Abstract
Eukaryotic organelles depend on nuclear genes to perpetuate their biochemical integrity. This is true for mitochondria in all eukaryotes and plastids in plants and algae. Then how do kleptoplasts, plastids that are sequestered by some sacoglossan sea slugs, survive in the animals’ digestive gland cells in the absence of the algal nucleus encoding the vast majority of organellar proteins? For almost two decades, lateral gene transfer (LGT) from algae to slugs appeared to offer a solution, but RNA-seq analysis, later supported by genome sequencing of slug DNA, failed to find any evidence for such LGT events. Yet, isolated reports continue to be published and are readily discussed by the popular press and social media, making the data on LGT and its support for kleptoplast longevity appear controversial. However, when we take a sober look at the methods used, we realize that caution is warranted in how the results are interpreted. There is no evidence that the evolution of kleptoplasty in sea slugs involves LGT events. Based on what we know about photosystem maintenance in embryophyte plastids, we assume kleptoplasts depend on nuclear genes. However, studies have shown that some isolated algal plastids are, by nature, more robust than those of land plants. The evolution of kleptoplasty in green sea slugs involves many promising and unexplored phenomena, but there is no evidence that any of these require the expression of slug genes of algal origin.
Keywords: lateral gene transfer, kleptoplasty, photosynthesis, plastid biology, photosynthetic sea slugs
Introduction
Sacoglossa are considered one of nature’s curiosities. Inside some of these sea slugs, plastids sequestered from algae can continue to photosynthesize for weeks, or even months, in the absence of algal nuclei (Greene 1970; Rumpho et al. 2001; Händeler et al. 2009). That is conspicuous, because when land plant plastids are isolated and removed from their cellular context they rapidly degrade (Leegood and Walker 1983; Seftor and Jensen 1986; Polanská et al. 2004; Green et al. 2005). With the description of endosymbiotic gene transfer (EGT; Martin et al. 1993) and the concomitant genome reduction the organelles experienced (Timmis et al. 2004), the prime cause for the instability of isolated plastids quickly became apparent: the majority of proteins working in plastids are nuclear-encoded and posttranslationally imported from the cytosol (McFadden 2014). Hence, the duration with which kleptoplasts are kept functional in animal cells in the absence of algae nuclei encoding a 1,000 + plastid proteins presents an obvious contradiction. This required an explanation and in 1996 (Pierce et al. 1996) it was proposed that slugs had acquired algal genes that encode proteins servicing the plastids through lateral gene transfer (LGT). Once the idea was presented, it was destined to be tested.
There Is No Evidence for Lateral Transfer of Algal Genes in Slugs
Let us first take a look at what we should expect if genes of algal origin were to play a role in kleptoplast maintenance. The slugs are sometimes referred to as “crawling leaves,” because the entire appearance of the species in question (e.g., Elysia chlorotica, Elysia timida, and Elysia viridis) is greenish and in a few cases indeed leaf-like. Scientists noticed this already more than 150 years ago. They were particularly intrigued by the digestive tubules that pervade almost the entire body, sometimes including the head, and whose cells house the kleptoplasts (fig. 1a). Based on what we know about the biology of photosynthesizing cells (in plants, algae, and cyanobacteria), we must predict that transcripts of algal origin, which are supposed to maintain kleptoplast integrity in the slugs, are abundant. Yet, among all RNA sequencing data available for several species (Rumpho et al. 2011; Wägele et al. 2011; Pierce et al. 2012; de Vries et al. 2015), sequencing reads pointing toward transcripts from genes of LGT origin remain close to zero. In fact, they remain well below the counts representing obvious contamination (fig. 2a and b).
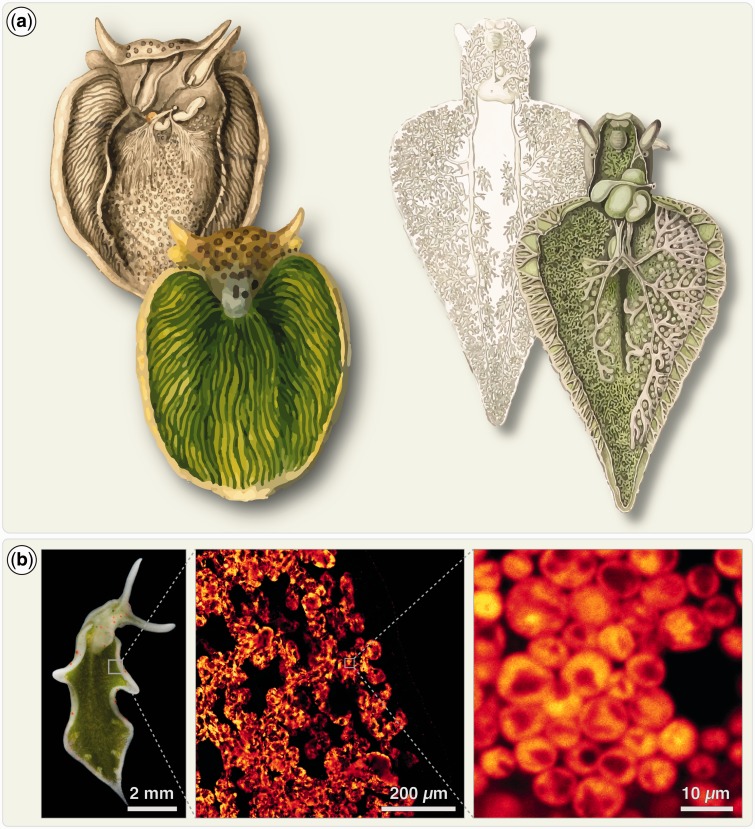
Fig. 1.—
Sacoglossan slugs can house millions of kleptoplasts. (a) Shown are two of the earliest depictions of sacoglossan slugs and their “green” digestive tubules that can pervade the entire body. On the left a drawing by van Hasselt from 1824 showing Plakobranchus ocellatus and on the right a drawing of Elysia viridis by J. Thomas from 1852. Both demonstrate that an extensive digestive system, able to house millions of kleptoplasts, is not limited to Elysia chlorotica. Note how the digestive tubules of E. viridis pervade even the head of the animal. (b) The extent of stored plastids becomes apparent when viewing the slugs (here Elysia timida) under the microscope and filtering for the chlorophyll autofluorescence of the kleptoplasts (red-orange hue). In the middle, a detail of a region of the parapodia, with the individual digestive tubules being visible through the kleptoplasts’ fluorescence. Zooming in further reveals the density with which the kleptoplasts are packed into the cytosol of the cells forming the digestive tubules.
Fig. 2.—
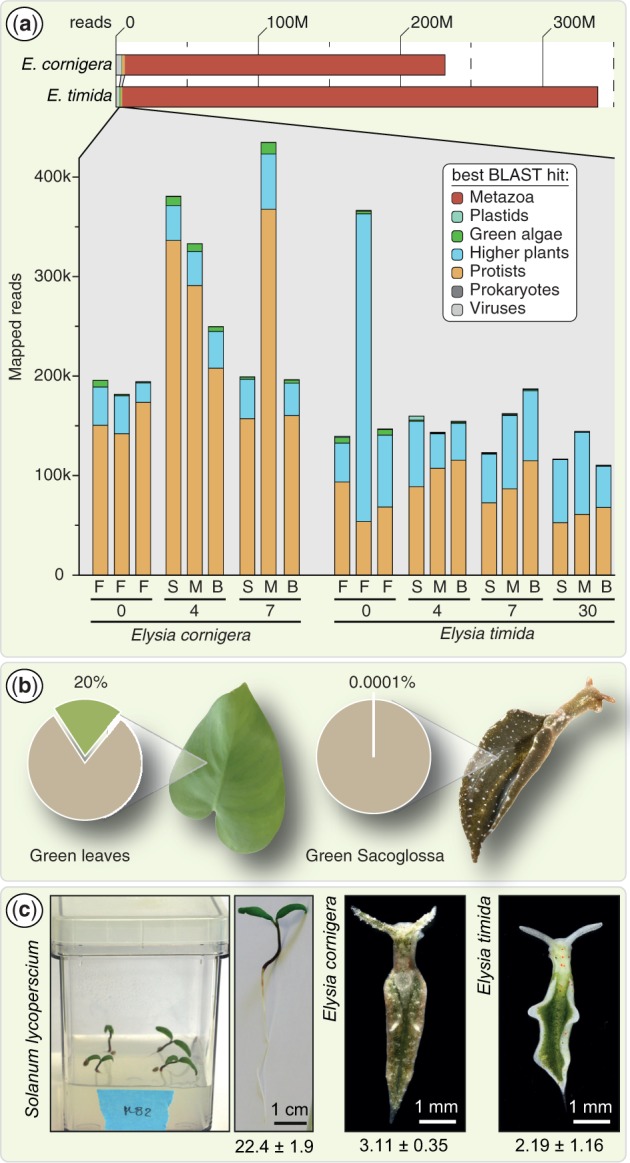
Among RNA-seq data, contaminating reads exceed reads of algal origin. (a) At the top the total number of reads (in million, M) recently sequenced for Elysia cornigera and Elysia timida (de Vries et al. 2015) are shown. Those reads were assembled into contigs and all contigs subjected to a BLAStx-based distribution analysis against RefSeq to determine their distribution among the taxonomic groups listed. Note that 1) the number of reads of protist origin in all cases exceeds those of green algal origin and that 2) in the LtR species E. timida, the amount of green algal reads declines with progressing starvation, while one would expect an elevated expression of genes supporting kleptoplasts. Slugs were freshly fed (F) and starved (S) for 4, 7, and 30 days under different conditions including monolinuron treatment blocking photosynthesis (M; 2 µg ml−1) and highlight bleaching (B; 1 h of 1,000 µE m−2s−1 once per day). (b) While the percentage of nuclear mRNA transcripts associated with photosynthesis in a green leaf ranges around 20% (Bhalerao et al. 2003), slug transcriptomes return on average around 0.0001%. If the 52 genes described by Pierce et al. (2012) were truly transferred to the slug nuclear genome, they are expressed at a level that is 200,000 times too low to support photosynthesis. (c) The chlorophyll a+b concentrations of two slug species (from de Vries et al. 2015) versus those of entire 10-day-old tomato seedlings in nmol/mg dry weight.
Single reads can easily be artifacts. In RNAseq analysis, it is common practice to filter for only those genes that are supported by a reliable number of independent reads. In the RNA-seq analysis by Pierce et al. (2012)—the last RNA-seq report published to claim expression of genes of LGT origin is relevant for kleptoplast performance—the highest read count for a single algal nuclear gene of potential LGT origin was two. Two among 98,238,204 reads. A favored argument to explain why only such few reads are detected is that “the symbiotic chloroplasts resides in only a few cells within the slugs” (Pierce et al. 2012) or that “only a relatively few cells in the slug contain plastids” (Pierce et al. 2015).
Both the appearance of the animal (fig. 1a and b) and factual numbers tell a different story. The chlorophyll content in a 6 mm long Elysia cornigera is around 3.1 nmol/mg dry weight and in a 10-day-old tomato seedling with fully developed green cotyledons it is about 22.4 nmol/mg dry weight (fig. 2c). A single chloroplast contains 2.5 × 108 chlorophyll (Chl) molecules (Stolz and Walz 1988). One nanomole of Chl thus corresponds to about 2 million chloroplasts, and hence about 6 million chloroplasts are found per mg dry weight in E. cornigera. Three nanomoles of Chl further translates into a mass of about 2.7 µg, and therefore accounts for about 3% of the total dry weight of slugs. Impressive, even if the estimations would be lower by a factor of 10. Also, if only a few cells of a photosynthetic sacoglossan slug would harbor kleptoplasts, then how would that match up with the concept that photosynthesis continues almost unabated for up to a year to support animal growth (Pierce et al. 2012)? Furthermore, among the RNA-seq data of 2012 from E. chlorotica, 4,234 reads for the plastid-encoded psbA were detected (Pierce et al. 2012). That is noteworthy, because the samples sequenced were enriched for poly(A)-tailed mRNA prior to sequencing and the plastid mRNA was copurified only as a contamination due to the high AT-content of the plastid transcripts. There is probably even more mRNA encoding psbA present than sequenced and even if not, the number of reads for this single psbA gene by far exceeds the total number of reads (111) found for the 52 genes of suggested LGT origin. Furthermore, all the reads interpreted to be of LGT origin are ≥99% identical in sequence to the algal transcripts and that would mean they are exempt from evolutionary codon adaptation in the slug’s nucleus.
Slugs analyzed are mostly collected from the wild and then grown on their food alga in open aquaria in the lab. The cultures are not axenic; they cannot be and they do not have to be for the kind of experiments that are currently performed. Contamination of the isolated RNA is unavoidable, but not a problem as long it is monitored. In the most recent transcriptomic analysis on two slug species (de Vries et al. 2015), the number of reads obtained for genes of heterotrophic protists was much higher than those for any algal nuclear gene and they were hence omitted from downstream analysis. We predict the amount of contamination in the data set of E. chlorotica (Pierce et al. 2012) to be comparable. The entire sequence data have never been made publicly available, rather only that of the few dozen genes discussed and therefore it was not possible to assess this issue in E. chlorotica. But a thought experiment is possible: we 1) do not screen the slug RNA-seq data for potential contamination and we 2) accept the presence of a few mRNA reads as evidence for functional LGT that support kleptoplast maintenance. We could then conclude that hundreds of ciliate genes support kleptoplasty in E. timida, but fail to do the same for the short-retaining E. cornigera albeit present (fig. 2a). The most rational conclusion that remains is that RNA-seq offers no support for the expression of slug nuclear genes that originate from the food alga. These slugs are not what they eat, and they eat a lot (Christa et al. 2014).
Recently, evidence for algal LGT in Sacoglossa other than sequencing data emerged: a study by Schwartz et al. (2014) used fluorescence in situ hybridization (FISH) to localize genes of algal origin among slug chromosome spreads. That report was quickly picked up by the popular press and it currently scores among the top 5% of all articles so far evaluated by Altmetric.com. Evidently, the public cares a lot about LGT in slugs. The public, however, is likely less aware that FISH analysis can be quite deceptive, for example consider the case of Chlamydomonas “basal body DNA” (Hall et al. 1989). The recent FISH analysis on E. chlorotica (Schwartz et al. 2014) also provided no controls for the specificity of the probes used (prk, actin, and rbcL) in form of Southern blots. Apart from these technical issues, FISH analysis is not a suitable tool for providing evidence for LGT. The only reliable evidence for LGT would be to demonstrate the integration of algal DNA into the context of slug chromosomes (through DNA sequencing), from where it is expressed to support the stolen organelles by the product being specifically targeted to kleptoplasts. And although independent genome data of E. chlorotica is available (Pierce et al. 2012) to the authors of the FISH analysis, it has not been used to support their concept and also challenges published slug genome data that found no evidence for algal LGTs in E. chlorotica (Bhattacharya et al. 2013).
As a last word on LGT, it should be mentioned that LGT to eukaryotes is manifest in two fundamentally different forms. First, there is gene transfer from organelles to the nucleus, or also called EGT. EGT is a continuous, ongoing process, and incontrovertibly documented in all sequenced genomes of eukaryotes (Timmis et al. 2004; Hazkani-Covo et al. 2010; Boto 2014). Second is outright LGT, where the donors are not chloroplasts or mitochondria. Newer findings show that latter, though it does occasionally occur, is extremely rare and does not manifest itself in the bigger picture of eukaryotic evolution (Ku et al. 2015). In this context, it is important to note that long-term retention of kleptoplasts evolved several times independently in sacoglossan sea slugs (Maeda et al. 2010; de Vries, Christa, et al. 2014; Christa et al. 2015). Hence, if the expression of nuclear genes of algal origin is the reason for robust kleptoplasts in one species, then the same should apply to other species as well. If all Sacoglossa retained functional kleptoplasts in a LGT-dependent manner, then they would have to be the record holders for LGT among animals, their LGT events outnumbering all other cases in animals thus far reported. Are Sacoglossa LGT magnets? Neither genome nor transcriptome data from these animals indicate that to be the case. Occam’s razor dictates favoring a less assumptive scenario.
Stable Kleptoplasts in the Absence of LGT
Since the 1970s, it is known that some plastids sequestered by the sea slugs show a remarkable independent robustness (Giles and Sarafis 1972; Trench and Ohlhorst 1976; Green et al. 2005). The best explanations we have for robust plastids are effective photoprotection mechanisms (Serôdio et al. 2014; Cruz et al. 2015), a different coding capacity of the plastid genomes in question (Rumpho et al. 2000; de Vries et al. 2013) and maybe an overall difference in the stability (half-life) of essential proteins. That plastids sequestered by the slugs are intrinsically robust is, based on current information, the most parsimonious scenario. It explains how such a broad range of slug species can perform kleptoplasty (Christa et al. 2015) and why plastids of the same source can behave identically in slug species that differ in their ability to survive food deprivation (de Vries et al. 2015). Slugs acquiring robust plastids will not automatically retain them long-term and endure starvation as recently interpreted (Pierce et al. 2015). It is of equal importance that the slugs are physiologically adapted and require to retain them functionally (de Vries, Rauch, et al. 2014). This likely depends on whether they experience food deprivation in their habitat due to seasonal variation or not (Cruz et al. 2013; Wägele and Martin 2013; de Vries et al. 2015).
The ability to sequester and maintain an entire heterologous structure of foreign origin is not restricted to sacoglossa and their plastids. For the purpose of using them as a defensive organ, some aeolidoidean sea slugs incorporate cnidocysts from their cnidarian prey to expose them on their surface (Obermann et al. 2012). Similar to the kleptoplasts, cnidocysts are first incorporated through oral feeding and as part of the regular diet. The peculiar thing is the release of the kleptoplasts from the phagosomes into the cytosol of the digestive epithelial cells. The latter appears more common when only organelles and not entire symbiotic organisms are retained by a host. The ciliate Myrionectra rubra releases transcriptionally active nuclei and plastids of its prey algae into the cytosol (Johnson et al. 2007), whereas symbiotic Chlorella algae of Paramecium bursaria or Hydra viridis remain inside a specialized digestive vacuole and isolated from the host’s cytosol (Nowack and Melkonian 2010; Fujishima and Kodama 2012). It is not known how the plastids are specifically sorted from other food particles and then released into the cytosol or really why. Does it facilitate the easier exchange of substrate and metabolites? These observations, together with how Sacoglossa deal with kleptoplast-produced toxins such as reactive oxygen species and the general differences in starvation tolerance, remain promising research topics. All of these, however, are not associated with LGTs of algal origin.
Acknowledgments
Financial support through the Deutsche Forschungsgemeinschaft to S.B.G. (GO1825/4-1) and H.W. (WA618/8 and WA618/12) and the DAAD to G.C. (P.R.I.M.E. program) is gratefully acknowledged. The authors thank the Library of the Naturalis Biodiversity Centre (Netherlands) for the rasterized scans of van Hasselt’s Plakobranchus.
Literature Cited
- Bhalerao R, et al. 2003. Gene expression in autumn leaves. Plant Physiol. 131:430–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Pelletreau KN, Price DC, Sarver KE, Rumpho ME. 2013. Genome analysis of Elysia chlorotica egg DNA provides no evidence for horizontal gene transfer into the germ line of this kleptoplastic mollusc. Mol Biol Evol. 30:1843–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boto L. 2014. Horizontal gene transfer in the acquisition of novel traits by metazoans. Proc R Soc Lond B Biol Sci. 281:20132450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christa G, et al. 2015. Phylogenetic evidence for multiple independent origins of functional kleptoplasty in Sacoglossa (Heterobranchia, Gastropoda). Org Divers Evol. 15:23–36. [Google Scholar]
- Christa G, Händeler K, Schäeberle TF, König GM, Wägele H. 2014. Identification of sequestered chloroplasts in photosynthetic and non-photosynthetic sacoglossan sea slugs (Mollusca, Gastropoda). Front Zool. 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz S, Calado R, Serôdio J, Cartaxana P. 2013. Crawling leaves: photosynthesis in sacoglossan sea slugs. J Exp Bot. 64:3999–4009. [DOI] [PubMed] [Google Scholar]
- Cruz S, et al. 2015. Photoprotection in sequestered plastids of sea slugs and respective algal sources. Sci Rep. 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Christa G, Gould SB. 2014. Plastid survival in the cytosol of animal cells. Trends Plant Sci. 19:347–350. [DOI] [PubMed] [Google Scholar]
- de Vries J, et al. 2013. Is FtsH the key to plastid longevity in sacoglossan slugs? Genome Biol Evol. 5:2540–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, et al. 2015. Comparison of sister species identifies factors underpinning plastid compatibility in green sea slugs. Proc R Soc Lond B Biol Sci. 282:20142519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Rauch C, Christa G, Gould SB. 2014. A sea slug’s guide to plastid symbiosis. Act Soc Bot Pol. 83:415–421. [Google Scholar]
- Fujishima M, Kodama Y. 2012. Endosymbionts in paramecium. Eur J Protistol. 48:124–137. [DOI] [PubMed] [Google Scholar]
- Giles KL, Sarafis V. 1972. Chloroplast survival and division in vitro. Nat New Biol. 236:56–58. [DOI] [PubMed] [Google Scholar]
- Green BJ, Fox TC, Rumpho ME. 2005. Stability of isolated algal chloroplasts that participate in a unique mollusc/kleptoplast association. Symbiosis 40:31–40. [Google Scholar]
- Greene RW. 1970. Symbiosis in sacoglossan opisthobranchs–functional capacity of symbiotic chloroplasts. Mar Biol. 7:138–142. [Google Scholar]
- Hall JL, Ramanis Z, Luck DJL. 1989. Basal body/centriolar DNA: molecular genetic studies in Chlamydomonas. Cell 59:121–132. [DOI] [PubMed] [Google Scholar]
- Händeler K, Grzymbowski YP, Krug PJ, Wägele H. 2009. Functional chloroplasts in metazoan cells—a unique evolutionary strategy in animal life. Front Zool. 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazkani-Covo E, Zeller RM, Martin W. 2010. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 6:e1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Oldach D, Delwiche C, Stoecker D. 2007. Retention of transcriptionally active cryptophyte nuclei by the ciliate Myrionecta rubra. Nature 445:426–428. [DOI] [PubMed] [Google Scholar]
- Ku C, et al. 2015. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524:427–432. [DOI] [PubMed] [Google Scholar]
- Leegood RC, Walker DA. 1983. Chloroplasts. In: Hall JL, Moore AL, editors. Isolation of membranes and organelles from plant cells. London: Academic Press; p. 85–210. [Google Scholar]
- Maeda T, Kajita T, Maruyama T, Hirano Y. 2010. Molecular phylogeny of the Sacoglossa, with a discussion of gain and loss of kleptoplasty in the evolution of the group. Biol Bull. 219:17–26. [DOI] [PubMed] [Google Scholar]
- Martin W, Brinkmann H, Savonna C, Cerff R. 1993. Evidence for a chimeric nature of nuclear genomes: eubacterial origin of eukaryotic glyceraldehyde-3-phosphate dehydrogenase genes. Proc Natl Acad Sci U S A. 90:8692–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. 2014. Origin and evolution of plastids and photosynthesis in eukaryotes. Cold Spring Harb Perspect Biol. 6:a016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack ECM, Melkonian M. 2010. Endosymbiotic associations within protists. Philos Trans R Soc Lond B Biol Sci. 365:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann D, Bickmeyer U, Wägele H. 2012. Toxicon Incorporated nematocysts in Aeolidiella stephanieae (Gastropoda, Opisthobranchia, Aeolidoidea) mature by acidi fi cation shown by the pH sensitive fl uorescing alkaloid Ageladine A. Toxicon 60:1108–1116. [DOI] [PubMed] [Google Scholar]
- Pierce SK, Biron SR, Rumpho M. 1996. Endosymbiotic chloroplasts in molluscan cells contain proteins synthesized after plastid capture. J Exp Biol. 199:2323–2330. [DOI] [PubMed] [Google Scholar]
- Pierce SK, Curtis NE, Middlebrooks ML. 2015. Sacoglossan sea slugs make routine use of photosynthesis by a variety of species-specific adaptations. Invert Biol. 10:1–13. [Google Scholar]
- Pierce SK, et al. 2012. Transcriptomic evidence for the expression of horizontally transferred algal nuclear genes in the photosynthetic sea slug, Elysia chlorotica. Mol Biol Evol. 29:1545–1556. [DOI] [PubMed] [Google Scholar]
- Polanská L, Vičánkován A, Dobrev PI, Cková IM, Vaňková R. 2004. Viability, ultrastructure and cytokinin metabolism of free and immobilized tobacco chloroplasts. Biotechnol Lett. 20:1549–1555. [DOI] [PubMed] [Google Scholar]
- Rumpho ME, Pelletreau KN, Moustafa A, Bhattacharya D. 2011. The making of a photosynthetic animal. J Exp Biol. 214:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho ME, Summer EJ, Green BJ, Fox TC, Manhart JR. 2001. Mollusc/algal chloroplast symbiosis: how can isolated chloroplasts continue to function for months in the cytosol of a sea slug in the absence of an algal nucleus? Zoology 104:303–312. [DOI] [PubMed] [Google Scholar]
- Rumpho ME, Summer EJ, Manhart JR. 2000. Solar-powered sea slugs. Mollusc/algal chloroplast symbiosis. Plant Physiol. 123:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JA, Curtis NE, Pierce SK. 2014. FISH labeling reveals a horizontally transferred algal (Vaucheria litorea) nuclear gene on a sea slug (Elysia chlorotica) chromosome. Biol Bull. 227:300–312. [DOI] [PubMed] [Google Scholar]
- Seftor RE, Jensen RG. 1986. Causes for the disappearance of photosynthetic CO2 fixation with isolated spinach chloroplasts. Plant Physiol. 81:81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serôdio J, Cruz S, Cartaxana P, Calado R. 2014. Photophysiology of kleptoplasts: photosynthetic use of light by chloroplasts living in animal cells. Philos Trans R Soc Lond B Biol Sci. 369:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz B, Walz D. 1988. The absorption spectrum of single plebs and the specific surface thylakoids. Mol Cell Biol. 7:83–88. [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Gen. 5:123–135. [DOI] [PubMed] [Google Scholar]
- Trench RK, Ohlhorst S. 1976. The stability of chloroplasts from siphonaceous algae in symbiosis with sacoglossan molluscs. New Phytol. 76:99–109. [Google Scholar]
- Wägele H, et al. 2011. Transcriptomic evidence that longevity of acquired plastids in the photosynthetic slugs Elysia timida and Plakobranchus ocellatus does not entail lateral transfer of algal nuclear genes. Mol Biol Evol. 28:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wägele H, Martin WF. 2013. Endosymbioses in sacoglossan sea slugs: plastid-bearing animals that keep photosynthetic organelles without borrowing genes. Endosymbiosis 291–324 [Google Scholar]