we experimentally validated 645 (611 intergenic and 34 exon targeted) polymorphic “young” Alu insertion events, absent from the human reference genome. Here, we report high resolution sequencing of 343 (322 unique) recent Alu insertion events, along with their respective target site duplications, precise genomic breakpoint coordinates, subfamily assignment, percent divergence, and estimated A-rich tail lengths. All the sequenced Alu loci were derived from the AluY lineage with no evidence of retrotransposition activity involving older Alu families (e.g., AluJ and AluS). AluYa5 is currently the most active Alu subfamily in the human lineage, followed by AluYb8, and many others including three newly identified subfamilies we have termed AluYb7a3, AluYb8b1, and AluYa4a1. This report provides the structural details of 322 unique Alu variants from individual human genomes collectively adding about 100 kb of genomic variation. Many Alu subfamilies are currently active in human populations, including a surprising level of AluY retrotransposition. Human Alu subfamilies exhibit continuous evolution with potential drivers sprouting new Alu lineages.
Keywords: retrotransposon, mobilome, structural variation
Introduction
The 1000 Genomes Project is an ongoing series of studies designed to comprehensively identify and characterize all forms of human genomic variation (Abecasis et al. 2010), as well as identify specific types of structural variation (SVs) and their origin and impact on human populations (Mills et al. 2011; Abecasis et al. 2012). The Pilot phase of the project was the first to employ advances in second-generation DNA sequencing technologies to perform population scale high-throughput genome-wide sequencing on multiple human individuals. The two primary second-generation strategies to detect SVs involved a “read-pair” (RP) method applied to Illumina paired-end short sequence reads and a complimentary “split-read” (SR) method applied to longer pyrosequencing reads generated by the Roche/454 platform (Stewart et al. 2011). The Pilot phase was comprised of three strategic approaches, low-coverage, high-coverage, and exon-targeted sequencing (Abecasis et al. 2010). The “low-coverage project” or “P1” consisted of 179 unrelated individuals who were sequenced with an average of 3.6× coverage, including 59 individuals from Yoruba, Nigeria (YRI), 60 individuals of European ancestry from Utah (CEU), 30 of Han ancestry from Beijing, and 30 of Japanese ancestry from Tokyo (Abecasis et al. 2010; Mills et al. 2011). The “high-coverage project,” also called “P2” or the “trio project” consisted of mother–father–offspring trios, one each from CEU and YRI populations, where each of the six individuals was sequenced to 42× coverage on average (Abecasis et al. 2010; Mills et al. 2011).
SVs can be balanced (i.e., inversions) or unbalanced (i.e., deletions, duplications, insertions). Unbalanced SVs are often referred to as copy number variants (CNVs) (Mills et al. 2011). Mobile element insertions (MEIs) are a type of unbalanced CNV known to be major contributors of structural variation (Cordaux and Batzer 2009; Xing et al. 2009, 2013; Abecasis et al. 2010; Beck et al. 2011; Mills et al. 2011; Stewart et al. 2011) with non-LTR (long terminal repeat) retrotransposons, L1 (long interspersed element 1), SVA, and Alu classes of MEIs having accumulated in such large copy numbers as to collectively account for one-third or more of the human genome (Lander et al. 2001; Cordaux and Batzer 2009; de Koning et al. 2011). Non-LTR retrotransposons have also been implicated in causing a variety of genetic diseases (Deininger and Batzer 1999; Callinan and Batzer 2006). Although most MEIs are ancient remnants in the genome, having lost their ability to replicate, their residual high sequence identity has contributed to genome instability (Sen et al. 2006; Han et al. 2007; Lee et al. 2008; Cook et al. 2011) and extensive genome rearrangements. Mobile elements are a source of genome instability both through insertion and postinsertion mutagenesis (Cordaux and Batzer 2009; Konkel and Batzer 2010; Deininger 2011; Ade et al. 2013). Younger non-LTR retrotransposons remain active in the human genome, propagating in a “copy and paste” mechanism leading to increased genomic diversity among humans (Xing et al. 2009; Beck et al. 2010; Hormozdiari et al. 2011; Stewart et al. 2011). Alu elements are nonautonomous and require the enzymatic machinery of L1 to mobilize (Dewannieux et al. 2003) yet they are the most prolific class of MEI in humans in terms of copy number, having accumulated greater than 1 million copies over the past 65 Myr (Lander et al. 2001; Batzer and Deininger 2002). The typical full-length human Alu element is about 300 bp long and has a dimeric structure in which the left monomer contains an RNA polymerase III (pol III) promotor (A and B boxes), followed by a middle A-rich region, right monomer and ending in an oligo (dA)-rich tail (Batzer and Deininger 2002; Deininger 2011; Wagstaff et al. 2012). Although most Alu copies have ceased to replicate, the current rate of Alu retrotransposition in humans is estimated to be one new insertion in every 20 live births (Cordaux et al. 2006), resulting in potentially 300 million recent Alu insertions in human populations globally (Bennett et al. 2008) with a potential for profound impact on human biology.
Although recent studies have shown that second-generation sequencing represents a powerful tool to identify SVs, including MEIs, with relatively low false positive detection rates, the need for detailed and wide-spread validations, especially in regions with high repeat content, has become evident (Mills et al. 2011; Stewart et al. 2011). During the Pilot phase of the 1000 Genomes Project, roughly 4,500 recent Alu insertion events absent from the human reference genome [hg18] were discovered (Stewart et al. 2011). Approximately 200 elements were randomly selected from each of the four insertion call sets (P1/RP, P1/SR, P2/RP, and P2/SR) for polymerase chain reaction (PCR) validation experiments and from these we experimentally validated 645 (611 intergenic and 34 exon targeted) recent polymorphic Alu insertion events, representing all three strategic approaches, low-coverage, high-coverage, and exon-targeted insertion events (for details see Stewart et al. 2011). Due to the nature of RP and SR second-generation sequencing technologies, Alu subfamily classification from this data set was performed by reconstruction of the supporting fragment reads to map each candidate insertion against the human reference genome, followed by RepeatMasker (Smit et al. 1996–2010) analysis to identify the Alu subfamily.
The goal of this project was to perform high-resolution Sanger chain termination DNA sequencing (Sanger et al. 1977) on a subset of at least 50% of these validated polymorphic Alu MEI events to report: 1) Complete Alu sequences including the variable middle A-rich region and immediate flanking sequence of the Alu element; 2) precise genomic insertion coordinates; 3) target site duplications (TSDs), and Alu subfamily analysis for each locus.
Materials and Methods
Following the original locus-specific PCR validation experiments reported in Stewart et al. (2011), all 34 validated Alu insertions located in exon-targeted regions were Sanger sequenced. Subsequent detailed analyses (Mills et al. 2011) concluded that the actual locations of these exon-targeted elements were almost exclusively intronic or untranslated region (UTR)-overlap with minimal affect to coding exons. Concurrently in the laboratory, Alu loci from the 611 validated intergenic insertion events were being randomly selected for Sanger sequencing. Once a locus was attempted to be sequenced it was considered “selected” and follow-up experiments were conducted until the locus was successfully Sanger sequenced with confidence. The process continued until we had roughly 50% of the data set completed. This was an arbitrary cut off point with the objective being to obtain a representative subset of ample sample size within a reasonable time frame.
DNA Samples
All DNA samples used for Sanger sequencing were selected from the PCR validation results showing a “filled site” or Alu present confirmation in a particular individual. The ID of the individual used for sequencing as well as the Alu genotype of that individual (+/+: homozygous present for the insertion; or +/−: heterozygous) is shown in supplementary file S3, tables S1 and S2, Supplementary Material online, for each locus. DNA samples for the original PCR validations consisted of a subset of 25 DNA samples from the 179 Pilot 1 low coverage samples: Ten European samples (CEPH/Utah USA), NA11881, NA12043, NA07346, NA07347, NA11894, NA07357, NA12003, NA11831, NA06986, and NA12828; five African samples (HAPMAP/YRI) NA18504, NA18870, NA18912, NA19210, and NA19201; five Chinese samples (HAPMAP/Han Chinese) NA18572, NA18577, NA18537, NA18563, and NA18542; and five Japanese samples (HAPMAP/Japanese, Tokyo) NA18942, NA18943, NA18944, NA18945, and NA18953. Pilot 2 PCR validation experiments included six DNA samples, CEU trio: NA12878 (daughter), NA12891 (father), and NA12892 (mother); and YRI trio: NA19238 (mother), NA19239 (father), and NA19240 (daughter). These Pilot 1 & 2 DNA samples were purchased from the Coriell Institute for Medical Research (Camden, NJ). In addition to the subset of 25 individuals used for the Pilot 1 validations, four more DNA samples from the low coverage Pilot 1 data set were obtained for subsequent experiments. DNA samples NA12872, NA12814, NA12815, and NA12044 (CEPH/Utah USA) were purchased from the Coriell Institute for Medical Research. Then, all 35 samples (25 + 6 + 4) were used for PCR validations associated with MEI events detected specifically from exon-targeted regions.
Oligonucleotide Primer Design
Most oligonucleotide primers used for PCR reactions in this study were designed for the original validation experiments as reported previously (Stewart et al. 2011). In cases in which the original primers were not acceptable for Sanger sequencing, primers for individual loci were redesigned as needed (supplementary file S3, table S3, Supplementary Material online) using Primer3 on-line software (Rozen 1998), (http://biotools.umassmed.edu/bioapps/primer3_www.cgi, last accessed September 1, 2015). We also designed five primers within the Alu sequence, three in the forward orientation and two in the reverse, that were used exclusively for sequencing experiments to obtain a high confidence consensus sequence in both directions for each Alu in the data set. The internal forward primers sequenced into the A-tail providing a confident estimation of A-tail length. The internal reverse primers sequenced out the “front” of each Alu confirming the 5′ flanking sequence upstream of the Alu element and the precise preintegration site. Virtual PCR was performed for each locus using the in silico function of BLAT (Kent 2002) to obtain the estimated PCR product size for the empty (no insertion) and the filled size (insertion present). Primers were ordered from Sigma Aldrich (Woodlands, TX).
PCR Analysis
PCR amplifications were performed in 25 µl reactions containing 15–50 ng of template DNA; 200 nM of each oligonucleotide primer; 1.5–3.0 mM MgCl2, 10× PCR buffer (1×:50 mM KCl; 10 mM Tris–HCl, pH 8.4); 0.2 mM dNTPs; and 1–2 U Taq DNA polymerase. PCR reactions were performed under the following conditions: Initial denaturation at 94 °C for 60 s, followed by 32 cycles of denaturation at 94 °C for 30 s, 30 s at optimum annealing temperature, and extension at 72 °C for 30 s. PCRs were terminated with a final extension at 72 °C for 2 min. 20 µl of each PCR product were fractionated in a horizontal gel chamber on a 2% agarose gel containing 0.2 µg/ml ethidium bromide for 60–70 min at 175 V. UV-fluorescence was used to visualize the DNA fragments and images were saved using a BioRad ChemiDoc XRS imaging system (Hercules, CA).
Sanger Sequencing
Four PCR fragments per locus were gel purified using a Wizard SV gel purification kit (Promega Corporation, Madison, WI, catalog A9282) according to the manufacturer’s instructions with the following modification. The 50 µl elution step was performed twice, resulting in 100 µl, which was then dried in a SpeedVac (ThermoSavant SPD 111 V). The DNA was reconstituted in 30 µl TVLE (tris very low ethylenediaminetetraacetic acid [EDTA], 10 mM Tris/0.05 mM EDTA) and 4 µl was used for chain termination cycle sequencing using BigDye Terminator v3.1. Cycle sequencing was performed under the following conditions: After an initial denaturation at 95 °C for 2 min, 40 cycles at 95 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min were performed followed by a hold at 4 °C. Sequencing reactions were cleaned by standard ethanol precipitation to remove any unincorporated dye terminators and then stabilized in 15 µl Hi-Di Formamide (Life Technologies, Inc.). Capillary electrophoresis was performed on an ABI 3130xl Genetic Analyzer (Applied Biosystems, Inc., Foster City, CA).
Cloning and Sequencing
In rare cases (<1%), sequencing using the PCR primers remained unresolved even after several attempts. When this occurred, gel-purified PCR products were cloned using a TOPO TA cloning kit for sequencing (Invitrogen Corporation, Carlsbad, CA, catalog K4575-40) according to the manufacturer’s instructions. For each candidate locus, at least three clones were picked from lysogeny broth (LB)/amp plates and verified for the presence of an insert of the expected size using colony PCR. Following overnight incubation in LB at 37°C, plasmid mini-preps were performed using the FastPlasmid Mini Kit (5 PRIME, Inc., Gaithersburg, MD) according to the manufacturer’s instructions. Generic M13 forward and reverse primers were used for cycle sequencing PCR as described above.
Sequence Analysis
Following capillary electrophoresis, sequence quality was evaluated using ABI software Sequence Scanner v1.0. Sequence alignment figures were constructed in BioEdit (Hall 1999) and a consensus sequence for each locus was determined from the multiple forward and reverse Sanger sequences obtained for each locus. Alu elements in the reverse orientation (minus strand) were reverse complimented to be viewed in BioEdit in the forward orientation. Once the consensus sequence for each locus was determined, data were recorded for TSDs, endonuclease cleavage site, and estimated A-tail length. Then, a query sequence which included some flanking sequence along with the Alu junction was screened in BLAT (Kent 2002) to determine the precise insertion coordinates. The integration site in the reference genome was identified as the nucleotide junction between the last base matching the reference genome upstream of the Alu and the adjacent base (typically the first base following the 3′ TSD). Using this convention, the insertion site coordinate was always at the last base pair of the 5′ TSD before the element. This is in contrast to some coordinates of the 1000 Genomes Pilot release data set which primarily focused on deletions and therefore the breakpoint even for MEI events was generally determined as the first base pair of the 5′ TSD, without regard for element orientation. We initially used human reference genome [hg18] coordinates for all our analyses to be consistent with the source data as reported in Stewart et al. (2011), but also report the [hg19] insertion coordinates for each locus converted using the LiftOver function of the UCSC genome browser (Kent et al. 2002).
The multiple alignment diagram of the new Alu subfamilies discovered in this study was constructed using the “view alignment report” option in MegAlign with the ClustalW algorithm followed by manual formatting of “alignment report contents” under Options (DNASTAR, Inc. Version 5.0 for Windows). The alignment report output was saved as a text file, followed by more manual refinement and labeling in Microsoft Word for Windows.
Results
We report Sanger sequencing results for 343 polymorphic Alu MEI events from the 1000 Genomes Pilot Project, 309 of the 611 intergenic insertion events representing each of the four insertion call sets (P1/RP, P1/SR, P2/RP and P2/SR), as well as all 34 validated Alu elements from exon-targeted regions which resided primarily within intronic and UTR-overlap regions (see Materials and Methods). These completed Alu consensus sequences (a consensus of multiple Sanger-sequenced amplicons for each locus) along with some genomic flanking sequence are available in FASTA format (supplementary file S1, Supplementary Material online), as GB files in BioEdit (Hall 1999) (supplementary file S2, Supplementary Material online) and have been deposited in GenBank under accession numbers KT305395–KT305737. A comprehensive summary table is available as supplementary file S3; tables S1 and S2, Supplementary Material online. Data for each locus include the original call set (P1 or P2 and Illumina [RP] or 454 [SR]), the subject identification number for the individual used for sequencing along with that individual’s population affiliation and genotype at the respective locus, the genomic insertion coordinates, the Alu subfamily, the percent divergence from the subfamily consensus sequence, estimated A-tail length, and TSDs. Redesigned and supplemental PCR primers used for Sanger sequencing which differed from the original validation experiments are listed in supplementary file S3, table S3, Supplementary Material online.
Deletions and Duplicate Calls
One criteria of the original MEI call sets was that all calls were absent from the human reference genome. Sequencing results identified six loci (of the 309 set; ∼2%) which appear to be lineage-specific deletions from the reference genome rather than novel Alu insertions. Five were classified as deletions based on sections of flanking sequence on at least one side of the Alu being deleted from the reference genome and by alignment with the chimpanzee genome [panTro4]. For the sixth event, locus #209, the beginning of the Alu sequence is present in the reference genome followed by a 550-bp deletion (supplementary files S1 and S3, table S1, Supplementary Material online).
For further analysis we removed the six loci determined to be deletions (highlighted in red in supplementary file S3, table S1, Supplementary Material online), and sorted our data set by insertion coordinates to identify any potential duplicate loci. As with the original validation sets some redundancy occurred due to the presence of the same Alu insertion candidate locus being detected in multiple call sets (Pilot 1 vs. Pilot2 or Illumina [RP] vs. 454 [SR]) followed by random selection of candidate loci for validation. Our sequenced loci included four duplicates from P1 which were called by both Illumina and 454 platforms (highlighted in blue in supplementary file S3, table S1, Supplementary Material online). As expected, the nucleotide sequence including the insertion site of the Alu was identical between these duplicates. In each case, we elected to remove the 454 (SR) duplicate. There were also multiple instances in which the same locus was in both the P1, low-coverage, and the P2, high-coverage trio data sets. Because P1 and P2 contained different human subjects it was important to record all the genotype and sequence data, but for the distribution of Alu subfamilies and subsequent analyses, it was important to retain only unique novel insertion events. Our sequenced Alu loci included 11 present in both P1 and P2 data sets (highlighted in yellow in supplementary file S3, table S1, Supplementary Material online). Once again, the nucleotide sequence including the preintegration site of the Alu insertion was identical between these duplicates. For consistency, we removed the P1 duplicate and retained the P2 locus.
Distribution of Active Alu Subfamilies
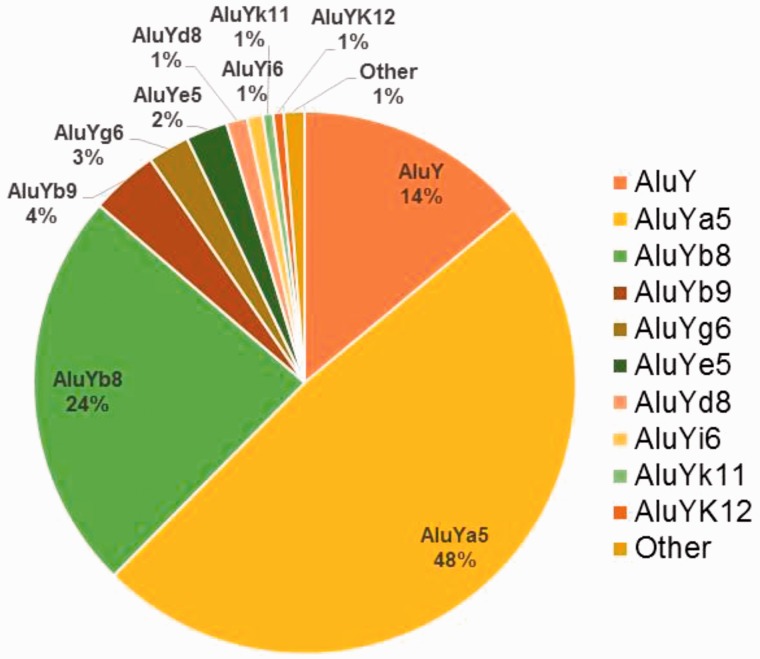
Following the removal of the 6 deletion events and 15 duplicate loci, 288 unique intergenic (P1/RP: N = 64, P1/SR: N = 124, P2/RP: N = 41, and P2/SR: N = 59), and 34 exon-targeted Alu MEI events remained in our data set. These insertions were randomly distributed across the genome based on the larger full set of MEI events reported previously (Stewart et al. 2011). Subfamily analysis using RepeatMasker (www.repeatmaskser.org, last accessed September 1, 2015) (Smit et al. 1996–2010) detected no appreciable difference in the Alu subfamily distribution between intergenic and exon-targeted elements and therefore the Alu subfamily distribution for the combined 322 loci is shown in figure 1. All 322 elements were derived from the AluY lineage with no evidence of older AluJ or AluS retrotransposition activity. The complete RepeatMasker output report is available as supplementary file S4, tables S4 and S5, Supplementary Material online. The most active human Alu subfamilies are AluYa5 and AluYb8 as reported previously (Carroll et al. 2001; Hormozdiari et al. 2011; Stewart et al. 2011) representing 48% and 24% of our data set, respectively. The ancestral AluY is considered the oldest of the “young” Alu subfamilies and the progenitor of all the subsequent subfamilies of the Y-lineage (Batzer et al. 1996). Yet about 14% of the young Alu elements we sequenced were identified as AluY, suggesting ongoing retrotransposition of this progenitor subfamily. We also observed moderate activity of the AluYb9 subfamily as well as lower levels of recent retrotransposition among ten other Alu subfamilies (fig. 1).
Fig. 1.—
Distribution of active Alu subfamilies. The percent membership to each Alu subfamily based on 322 unique Alu elements in the RepeatMasker subfamily analysis. The category “Other” is comprised one Alu element each from subfamilies Ya8, Yc1, Yh7, and Yj4.
Characterization of Confirmed Novel Alu Insertions
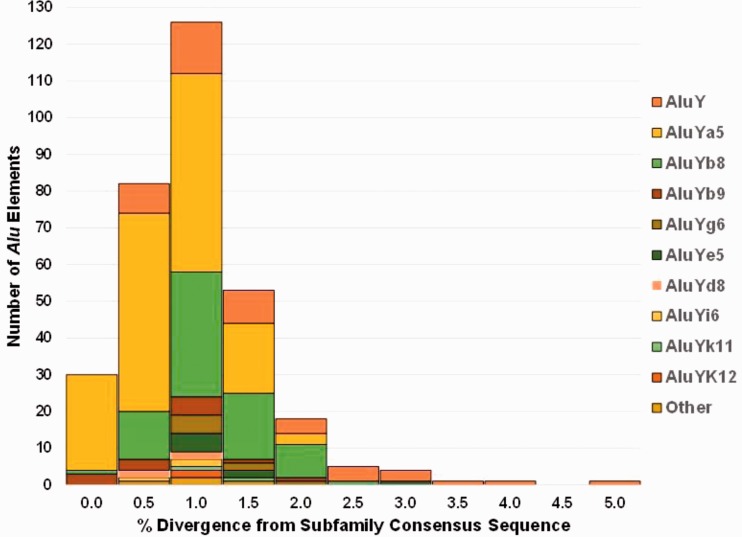
The percent divergence from each subfamily consensus sequence for all 322 novel Alu insertions ranged from 0.0% to 6.6% with an average of 0.8% and a standard deviation of 0.7%. The maximum value of 6.6% is for exon-targeted Alu #51 which is 5′ truncated by 98 bp and represents an extreme outlier in the data set by being more than 8 SD away from the mean, within the AluYb8 subfamily which otherwise has a range of 0.0–2.1% divergence. With this locus removed from the calculation, the maximum value is 4.8% divergence (for a full-length AluY), the average is still 0.8%, and the standard deviation is 0.6%. The distribution of percent divergence from each subfamily consensus sequence is shown in figure 2. A total of 30 elements (N = 26 Ya5, N = 3 Yb9, and N = 1 Yb8) were scored by RepeatMasker (Smit et al. 1996–2010) as having 0.0% divergence from their respective consensus sequences (about 9.3% of the data set). All 156 AluYa5 elements (yellow) and 75 of 76 AluYb8 elements (green) were ≤2% diverged from their respective consensus sequence, providing strong support for their recent genomic integration. As expected the ancestral AluY subfamily (orange) showed the broadest distribution of percent divergence from the AluY consensus sequence. Typically, as percent divergence increases the estimated age of the insertion event also increases, as older elements have more time to accumulate random mutations. However, because this data set is comprised exclusively of proven young polymorphic Alu elements, the broad range in divergence is suggestive of the amplification of secondary source elements, or multiple members within the AluY subfamily capable of making new copies, thus increasing diversity.
Fig. 2.—
Distribution of the percent divergence from the subfamily consensus sequence for each Alu element based on RepeatMasker. All Ya5 elements (yellow) and over 96% of all the Alu elements have ≤2% divergence from the respective consensus sequences. Alu elements with greater than 2% divergence are primarily AluY elements in addition to one each Yb8 and Ye5 elements.
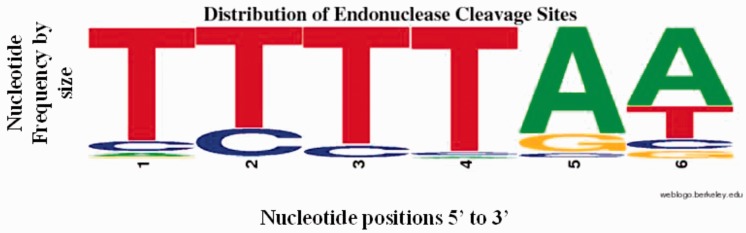
The distribution of endonuclease cleavage sites for the 322 Sanger-sequenced Alu insertions is graphically displayed in WebLogo format (Schneider and Stephens 1990; Crooks et al. 2004) (fig. 3) and listed individually for each locus in supplementary file S3, tables S1 and S2, Supplementary Material online. Although the most common nucleotide at each position, as depicted by letter size in figure 3, corresponds to the typical 5′-TTTT/AA-3′ recognition sequence for L1 endonuclease (Feng et al. 1996; Jurka 1997; Konkel et al. 2010), only 72 elements (22.5%) contained an exact match to that sequence, whereas the majority did not, further underlining that the human/primate-specific L1 endonuclease is not entirely restricted to the canonical 5′-TTTT/AA-3′ endonuclease cleavage site. In particular, the second nucleotide of the endonuclease cleavage site frequently (21.3% of elements) contained a cytosine instead of a tyrosine. In fact, 5′-TCTT/AA-3′ has been reported previously as the second most frequently observed hexa-nucleotide pattern adjacent to the primary nick site associated with retrotransposon insertions in the genome (Jurka 1997) and in vitro (Morrish et al. 2002), consistent with our data set.
Fig. 3.—
Distribution of endonuclease cleavage sites for 322 unique Alu elements as a WebLogo diagram. The nucleotide frequency at each of the six positions is graphically proportional to the height of each letter.
All 322 analyzed Alu elements were 3′ intact displaying A-rich tails of varying lengths, and nearly all (320) were surrounded by TSDs consistent with the target-primed reverse transcription (TPRT) integration mechanism using L1 endonuclease and reverse transcriptase (Luan et al. 1993). We found no evidence that any of these Alu insertions integrated through a recombination event, indicating that the original calling algorithm selected for TPRT events. Two exon-targeted Alu insertions, #30 and #40 (see supplementary file S1, Supplementary Material online), lacked clearly defined TSDs, but were also severely 5′ truncated, 67 and 160 bp, respectively. The most likely endonuclease cleavage site was determined to be 5′-TTTT/AA-3′ in both cases. Thus, all Alu insertions, including the truncated ones, in the data set are consistent with proper insertion events generated through the enzymatic machinery of L1. Compared with the chimpanzee genome [panTro4], the integration of exon-targeted Alu elements #30 and #40 appears to be combined with short deletions of A-T rich stretches of approximately 19 and 10 bp, respectively, obscuring the TSDs during these 5′ truncated insertion events. In total, 14% (45 of 322) of the sequenced Alu insertions were 5′ truncated as defined here as having a start position after the third base pair of the Alu sequence. The severity of 5′ truncation ranged from a start position of 4 bp to a start position of 193 bp within the Alu sequence, N = 14 elements 4–25 bp; N = 20 elements 26–50 bp; N = 5 elements 21–100 bp; and N = 6 elements 101–193 bp (see supplementary files S1 and S3, Supplementary Material online).
Of the 320 sequenced Alu elements with intact TSDs, 93% (297) contained perfect TSDs, matching exactly on both the 5′- and 3′-ends of the element. Only 7% of the analyzed Alu elements displayed TSDs with mismatches between the 5′- and 3′-ends and almost all of these were single nucleotide potential mismatches based on the sequencing results. This is consistent with very recent Alu insertion events with insufficient time for decay. The TSD lengths of the analyzed elements were generally within the expected range of 6–21 bp (Moran et al. 1996; Konkel et al. 2010), with the range across our loci being 7–22 bp with the average and median both being 14 bp.
The vast majority of A-rich tails were characterized as perfect homopolymeric stretches without interruptions. Only 7% of the insertions contained A-tails with one or more nucleotide substitutions. Because nearly all Alu insertions were sequenced from PCR products, the exact size of each A-tail was impossible to determine as forward and reverse sequences typically terminated within the homopolymeric stretch of adenosines. However, we estimated the approximate size of each A-tail on the basis of the Sanger sequencing (see Materials and Methods). Sequence alignments estimated that the smallest A-tail was 13 bp and the largest was 62 bp, with an average length of approximately 29 bp. The intactness of the A-tails in addition to the relatively long size of the A-tails further supports the relatively young age of these insertions (Roy-Engel et al. 2002). Longer A-tails free of nucleotide substitutions are among the known characteristics of active source elements (Roy-Engel et al. 2002; Dewannieux and Heidmann 2005).
Another factor critical for Alu replication is the structural integrity of internal RNA Pol III promotor A and B boxes located in the left monomer (Mills et al. 2007; Bennett et al. 2008; Comeaux et al. 2009). Also important is the distance between the 3′ A-tail of the element and the first downstream Pol III TTTT termination signal, where a distance of about 15 bp or greater results in a strong decrease in retrotransposition ability (Comeaux et al. 2009). Our data set contained 23 Alu insertions (7%) in which the TTTT termination was within the 3′ TSD or immediately after, and an additional 20 loci where the first downstream termination signal was within 15 bp. Filtering these 43 loci for only full-length elements with intact left monomers (no 5′ truncation) that also have an intact A-tail greater than 20 bp in length, resulted in 28 Alu elements (about 8.7% of the data set) from seven different subfamilies having all the traditional hallmarks of source elements with the potential ability to generate new insertions. These are highlighted in green in supplementary file S3, tables S1 and S2, Supplementary Material online. We certainly do not mean to imply that other elements in the data set are necessarily unable to replicate, only that the identification of true Alu source elements is complicated and imprecise and these 28 represent our most likely source candidates from this data set.
Evolution of Alu Subfamilies
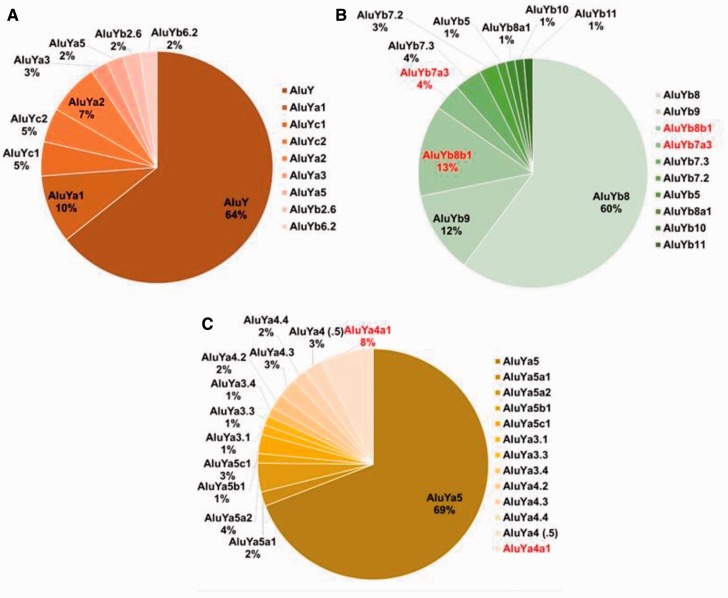
Our data set contained two Alu elements initially identified as belonging to subfamily AluYa8, but upon alignment with the consensus sequences for AluY, Ya5 and Ya8 were found to be 5′ truncated to the extent that the three diagnostic substitutions defining an Ya8 as different from an Ya5 were not available. As such, these two loci could not be authenticated as belonging to the AluYa8 subfamily. Locus 345 was the only full-length AluYa8 element in our sequenced data set and it also possesses all the known hallmarks of being retrotransposition competent, as described above. This is just one example of the benefit of careful sequence analysis using known subfamily consensus sequences. Recognizing the potential for such confounding factors, we constructed Alu sequence alignments for the three primary subfamilies identified in our data set, AluY, AluYb8 and AluYa5, comparing our Sanger-generated sequences to each subfamily consensus sequence (Jurka 2000; Jurka et al. 2005). These alignments unveiled an abundance of variation within each subfamily providing evidence for a dynamic and continuous evolution of human Alu subfamilies (fig. 4A–C).
Fig. 4.—
High-resolution distribution of Alu subfamilies following sequence alignment analysis for the three primary subfamilies identified in our data set. (A) 42 AluY elements, (B) 67 AluYb8 and 11 Yb9 elements and (C) 149 AluYa5 elements. Red font indicates subfamilies discovered in this study.
Of the 45 AluY elements sequenced, 42 were considered full length for the purpose of subfamily determination (at least 275 bp). Of these 42, nearly 80% (N = 33) were ≤2% diverged from the ancestral AluY consensus sequence, consistent with being relatively young. Although none was exact match to the AluY consensus sequence in this data set, several (N = 6) only had one substitution when the variable middle A-rich region was excluded, a G to A transition at either position 145 or 148. The four AluY loci with the G to A substitution at position 145 were loci 63, 204, 280, and 445. Based on the reported evolution of Ya-lineage diagnostic nucleotides, these four elements can be considered members of the Ya1 subfamily (Shen et al. 1991; Roy et al. 2000). The two AluY loci with a single G to A substitution at position 148 were loci 234 and 616. This single change from the AluY consensus sequence corresponds with an AluYc1 as defined previously by Roy-Engel et al. (2001) and others (Garber et al. 2005). In total, our AluY data set contained 17 full-length elements that shared this substitution, the two previously mentioned, eight with one additional substitution, including loci 156 and 357 matching the AluYc2 consensus sequence (Jurka et al. 2002) and seven with two or more additional mutations. Locus 357 also possesses all the known hallmarks of being a potential source element as described above.
We also identified four other AluY loci in our data set with evidence of subfamily evolution along the Ya-lineage. Locus 674 and exon-targeted locus 34 both have two of the five AluYa5 diagnostic substitutions, the T to C transition at position 89 (1st of 5) and the C to T GpG mutation at position 174 (4th of 5). Locus 429 also has two of the five AluYa5 diagnostic nucleotides, the 1st of 5 as described above, and the C to A transversion at position 96 (2nd of 5). Therefore, these are variants of the Ya2 lineage. In addition, Alu locus 92 is a member of the Ya3 lineage and contains three of the five Ya5 diagnostic changes, the T to C transition at position 89, the G to A at position 145, and the C to T GpG mutation at position 174. The forth, Alu locus 143 contains all five of the AluYa5 diagnostic nucleotide substitutions (supplementary file S5, Supplementary Material online, alignment file). Alu locus 143 was initially classified as a member of the AluYk3 subfamily, but we were unable to identify a known consensus sequence available for this subfamily for comparison. Also noting that this sequence did not appear to be intermittent between an AluY and an Yk4 (for which there is a consensus sequence available), we determined it was prudent to classify this locus as an AluY until further analysis, even though it was 2.9% diverged from the AluY consensus sequence. Upon alignment, Alu locus 143 contains all five of the AluYa5 diagnostic nucleotide substitutions. This has been noted in supplementary file S3, table S1, Supplementary Material online.
AluY sequence alignments also provided evidence for evolution along the Yb-lineage. Alu locus 269 has the first and third diagnostic substitutions of the Yb8 lineage, consistent with the Yb2.6 subfamily (Carroll et al. 2001). Also, locus 733, at 3% diverged from the AluY consensus sequence, contains the first six of the eight Yb8 diagnostic changes, lacking only the C to G transversion for the seventh substitution and the duplication as the eighth change near the 3′-end of the element. This is consistent with the Yb6.2 subfamily (Carroll et al. 2001). A sequence alignment of our AluY loci is available in BioEdit (Hall 1999) as supplementary file S5, Supplementary Material online. Therefore, at higher resolution the 42 full-length AluY elements obtained from Sanger sequence alignments show gradients of subfamily substructure among young AluY insertions (fig. 4A).
We also performed sequence alignments of all the AluYb8 (N = 77) and AluYb9 (N = 13) elements from our data set (supplementary file S6, Supplementary Material online). Of the 77 Yb8 elements, 67 were full length, and of the 13 Yb9 elements, 11 were considered full length (at least 275 bp) for subfamily determination. Loci 36 and 598 lack the Yb8 diagnostic T to C substitution at position 144, consistent with the previously described Yb7.2 subfamily (Carroll et al. 2001). We recovered seven Yb8 elements lacking the diagnostic CpG mutation at position 64 (loci 8, 116, 216, 262, 327, 371, and 381), consistent with the Yb7.3 subfamily (Carroll et al. 2001). Of these, locus 216 was also lacking two additional diagnostic nucleotides of the Yb8 subfamily, giving the appearance of an intermediate Yb5 element along the lineage. Locus 216 is also one of our 28 potential source elements in the data set. In addition to lacking the CpG mutation at position 64, loci 8, 116, and 327 of the Yb7 elements also shared three additional unique modifications (G to A at position 207, C to T at position 243, and G to A at position 268) which do not appear to match the consensus sequence of any previously characterized Alu subfamily. For loci 8 and 327, these are the only other substitutions. We have named these Yb7a3 following the standardized nomenclature for Alu repeats (Batzer et al. 1996) (fig. 5). A BLAT (Kent 2002) search using the locus 327 consensus sequence reveals eight exact matches in [hg19] (table 1) and zero exact matches in chimpanzee [panTro4] indicating this is a human-specific independent subfamily. These eight loci from the reference genome are generally located in high repeat regions with four of the eight insertions occurring directly into another repeat, such as an MIR, (mammalian interspersed repeat) or L1MC (an ancient L1 subfamily), they are relatively young in appearance (1.6% average divergence from theYb8 consensus sequence) and all were confirmed by sequence alignments to be exact matches to locus 327 (data not shown).
Fig. 5.—
An alignment diagram showing the consensus sequence of the three new subfamilies discovered in this study, AluYb7a3, Yb8b1, and Ya4a1. The consensus sequence for AluY is shown on the top row along with a base pair ruler. The dots represent the same nucleotide as the consensus AluY. Diagnostic mutations of AluYb8 are highlighted in yellow and the five diagnostic mutations of AluYa5 are highlighted in green. Diagnostic nucleotide changes of the new subfamilies, Yb7a3, Yb8b1 and Ya4a1 are highlighted in pink, blue and gray, respectively.
Table 1.
[hg19] Coordinates of Yb7a3 Exact Matches
| CHR | STR | Start | End | Span | Identity (%) |
|---|---|---|---|---|---|
| 6 | − | 158876118 | 158876405 | 288 | 100.00 |
| 4 | − | 64442586 | 64442873 | 288 | 100.00 |
| 17 | − | 75236003 | 75236290 | 288 | 100.00 |
| X | + | 81065458 | 81065745 | 288 | 100.00 |
| 7 | + | 31070590 | 31070877 | 288 | 100.00 |
| 2 | + | 87339719 | 87340006 | 288 | 100.00 |
| 14 | + | 33520199 | 33520486 | 288 | 100.00 |
| 1 | + | 66079155 | 66079442 | 288 | 100.00 |
Ahmed et al. (2013) recently reported the identification of three new AluYb subfamilies they termed, Yb8a1, Yb10 and Yb11. Using the consensus sequences provided in that report, we screened our data set and identified one locus corresponding to each of these three new subfamilies (see supplementary file S6, Supplementary Material online, sequence alignment). Our locus 58 (Yb8) contains the G to A substitution at position 259, consistent with Yb8a1 as defined by Ahmed et al. (2013). Our locus 325 shares this same change as well as having the G at position 174, the diagnostic ninth mutation defining Yb9 from Yb8, together now termed AluYb10 (Ahmed et al. 2013). Our locus 613 has both these mutations and in addition has the T insertion at position 200-1, diagnostic changes of the recently reported Yb11 subfamily (Wang et al. 2006; Ahmed et al. 2013). Our Yb10 and Yb11 loci are also reported in the supplementary data of Ahmed et al. (2013), Additional file 3, table S2, Supplementary Material online, as ID P1_MEI_27 and ID P1_MEI_275, respectively. However, our identification of locus 58 as belonging to the newly defined Yb8a1 subfamily does not appear to have been reported previously.
Our Yb8 sequence alignments also revealed ten other Alu insertion events, containing all eight diagnostic changes, plus a shared G to A transition at position 260 (loci 100, 264, 275, 298, 323, 348, 352, 384, 413, and exon-targeted locus 44). For loci 100, 323, 352, 384 and exon-targeted locus 44, this is the only additional substitution (supplementary file S6, Supplementary Material online). We have named these Yb8b1 (fig. 5) following the standardized nomenclature (Batzer et al. 1996) because Yb8a1 was recently used by Ahmed et al. (2013) and this represents a different single variant of Yb8. A BLAT (Kent 2002) search using locus 384 finds 25 exact matches in [hg19] (table 2) and zero exact matches in chimpanzee [panTro4], further evidence that this is a separate human-specific subfamily. As with Yb7a3, these exact matches from the reference genome are generally located in high repeat regions with 7 of the 25 insertions occurring directly into another repeat, they are relatively young in appearance (0.7% average divergence from Yb8), and all were confirmed by sequence alignments to be exact matches to locus 327 (exceptions: chr8:113225903 has an extra adenosine in the middle A-rich region; chr3:155410974 is missing the first G of the Alu element at position 1) (data not shown). A more refined breakdown of the AluYb8/9 subfamily evolution in our data set is shown in figure 4B.
Table 2.
[hg19] Coordinates of AluYb8b1 Exact Matches
| CHR | STR | Start | End | Span | Identity (%) |
|---|---|---|---|---|---|
| 4 | − | 190253873 | 190254160 | 288 | 100.00 |
| 4 | − | 167108142 | 167108429 | 288 | 100.00 |
| 4 | − | 81302578 | 81302865 | 288 | 100.00 |
| 3 | − | 54843357 | 54843644 | 288 | 100.00 |
| 3 | − | 35051698 | 35051985 | 288 | 100.00 |
| 2 | − | 155891208 | 155891495 | 288 | 100.00 |
| 19 | − | 57111577 | 57111864 | 288 | 100.00 |
| 18 | − | 14823017 | 14823304 | 288 | 100.00 |
| 16 | − | 74601478 | 74601765 | 288 | 100.00 |
| 12 | − | 114215012 | 114215299 | 288 | 100.00 |
| 12 | − | 84484513 | 84484800 | 288 | 100.00 |
| 11 | − | 113940842 | 113941129 | 288 | 100.00 |
| 1 | − | 220285488 | 220285775 | 288 | 100.00 |
| 1 | − | 8493632 | 8493919 | 288 | 100.00 |
| 7 | + | 73190208 | 73190495 | 288 | 100.00 |
| 7 | + | 38513023 | 38513310 | 288 | 100.00 |
| 5 | + | 24452853 | 24453140 | 288 | 100.00 |
| 3 | + | 3873218 | 3873505 | 288 | 100.00 |
| 21 | + | 16257078 | 16257365 | 288 | 100.00 |
| 20 | + | 33621667 | 33621954 | 288 | 100.00 |
| 18 | + | 37061647 | 37061934 | 288 | 100.00 |
| 11 | + | 58809303 | 58809590 | 288 | 100.00 |
| 1 | + | 81687798 | 81688085 | 288 | 100.00 |
| 8 | − | 113225903 | 113226191 | 289 | 100.00 |
| 3 | + | 155410974 | 155411260 | 287 | 100.00 |
The most abundant subfamily in our data set was AluYa5 (N = 156) (Batzer et al. 1990, 1996) comprising about 48% of the elements we Sanger sequenced. Of the 156 Ya5 loci, 149 were considered full length for the purpose of subfamily determination (at least 275 bp). Sequence alignments (supplementary file S7, Supplementary Material online) identified considerable substructure within the Ya5 data set suggestive of continuous ongoing evolution of Alu subfamilies. Not unexpectedly, six loci were identified as Ya5a2 elements, 251, 291, 339, 373, 583, and 729. Of these six, loci 251, 291, and 583 were exact matches to the consensus sequence for theYa5a2 subfamily as characterized by Roy et al. (2000) (supplementary file S7, Supplementary Material online). Most notably our Ya5 data set contained a total of 11 elements missing the fifth diagnostic Ya5 substitution (G to C at position 237) and instead shared a C to T CpG mutation at adjacent position 236. Six of these, loci 168, 394, 283, 350, 671, and 742 had no other random mutations, whereas the other five, loci 214, 270, 353, 368 and exon-targeted locus 52 contained additional substitutions. This does not match the consensus sequence of any previously characterized Alu subfamily. We have named these AluYa4a1 for the four diagnostic changes of an Ya5 plus one additional substitution (fig. 5). A BLAT search using locus 168 provides 13 exact matches for this consensus sequence in [hg19] (table 3) and zero exact matches in chimpanzee [panTro4] indicating this is a human-specific subfamily. In addition, loci 350 and 671, exact matches to this consensus sequence, also have the TTTT pol III termination signal within 15 bp of the A-tail, and are among our 28 candidate source elements with the potential to generate new copies. Furthermore, [hg19] locus chr10:64784930 displays a perfect A-tail 38 bp long and the TTTT termination signal within the 3′ TSD (data not shown). We also identified numerous other matches to previously identified subfamilies of the Ya-lineage (Shen et al. 1991; Roy et al. 2000; Jurka et al. 2002) summarized in table 4. A more refined illustration of the subfamily evolution within our Ya5 elements is shown as figure 4C. When we redraw the distribution of active human Alu subfamilies in our data set to incorporate the combined findings of this study, a more complex network emerges (fig. 6) providing a more realistic illustration of the dynamic evolution of Alu subfamilies.
Table 3.
[hg19] Coordinates of Ya4a1 Exact Matches
| CHR | STR | Start | End | Span | Identity (%) |
|---|---|---|---|---|---|
| X | − | 108225804 | 108226084 | 281 | 100.00 |
| 7 | − | 102476075 | 102476355 | 281 | 100.00 |
| 5 | − | 33200890 | 33201170 | 281 | 100.00 |
| 21 | − | 17522260 | 17522540 | 281 | 100.00 |
| 17 | − | 5933644 | 5933924 | 281 | 100.00 |
| 10 | − | 64784930 | 64785210 | 281 | 100.00 |
| 7 | + | 56353698 | 56353978 | 281 | 100.00 |
| 7 | + | 25044613 | 25044893 | 281 | 100.00 |
| 5 | + | 24091623 | 24091903 | 281 | 100.00 |
| 3 | + | 174705078 | 174705358 | 281 | 100.00 |
| 3 | + | 148844387 | 148844667 | 281 | 100.00 |
| 18 | + | 32661973 | 32662253 | 281 | 100.00 |
| 8 | − | 7773890 | 7774171 | 282 | 100.00 |
Table 4.
Distribution of AluYa5 Elements Based on Sequence Alignments
| Set 1 | Set 2 | Set 3 | Set 4 | Total | Reference | |
|---|---|---|---|---|---|---|
| % Divergence from Ya5 consensus | 0.0 | 0.3 | 0.4 to 0.9 | 1.0 to 1.9 | RepeatMasker, Smit et al. (1996–2010) | |
| Ya5 | 24 | 31 | 17 | 31 | 103 | Batzer et al. (1990); Batzer et al. (1996) |
| Ya5a1 | 0 | 0 | 2 | 1 | 3 | Roy et al. (2000) |
| Ya5a2 | 0 | 5 | 1 | 0 | 6 | Roy et al. (2000) |
| Ya5b1 | 0 | 2 | 0 | 0 | 2 | Roy et al. (2000) |
| Ya5c1 | 0 | 4 | 0 | 0 | 4 | Roy et al. (2000) |
| Ya3.1 | 0 | 0 | 0 | 2 | 2 | Shen et al. (1991); Roy et al. (2000) |
| Ya3.3 | 0 | 0 | 0 | 2 | 2 | Shen et al. (1991); Roy et al. (2000) |
| Ya3.4 | 0 | 0 | 0 | 2 | 2 | Shen et al. (1991); Roy et al. (2000) |
| Ya4.2 | 0 | 3 | 0 | 0 | 3 | Shen et al. (1991); Roy et al. (2000) |
| Ya4.3 | 0 | 2 | 2 | 0 | 4 | Shen et al. (1991); Roy et al. (2000) |
| Ya4.4 | 0 | 0 | 3 | 0 | 3 | Shen et al. (1991); Roy et al. (2000) |
| Ya4.5 (Ya4) | 0 | 4 | 0 | 0 | 4 | Shen et al. (1991); Roy et al. (2000); Jurka et al. (2002) |
| Ya4a1 | 0 | 0 | 5 | 6 | 11 | This study |
| Total | 24 | 51 | 30 | 44 | 149 |
Fig. 6.—
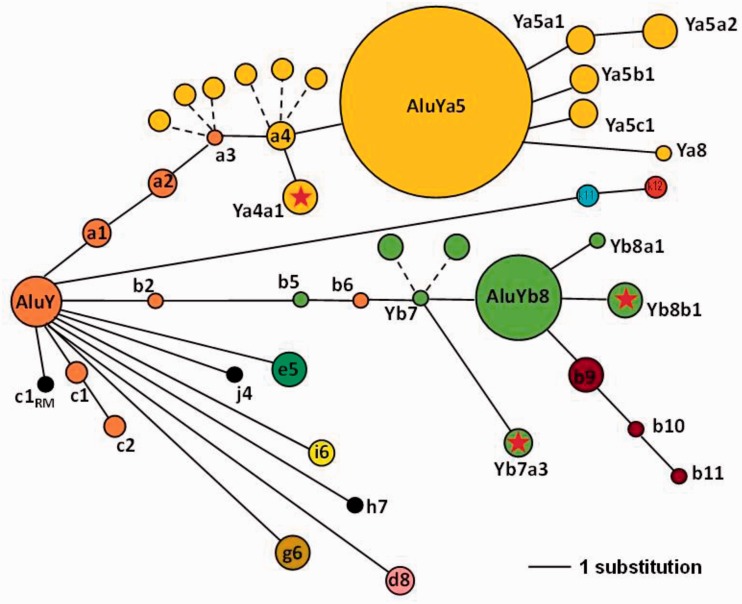
A schematic drawing of a network diagram for the high-resolution distribution of Alu subfamilies analyzed in this study to illustrate the “bush-like” evolutionary tree. The diameter of each primary node is roughly proportional to the number of elements within the subfamily (AluY: N = 27; Yb8: N = 47 and Ya5: N = 103), whereas the smallest node equals a single element (i.e., Ya8, Yb10, Yj4, etc.). The distance between nodes represents the number of nucleotide substitutions from the most recent ancestral node (solid lines). For example, Yb7a3 is three substitution lengths from the Yb7 node. Dashed lines indicate nodes with different combinations of the same number of diagnostic variants, without an additional substitution. For example, the dashed lines branching from theYa3 and Ya4 nodes depict the Ya3.1, Ya3.3, Ya3.4 or Ya4.2, Ya4.3 and Ya4.4 elements in our data set, as defined by Shen et al. (1991) and Roy et al. (2000). The dashed lines branching from the Yb7 node depict Yb7.2 and Yb7.3 elements in our data set, as defined by Carroll et al. (2001). The AluY lineage is shown in orange, Yb8 in green, and Ya5 in yellow. The RM subscript at the base of the AluYc1 node shown in black denotes this as a RepeatMasker-defined consensus sequence, as opposed to Yc1 as defined by Roy-Engel et al. (2001) and others. A red star within the node represents the three new subfamilies discovered in this study.
Discussion
The purpose of this study was not necessarily to identify new Alu subfamilies or to potentially introduce more confusion with regard to Alu nomenclature. Presently, there are instances in the literature where more than one consensus sequence has been reported for a given subfamily name, such as AluYa1 (Roy et al. 2000; Jurka et al. 2002) and Yb10 (Ahmed et al. 2013; Teixeira-Silva et al. 2013), or conversely for which a single consensus sequence has been given different names, such as Yc (Smit et al. 1996–2010) and Yd (Jurka 2000; Xing et al. 2003) as subfamilies are discovered by multiple investigators at nearly the same time. However, a systematic evaluation of our Sanger-sequenced Alu elements revealed a dynamic and continuous process of Alu subfamily evolution worthy of report and discussion. It has long been recognized that the evolution of Alu subfamilies is a complex proliferation in which the “tree” of subfamilies is more “bush-like” in appearance with many active secondary source elements sprouting new lineages (Cordaux et al. 2004; Price et al. 2004).
The findings of this study support a “bush-like” evolutionary model and are consistent with the modified “master gene” model of Alu amplification, or “stealth model” of Alu amplification where a few members remain active over time to preserve the lineage (Deininger et al. 1992; Han et al. 2005). Sequence alignment analyses of the three most prolific subfamilies in our data set, AluY, Yb8/9 and Ya5, revealed the existence of at least three new human-specific Alu subfamilies actively propagating new copies in human populations. Traditionally, a single CpG mutation, such as the diagnostic variant defining AluYa4a1, would not warrant naming a new Alu subfamily simply because CpG sites have six to ten times faster mutation rates than non-CpG sites (Labuda and Striker 1989; Batzer et al. 1990; Xing et al. 2004), increasing the potential for independently occurring random mutation events rather than authentic diagnostic variants. However, given that we identified 24 independent insertion events matching this variation of the AluYa5 lineage and at least the 11 from our data set are confirmed to be young polymorphic events, we are confident in reporting this as a new actively propagating human-specific Alu subfamily.
All the Alu insertion events identified in this study were derived from the AluY lineage. We did not find any evidence of older AluJ or AluS subfamily amplification in these human populations. This is in contrast to some previous reports (Mills et al. 2006; Hormozdiari et al. 2011). However, in these previous studies the loci were computationally ascertained using nonoverlapping data sets. Although it has been well established that Alu subfamilies greater than 20 Myr old can still have active members (Johanning et al. 2003; Salem et al. 2005), our findings suggest that in vivo retrotransposition of AluS is minimal in humans. Bennett et al. (2008) demonstrated AluS activity using a plasmid-based mobilization assay but concluded that sequence decay of older AluS elements in vivo occurred more rapidly than the propagation of new copies, supporting a model for their extinction. In general, these findings are consistent with cell culture Alu mobilization assays. In a recent study, Alu elements that were pol III-bound lacked the sequence characteristics important for retrotransposition and the majority of these pol III-bound Alu loci belonged to the older AluS and AluJ subfamilies (Oler et al. 2012). Whereas, the plasmid-based mobilization assay reported that AluY and all of the younger AluY subfamilies demonstrated activity (Bennett et al. 2008). The sequence features of the Alu insertion events identified in this study are also comparable to those recovered from tissue culture assays (Wagstaff et al. 2012).
Can the results of this study help us to estimate the number of source Alu driver elements in the human genome? We identified 28 potential source elements from our data set alone based on the traditional hallmarks associated with retrotransposition activity, two of which belong to the new AluYa4a1 subfamily. We also report the bush-like proliferation of at least 42 active Alu subfamilies from our data set (as shown in fig. 6). Given that each active Alu subfamily must have at least one source element by definition, then the human genome must contain a minimum of 42 active driver Alu elements. Further, it is estimated that about 15% of Alu subfamily members can remain active as secondary source elements continuing to generate new subfamily members (Cordaux et al. 2004). Extrapolating these numbers means that an individual human genome could realistically harbor several hundred source driver elements and potentially many more.
The purpose of this study was to report the complete sequences for a broad subset of validated Alu insertions from the Pilot 1000 Genomes Project. A comprehensive analysis of the sequence structure for 322 unique polymorphic Alu MEI events illustrates that their impact on human genome structural variation is dynamic and ongoing. Separating AluY elements into smaller and more refined subfamilies with evidence of active proliferation is undoubtedly far from complete. We can expect this to continue into the foreseeable future as detection algorithms continue to improve. Enhanced sensitivity and accuracy of MEI detection methods will undoubtedly reveal a greater number of rare population-specific and novel Alu insertions from the ongoing 1000 Genomes Project as well as from other strategies which take advantage of emerging technology in high-throughput-targeted sequencing. By understanding more about the complex patterns of Alu proliferation we can gain further insight into their impact on structural variation in human populations.
Supplementary Material
Supplementary files S1–S8 including tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Authors’ Information
J.S. conducted experiments for this project in the Department of Biological Sciences, LSU-Baton Rouge as a participant in the Howard Hughes Medical Institute (HHMI) Summer Research Program while completing a degree in Biological Sciences at Providence College in Rhode Island. J.S. is currently a graduate student in the Department of Molecular, Cellular and Developmental Biology at The Ohio State University.
Supplementary Material
Acknowledgments
Membership of the 1000 Genomes Project is listed in supplementary file S8, Supplementary Material online. The authors thank all the members of the Batzer Lab for their helpful suggestions and the 1000 Genomes Consortium. This research was supported by the National Institute of Health R01 GM59290 (M.A.B.) and U41 HG007497 (M.A.B. and M.K.K). A.B.H. and J.S. were supported in part by a grant to Louisiana State University from the Howard Hughes Medical Institute (HHMI) through the Precollege and Undergraduate Science Education Program. The authors also honor the memory of Dr Jerzy Jurka, founder of Repbase Update, for his lifelong contribution to the study of repetitive DNA. The authors declare that they have no competing interests.
Literature Cited
- Abecasis GR, et al. 2010. A map of human genome variation from population-scale sequencing. Nature 467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, et al. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ade C, Roy-Engel AM, Deininger PL. 2013. Alu elements: an intrinsic source of human genome instability. Curr Opin Virol. 3:639–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M, Li W, Liang P. 2013. Identification of three new Alu Yb subfamilies by source tracking of recently integrated Alu Yb elements. Mob DNA. 4:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. 2002. Alu repeats and human genomic diversity. Nat Rev Genet. 3:370–379. [DOI] [PubMed] [Google Scholar]
- Batzer MA, et al. 1990. Structure and variability of recently inserted Alu family members. Nucleic Acids Res. 18:6793–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, et al. 1996. Standardized nomenclature for Alu repeats. J Mol Evol. 42:3–6. [DOI] [PubMed] [Google Scholar]
- Beck CR, et al. 2010. LINE-1 retrotransposition activity in human genomes. Cell 141:1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV. 2011. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 12:187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EA, et al. 2008. Active Alu retrotransposons in the human genome. Genome Res. 18:1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan PA, Batzer MA. 2006. Retrotransposable elements and human disease. Genome Dyn. 1:104–115. [DOI] [PubMed] [Google Scholar]
- Carroll ML, et al. 2001. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J Mol Biol. 311:17–40. [DOI] [PubMed] [Google Scholar]
- Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. 2009. Diverse cis factors controlling Alu retrotransposition: what causes Alu elements to die? Genome Res. 19:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook GW, et al. 2011. Alu pair exclusions in the human genome. Mob DNA. 2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Hedges DJ, Batzer MA. 2004. Retrotransposition of Alu elements: how many sources? Trends Genet. 20:464–467. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Hedges DJ, Herke SW, Batzer MA. 2006. Estimating the retrotransposition rate of human Alu elements. Gene 373:134–137. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7:e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. 2011. Alu elements: know the SINEs. Genome Biol. 12:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. 1999. Alu repeats and human disease. Mol Genet Metab. 67:183–193. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA, Hutchison CA, 3rd, Edgell MH. 1992. Master genes in mammalian repetitive DNA amplification. Trends Genet. 8:307–311. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 35:41–48. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Heidmann T. 2005. Role of poly(A) tail length in Alu retrotransposition. Genomics 86:378–381. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905–916. [DOI] [PubMed] [Google Scholar]
- Garber RK, Hedges DJ, Herke SW, Hazard NW, Batzer MA. 2005. The Alu Yc1 subfamily: sorting the wheat from the chaff. Cytogenet Genome Res. 110:537–542. [DOI] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98. [Google Scholar]
- Han K, et al. 2005. Under the genomic radar: the stealth model of Alu amplification. Genome Res. 15:655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, et al. 2007. Alu recombination-mediated structural deletions in the chimpanzee genome. PLoS Genet. 3:1939–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, et al. 2011. Alu repeat discovery and characterization within human genomes. Genome Res. 21:840–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanning K, et al. 2003. Potential for retroposition by old Alu subfamilies. J Mol Evol. 56:658–664. [DOI] [PubMed] [Google Scholar]
- Jurka J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci U S A. 94:1872–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. 2000. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16:418–420. [DOI] [PubMed] [Google Scholar]
- Jurka J, et al. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110:462–467. [DOI] [PubMed] [Google Scholar]
- Jurka J, Krnjajic M, Kapitonov VV, Stenger JE, Kokhanyy O. 2002. Active Alu elements are passed primarily through paternal germlines. Theor Popul Biol. 61:519–530. [DOI] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel MK, Batzer MA. 2010. A mobile threat to genome stability: the impact of non-LTR retrotransposons upon the human genome. Semin Cancer Biol. 20:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel MK, Walker JA, Batzer MA. 2010. LINEs and SINEs of primate evolution. Evol Anthropol. 19:236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuda D, Striker G. 1989. Sequence conservation in Alu evolution. Nucleic Acids Res. 17:2477–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. [DOI] [PubMed] [Google Scholar]
- Lee J, Han K, Meyer TJ, Kim HS, Batzer MA. 2008. Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PLoS One 3:e4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Devine SE. 2007. Which transposable elements are active in the human genome? Trends Genet. 23:183–191. [DOI] [PubMed] [Google Scholar]
- Mills RE, et al. 2006. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet. 78:671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, et al. 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, et al. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917–927. [DOI] [PubMed] [Google Scholar]
- Morrish TA, et al. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 31:159–165. [DOI] [PubMed] [Google Scholar]
- Oler AJ, et al. 2012. Alu expression in human cell lines and their retrotranspositional potential. Mob DNA. 3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Eskin E, Pevzner PA. 2004. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 14:2245–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AM, et al. 2000. Potential gene conversion and source genes for recently integrated Alu elements. Genome Res. 10:1485–1495. [DOI] [PubMed] [Google Scholar]
- Roy-Engel, et al. 2001. Alu insertion polymorphisms for the study of human genomic diversity. Genetics 159:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel AM, et al. 2002. Active Alu element “A-tails”: size does matter. Genome Res. 12:1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. 1998. Primer3. Code available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html [DOI] [PubMed] [Google Scholar]
- Salem AH, Ray DA, Hedges DJ, Jurka J, Batzer MA. 2005. Analysis of the human Alu Ye lineage. BMC Evol Biol. 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 74:5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen SK, et al. 2006. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 79:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MR, Batzer MA, Deininger PL. 1991. Evolution of the master Alu gene(s). J Mol Evol. 33:311–320. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 1996. –2010. RepeatMasker Open-3.0 [Internet]. Available from: http://www.repeatmasker.org [Google Scholar]
- Stewart C, et al. 2011. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 7:e1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira-Silva A, Silva RM, Carneiro J, Amorim A, Azevedo L. 2013. The role of recombination in the origin and evolution of Alu subfamilies. PLoS One 8:e64884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff BJ, et al. 2012. Rescuing Alu: recovery of new inserts shows LINE-1 preserves Alu activity through A-tail expansion. PLoS Genet. 8:e1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. 2006. Whole genome computational comparative genomics: a fruitful approach for ascertaining Alu insertion polymorphisms. Gene 365:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, et al. 2003. Comprehensive analysis of two Alu Yd subfamilies. J Mol Evol. 57(Suppl 1):S76–S89. [DOI] [PubMed] [Google Scholar]
- Xing J, et al. 2004. Alu element mutation spectra: molecular clocks and the effect of DNA methylation. J Mol Biol. 344:675–682. [DOI] [PubMed] [Google Scholar]
- Xing J, et al. 2009. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 19:1516-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Witherspoon DJ, Jorde LB. 2013. Mobile element biology: new possibilities with high-throughput sequencing. Trends Genet. 29:280–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.