Abstract
As they belong to the most species-rich class of tetrapod vertebrates, birds have long been believed to possess an inferior taste system. However, the bitter taste is fundamental in birds to recognize dietary toxins (which are typically bitter) in potential food sources. To characterize the evolution of avian bitter taste receptor genes (Tas2rs) and to test whether dietary toxins have shaped the repertoire size of avian Tas2rs, we examined 48 genomes representing all but 3 avian orders. The total number of Tas2r genes was found to range from 1 in the domestic pigeon to 12 in the bar-tailed trogon, with an average of 4, which suggested that a much smaller Tas2r gene repertoire exists in birds than in other vertebrates. Furthermore, we uncovered a positive correlation between the number of putatively functional Tas2rs and the abundance of potential toxins in avian diets. Because plant products contain more toxins than animal tissues and insects release poisonous defensive secretions, we hypothesized that herbivorous and insectivorous birds may demand more functional Tas2rs than carnivorous birds feeding on noninsect animals. Our analyses appear to support this hypothesis and highlight the critical role of taste perception in birds.
Keywords: bitter taste, Tas2r, diet, birds, feeding ecology
Introduction
Sensing the external environment is of critical importance for the survival of animals. The five traditional senses in vertebrates of taste, sight, smell, sound, and touch recognize environmental cues that trigger or adjust animal behaviors accordingly. The sense of taste is specialized to evaluate the chemical components in potential food resources, which evoke appetitive or aversive reactions to ensure the ingestion of nutrients rather than poisonous substances (Yarmolinsky et al. 2009). The five basic taste modalities in vertebrates are bitter, sweet, umami, sour, and salty (Lindemann 1996; Bachmanov and Beauchamp 2007). Of them, bitter taste is dedicated to identifying bitter-tasting chemicals, such as plant alkaloids and insect defensive secretions (Garica and Hankins 1975; de Jong et al. 1991; Glendinning 1994), which are potentially poisonous to animals. Thus, bitter taste is a critical natural defense preventing the ingestion of toxic or harmful substances, which are typically bitter in nature (Garica and Hankins 1975; Glendinning 1994).
Bitter taste is conferred by the physical interaction of bitter chemicals with a group of G protein-coupled receptors (Tas2rs) that are encoded by members of the type 2 taste receptor genes (Tas2rs) (Adler et al. 2000; Chandrashekar et al. 2000; Matsunami et al. 2000). It is generally believed that the repertoire size of taste receptors is intimately associated with the external environment that animals inhabit. Indeed, the total number of Tas2rs, varying substantially from 3 in the chicken to 69 in the guinea pig, and the number of putatively functional Tas2rs, ranging from 0 in the dolphin to 51 in the frog, are positively correlated with the amount of plant materials in diets across vertebrates (Li and Zhang 2014). In addition, frequent expansions of Tas2rs in some primate lineages were also assumed to link with the development of plant feeding (Hayakawa et al. 2014). These findings agreed with the assumption that plant materials contain more bitter compounds than animal tissues (Glendinning 1994; Wang et al. 2004) and supported the hypothesis that bitter tastants have driven the evolution of the Tas2r gene repertoire in vertebrate animals (Li and Zhang 2014). Thus, bitter taste is a good model to evaluate how the chemosensory receptor gene repertoire was shaped by dietary or environmental factors. However, within vertebrates, birds were reported to possess a smaller Tas2r gene repertoire compared with mammals (Go 2006; Li and Zhang 2014; Zhao et al. 2015). Specifically, the members of Tas2rs have been examined thus far in 16 birds with fully sequenced genomes, ranging from 1 in the dove (i.e., domestic pigeon) to 12 in the hummingbird (Zhao et al. 2015), with an average of 5, whereas the Tas2rs of mammals vary in number from 10 in the dolphin (or platypus) to as many as 69 in the guinea pig, with an average of 31 (Li and Zhang 2014). Similarly, the number of putatively functional Tas2rs in birds (a mean of 3.9 and a range of 0–10) is generally lower than that in mammals (a mean of 19.5 and a range of 0–36) (Li and Zhang 2014; Zhao et al. 2015). The taste receptor genes in birds have not been well characterized thus far because the avian taste system has long been believed to be largely reduced, as inferred from a low number of taste buds and the absence of teeth (Roura et al. 2013). Indeed, chickens show an indifference to sweet stimuli in behavioral tests (Ganchrow et al. 1990), and the gene encoding the sweet taste receptor is missing from its genome (Shi and Zhang 2006). Intriguingly, some birds, such as the white-throated sparrow (Zonotrichia albicollis), were found to have 18 putatively functional Tas2rs, a number that is comparable with many mammals (Davis et al. 2010). Such a dramatic change in the Tas2r repertoire size may not be uncommon in birds with the increasing number of additional avian genomes being deciphered. To characterize the origin and evolution of avian Tas2rs and to test whether dietary toxins have shaped the repertoire size of avian Tas2rs, we examined 48 avian genomes, representing nearly all avian orders (Jarvis et al. 2014; Zhang et al. 2014), to understand the evolution of Tas2rs in birds. We found that the putatively functional Tas2r repertoire size in birds is positively correlated with the abundance of potential toxins in their diets, although birds generally carry a small number of Tas2rs.
Materials and Methods
Diet Classification
Very few bird species feed on a single type of food; the composition of avian diets is significantly influenced by food availability, seasonal changes, age, and other factors (DeGolier et al. 1999). To be consistent with earlier studies, we followed the method of Wilson (1974), which is based on the stomach contents: When a food type predominated in the stomachs of 51% or more of samples, the bird species was assigned to that food category (DeGolier et al. 1999). We did our best to search the quantitative data regarding diet composition in the literature (supplementary table S1, Supplementary Material online) and the Animal Diversity Web (http://animaldiversity.org, last accessed June 30, 2015); when such data were not available but a food type was the most abundant component in the description of food habits, we assigned the species to that food category. Because plant products may contain more toxins than animal tissues and insects release poisonous defensive secretions, we did not differentiate qualitatively among birds that feed on different plant products, whereas birds that eat animals were divided into insectivores (insect eaters) and carnivores (noninsect animal eaters). As a result, we classified birds into seven categories according to their food habits (supplementary table S1, Supplementary Material online): 1) Folivores (referring to bird species that mostly eat leaves); 2) frugivores (referring to those birds that mostly feed on fruits); 3) granivores (referring to birds that predominantly eat the seeds of plants); 4) nectarivores (referring to those that mainly feed on the sugar-rich nectar); 5) insectivores (referring to any bird species that predominantly feeds on insects); 6) carnivores (referring to any bird species that predominantly eats noninsect animals, such as piscivores); and 7) omnivores (referring to any bird species that eats insects, noninsect animals, and plant products without quantitative records).
Genome Data
A total of 48 avian genome sequences, including 45 that were recently released (Jarvis et al. 2014; Zhang et al. 2014) and 3 that were published earlier (Hillier et al. 2004; Dalloul et al. 2010; Warren et al. 2010), were retrieved from the Avian Phylogenomics Project (http://avian.genomics.cn/en/, last accessed January 30, 2015). The three genome sequences of crocodilians, representing the closest outgroup of all extant birds (Green et al. 2014), were obtained from the National Center for Biotechnology Information database with the following accession numbers: AKHW00000000 (Alligator mississippiensis), JRXG00000000 (Crocodylus porosus), and JRWT00000000 (Gavialis gangeticus).
Gene Identification
Vertebrate Tas2rs are single-exon genes that encode bitter taste receptors characterized by seven transmembrane domains (Adler et al. 2000; Chandrashekar et al. 2000; Matsunami et al. 2000). To identify the Tas2r repertoire in each of the 48 birds and in 3 outgroup species of crocodilians, we followed an earlier study (Shi and Zhang 2006) with minor modifications. First, we used full-length Tas2r protein sequences from human, mouse, zebra finch, chicken, lizard, frog, and zebra fish as queries to conduct TBLASTN searches against each of the 51 genomes, with a cutoff e-value of 1 × 10−10. Second, we filtered the redundant sequences that hit on the same genomic regions and discarded the blast hits that were shorter than 200 nt. Third, the remaining blast hits were extracted from the genomes and extended in both 5′ and 3′ directions. Those with more than 270 codons and a putative start and stop codon are intact genes; those with more than 200 nt and a putative start codon (or a putative stop codon) were considered to be partial genes, which were characterized by a truncated open reading frame (ORF) resulting from either incomplete genome sequencing or poor genomic assembly; those with more than 200 nt and an interrupted reading frame were regarded to be pseudogenes. Fourth, we used newly obtained intact genes as queries to conduct TBLASTN searches against the genomes and attempted to identify additional Tas2rs. Fifth, all full-length genes were checked to predict whether the seven transmembrane domains were intact using the TMHMM method (Sonnhammer et al. 1998), and those without any of the domains were considered to be partial genes. We additionally assessed whether the partial genes are from independent loci or not, which is particularly necessary for low-coverage genomes. If multiple partial genes from a given species share a same orthology but do not overlap, these partial genes could be a single gene due to poor genomic assembly (supplementary table S2, Supplementary Material online), as suggested in a previous study (Hayakawa et al. 2014). Synteny analysis is also helpful to assess whether partial genes are unique, as shown in supplementary table S3, Supplementary Material online. All candidate genes were ultimately verified by BLASTN searches against the GenBank database, with the best hits being the known Tas2r genes. The deduced protein sequences of all newly identified intact genes are provided in the supplementary data set S1, Supplementary Material online.
Phylogenetic Analysis
A total of 116 avian and 20 crocodilian intact Tas2rs were analyzed with an alligator V1r1 gene (GenBank: XM_006031313) as the outgroup because vertebrate V1r genes are closely related to Tas2rs among the G protein-coupled receptor genes (Matsunami et al. 2000; Shi and Zhang 2006). The 136 Tas2rs and 1 V1r1 were translated into protein sequences and were subsequently aligned with the MUSCLE program (Edgar 2004), and the resulting alignment was subjected to manual inspection in MEGA6 (Tamura et al. 2013). Phylogenetic analyses were conducted by both Neighbor-Joining (NJ) (Saitou and Nei 1987) and Bayesian Inference (BI) (Yang and Rannala 1997) approaches. The NJ phylogenetic tree was reconstructed using the protein Poisson distances (Nei and Kumar 2000) and the pairwise deletion of gap sites implemented in MEGA6 and was evaluated with 1,000 bootstrap replicates (Felsenstein 1985a). The BI tree was constructed by MrBayes version 3.1.2 (Ronquist and Huelsenbeck 2003) with 6 million generations after the best-fitting substitution model was determined by the jModelTest2 program (Darriba et al. 2012), following Bayesian information criterion (Posada and Buckley 2004).
Evolutionary Analysis
To infer the processes of gains and losses of Tas2rs across the bird phylogeny, we carried out a reconciliation analysis in NOTUNG 2.6 program (Chen et al. 2007) by comparing the species tree with the gene tree. This method works with a nonbinary gene tree where some nodes are collapsed due to weak support. The gene gains and losses were predicted by the incongruence between the species and gene trees on the basis of the parsimony principle. The species tree topology was taken from a recent study (Zhang et al. 2014), while the gene tree topology was from our BI tree (supplementary fig. S1, Supplementary Material online) where nodes with Bayesian posterior probability below 50% were collapsed, as shown in the supplementary fig. S2, Supplementary Material online.
To determine the potential impact of the feeding ecology on the evolution of the Tas2r gene repertoire size in birds, we coded each bird as 0 (carnivore) and 1 (insectivore/folivore/frugivore/granivore/nectarivore) according to the abundance of plant products or insect tissues in their diets because plant and insect tissues may have the most abundant potential toxins, whereas noninsect animals have the least. With one exception, all the studied omnivorous birds were described in the relevant literature (supplementary table S1, Supplementary Material online). The diet of each omnivorous species appeared to contain 51% or more plant and insect tissues; we therefore coded each omnivorous bird as 1. The only exception is the carnivorous red-legged seriema (del Hoyo et al. 1992), for which we were unable to determine the amount of noninsect, insect, and plant tissues in its diet; we therefore coded the red-legged seriema as 0 and 1 separately to verify the analysis. A regression analysis of Tas2r gene repertoire size against diet codes was conducted. Because our data do not fit the standard normal distribution (P < 0.05, Kolmogorov–Smirnov test), the nonparametric Spearman’s rank correlation coefficient (ρ) was used to assess the correlation. As described earlier, we used two sets of Tas2r genes to test the consistency: The first consisted of all putatively functional Tas2rs (intact and partial genes), and the second comprised all Tas2rs (intact, partial, and pseudogenes). Functional genes can reflect the physiological needs, and identifiable pseudogenes that were recently lost may also reflect the physiological needs. Indeed, both the total number and the proportion of functional olfactory receptor genes were found to be positively correlated with olfactory acuity in mammals (Rouquier et al. 2000; Gilad et al. 2004). Because the phylogenetic inertia can potentially confound comparative analyses across a group of species (Fisher and Owens 2004), we performed a phylogenetically independent contrast (PIC) analysis implemented in the package Analyses of Phylogenetics and Evolution (Paradis et al. 2004). The input tree is the species tree (Zhang et al. 2014), and the branch lengths were estimated from the divergence times among species according to a recent study (Jarvis et al. 2014) and the TimeTree database (http://www.timetree.org/, last accessed June 30, 2015). We did not include the white-throated sparrow because its divergence time from other birds is unknown.
Results
Identification of Tas2rs
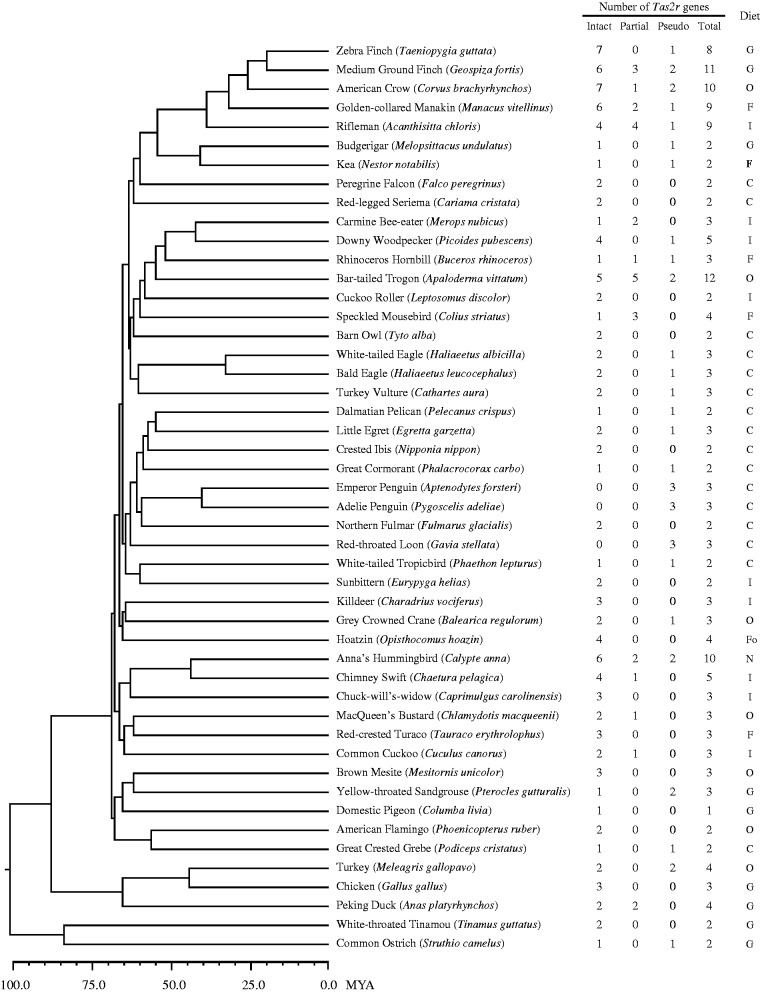
The avian Tas2r gene repertoire was characterized in few species due to the scarcity of available genome sequences (Li and Zhang 2014; Zhao et al. 2015). Recently, a total of 48 avian genome sequences were reported (Jarvis et al. 2014; Zhang et al. 2014). By using the published vertebrate Tas2rs as queries, we performed TBLASTN searches and identified Tas2rs from the genome sequences of 48 birds (fig. 1 and supplementary table S4, Supplementary Material online), representing all but three orders in the class Aves (Jarvis et al. 2014; Zhang et al. 2014). For convenience, we classified the identified Tas2rs into three categories: Intact genes (with an intact ORF and complete coding region), partial genes (with an intact ORF but partial coding region due to incomplete genome sequencing), and pseudogenes (with a disruptive ORF resulting from nonsense or frame-shifting mutations). The intact and partial genes are putatively functional, whereas the pseudogenes are possibly nonfunctional. We detected 0–7 intact genes (mean 2.4, median 2), 0–5 partial genes (mean 0.6, median 0), and 0–3 pseudogenes (mean 0.8; median 1) (fig. 1). The number of putatively functional Tas2rs in each species varied from 0 in the red-throated loon and the two penguins to 10 in the bar-tailed trogon, with a mean of 3 (fig. 1). Although all three categories of Tas2rs were counted, the gene number ranged from 1 in the domestic pigeon to 12 in the bar-tailed trogon, with an average of 4 (fig. 1). Overall, the Tas2r gene repertoire size in bird species is much smaller than that in mammals (Li and Zhang 2014). To detect whether avian Tas2rs resulted from tandem duplication as did mammalian Tas2rs, we checked the genomic location for each Tas2r gene. Indeed, some Tas2rs were found to be aligned in arrays (supplementary table S5, Supplementary Material online). Furthermore, we found that longer scaffolds tend to have more tandem duplicates of Tas2r genes (supplementary table S6, Supplementary Material online; R = 0.617, P = 0.025, Pearson correlation test), which is also a signature of tandem duplication. In addition, we similarly searched the genome sequences of 3 crocodilians, which are the closest outgroup of all extant birds, and identified 10, 6, and 11 Tas2rs (supplementary table S4, Supplementary Material online), suggesting that these reptiles have lower Tas2r gene numbers than other reptiles, such as the lizard (50 in total) (Li and Zhang 2014).
Fig. 1.—
The bitter taste receptor gene repertoires of 48 birds and their dietary preferences. Species tree and divergence times were taken from a recent study (Jarvis et al. 2014). Dietary information was from the literature and the Animal Diversity Web (supplementary table S1, Supplementary Material online). C, carnivore; I, insectivore; F, frugivore; Fo, folivore; G, granivore; N, nectarivore; O, omnivore.
Phylogenetic Reconstruction
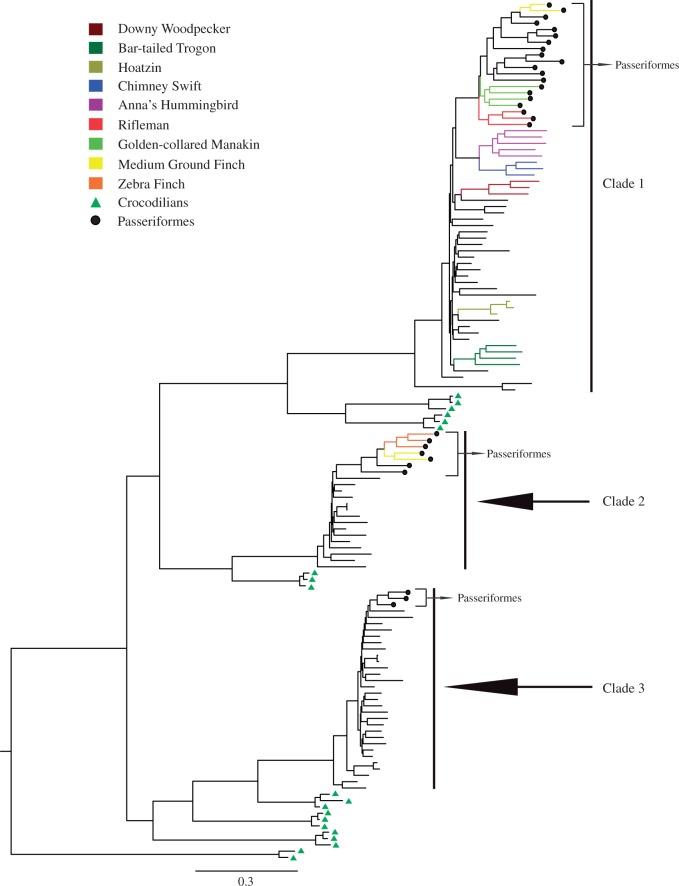
We aligned the deduced protein sequences of 136 intact Tas2rs from 45 birds (the red-throated loon and the 2 penguins have no intact Tas2r) and 3 crocodilians. The resulting alignment was used to construct phylogenetic trees with the NJ and BI approaches, and a crocodilian V1r gene was used as an outgroup. The partial genes and pseudogenes were not included in our phylogenetic analyses because most were too short to be aligned. The BI phylogenetic tree showed that all avian Tas2r genes formed three clades (fig. 2 and supplementary fig. S1, Supplementary Material online). Each of the three avian clades was allied with a group of crocodilian genes (fig. 2), suggesting that these genes appeared to have originated prior to the divergence of archosaurs (including crocodilians, dinosaurs, and birds). The first clade of avian genes was found to be enriched with putatively species-specific duplications, which are indicated by various colors (fig. 2). For example, the bar-tailed trogon has a cluster of four genes and Anna’s hummingbird is characterized by a cluster of five genes. Tests of gene conversion among paralogous genes were conducted using Sawyer’s method, as implemented in the software GENECONV (Sawyer 1989). Only two possible events of gene conversion were detected (supplementary table S7, Supplementary Material online), suggesting that such events may not have played a major role in avian Tas2r evolution. In addition, avian species from clades 2 and 3 have no species-specific gene duplications, except the medium ground finch and zebra finch (fig. 2). After removing gaps with the pairwise-deletion option, a total of 338 informative positions were used to build NJ tree. The NJ tree shows an overall topology similar to the BI tree (supplementary fig. S2, Supplementary Material online), although many more nodes of NJ tree were weakly supported. For comparison, we also selected the complete deletion option to remove gaps in building the NJ tree, and a total of 210 codons were used. Both deletion options resulted in nearly identical tree topologies (supplementary figs. S3 and S4, Supplementary Material online).
Fig. 2.—
Evolutionary relationships of all 136 intact Tas2r genes from 48 birds and 3 crocodilians. The tree was reconstructed using the Bayesian approach with the best fitting model of GTR+I+G. Branch lengths were drawn to the scale. Putative species-specific gene duplications were marked in the branches with various colors, and members from Passeriformes were bracketed. The detailed information about species and gene names and Bayesian posterior probabilities was shown in supplementary fig. S1, Supplementary Material online, and the NJ tree showing a similar topology to this tree was provided in supplementary fig. S3, Supplementary Material online.
Evolution of the Tas2r Gene Repertoire
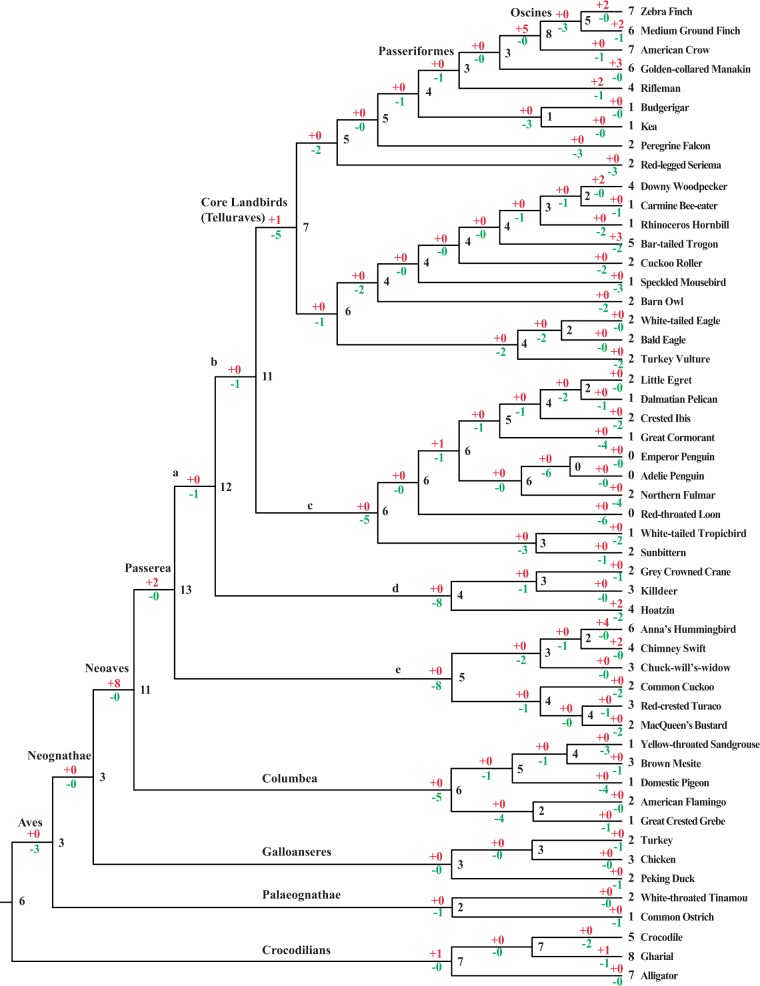
To recover the evolutionary history of intact Tas2r gene repertoire among avian species, we predicted the numbers of intact Tas2rs in avian ancestors and inferred the evolutionary changes of intact Tas2r gene numbers in the ancestral and extant species by comparing the gene tree with the species tree using the reconciliation analysis (Chen et al. 2007). We used the BI tree (fig. 2 and supplementary fig. S1, Supplementary Material online) to predict gene number changes, because it appears to be better supported than the NJ tree (supplementary fig. S3, Supplementary Material online). Weakly supported branches with Bayesian posterior probability below 50% were collapsed, and the resulting gene tree was shown in supplementary fig. S2, Supplementary Material online. We found that the number of intact Tas2rs (6 genes) in the common ancestor of birds and crocodilians was small (fig. 3). Because a reduction occurred in the turtle (11 intact Tas2rs) compared with the lizard (36 intact Tas2rs) (Li and Zhang 2014), our data suggested that the reduction of Tas2rs may have occurred before the divergence between turtles and archosaurs (including crocodilians, dinosaurs, and birds) approximately 265 Ma (Janke and Arnason 1997; Green et al. 2014). Moreover, we observed a further reduction (n = 3) in the ancestral branch of all extant birds, which resulted in only three intact Tas2rs in the common ancestor of birds (fig. 3). The majority of ancestral lineages of birds carried a small intact Tas2r gene repertoire, while the branch a, branch b, and lineages leading to Neoaves and Passerea were estimated to have an intact gene number exceeding 10 (fig. 3). Substantial reductions (n ≥ 5) were observed in the lineage leading to Telluraves, branch c, branch d, branch e, and lineages leading to Columbea, the red-throated loon, and the common ancestor of penguins. In contrast, a substantial gene gain (n = 5) was inferred in the lineage leading to Oscines, suggestive of a slightly larger number of intact Tas2rs in Oscines compared with other birds (fig. 3). Evolutionary changes of the Tas2r gene number in chickens were controversial, with both an extensive gene loss (Go 2006) and no change (Dong et al. 2009) being proposed. In our analysis, a gene number change was not observed in chickens since their separation from turkeys (fig. 3).
Fig. 3.—
Evolutionary changes of intact Tas2r gene numbers in 48 birds and 3 crocodilians. The estimated Tas2r gene numbers for ancestral lineages were shown with black, whereas the numbers of gene gains and gene losses were indicated with purple and green, respectively.
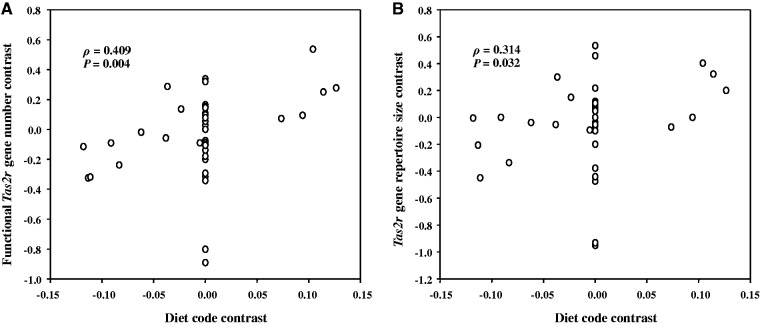
To examine whether dietary preferences influenced the evolution of avian Tas2r gene repertoires, which are generally small in size, we divided birds into carnivores, insectivores, folivores, frugivores, granivores, nectarivores, and omnivores (fig. 1) according to the Animal Diversity Web (http://animaldiversity.org/, last accessed June 30, 2015) and other references (supplementary table S1, Supplementary Material online). Because insectivorous birds feed on insects, of which many species release defensive secretions that are toxic to their predators (Weatherston and Percy 1970; Blum 1981; de Jong et al. 1991), we assumed that insectivorous birds may confront a similar amount of toxins as their herbivorous relatives, although it is well known that plant tissues tend to contain more toxins than animal tissues (Glendinning 1994; Wang et al. 2004; Li and Zhang 2014). Indeed, this assumption was supported by multiple Tas2r gene expansions in insect-eating bats rather than fruit-eating bats (Zhou et al. 2009). We predicted that carnivorous birds carry smaller Tas2r gene repertoires than other birds. We coded the dietary preference in a bird as 0 (carnivore) or 1 (insectivore/folivore/frugivore/granivore/nectarivore) under the assumption that other birds consuming more plant and insect tissues encounter more toxins than do carnivorous birds. After converting the diet codes and the Tas2r gene numbers into PICs (Felsenstein 1985b), we conducted a regression analysis. We observed a significant positive correlation between the PICs of the functional Tas2r gene numbers and those of diet codes (Spearman’s ρ = 0.409, P = 0.004; fig. 4). The same trend was revealed when the PICs of the total Tas2r numbers were correlated with the PICs of the diet codes (ρ = 0.314, P = 0.032; fig. 4). We repeated the PIC analysis while coding the red-legged seriema as 1 because this bird may be omnivorous (del Hoyo et al. 1992). The repeated analysis confirmed the correlation between diet codes and functional Tas2r gene numbers (ρ = 0.335, P = 0.021; supplementary fig. S5, Supplementary Material online) and revealed a same trend between the PICs of diet codes and those of total Tas2r gene numbers, although it was not significant (ρ = 0.235, P = 0.111; supplementary fig. S5, Supplementary Material online). To compare with an earlier vertebrate-wide study (Li and Zhang 2014), we also coded insectivores as 0, the positive correlation between the PICs of diet codes and those of functional gene numbers remains significant (ρ = 0.319, P = 0.029). Our findings clearly showed a significant positive correlation between the number of functional Tas2rs in birds and the amount of potential toxins in their diet.
Fig. 4.—
Dietary preferences impact the avian Tas2r gene repertoires. (A) PIC in putatively functional Tas2r gene number is positively correlated with that in diet preference; (B) PIC in total Tas2r gene number remains an increasing trend as PIC in diet codes increases, although it was only marginally significant. According to the amount of potential toxins in its diet, each bird was coded as 0 (carnivore), 1 (folivore), 1 (insectivore), 1 (frugivore), 1 (granivore), 1 (nectarivore), and 1 (omnivore). The Spearman’s rank correlation coefficient (ρ) with a two-tailed P value was used to evaluate the association.
Discussion
With the advent of 45 recently released avian genome sequences, we identified 215 Tas2rs from 48 avian and 3 crocodilian genomes and characterized the evolutionary history of avian Tas2rs spanning approximately 100 My (Jarvis et al. 2014). The avian Tas2r gene repertoire contains approximately 4 members, on average, ranging from 1 to 12 (fig. 1 and supplementary table S4, Supplementary Material online). Relative to most other vertebrates (Li and Zhang 2014), bird species exhibit a dramatic reduction in the Tas2r repertoire size. Furthermore, we found that carnivorous birds carry a smaller Tas2r repertoire than do other birds and observed a positive correlation between the number of putatively functional Tas2rs and the amount of potential toxins in the diet, supporting the hypothesis that dietary toxins have driven the evolution of bitter taste receptor genes, even in bird species carrying diminutive Tas2r repertoires.
The 48 avian species with whole genome sequences represent all but 3 orders of birds (Zhang et al. 2014), providing an excellent opportunity to recover an overall evolutionary history of Tas2rs across bird species. Our study unambiguously revealed a general pattern that bird species carry a small Tas2r gene repertoire relative to other vertebrates (Li and Zhang 2014). This finding is consistent with anatomical evidence, which showed fewer taste buds and a lower number of taste receptors in bird species compared with other vertebrates (Berkhoudt 1985; Mason and Clark 2000). Notably, the red-throated loon, Adelie penguin, and emperor penguin possess no functional Tas2r, which suggested a loss of bitter taste perception. Other than the 3 bird species mentioned, the remaining bird species carry at least one functional Tas2r, indicating that the bitter taste function is retained in 45 birds. We also observed a few gene clusters consisting of 2–5 genes, but we did not detect a large expansion in any bird comparable with the white-throated sparrow (Z. albicollis), which was found to possess a gene cluster encoding 18 functional Tas2r receptors (Davis et al. 2010). The lineage-specific expansion in the white-throated sparrow may not be an isolated case because we identified a gene gain (n = 5) in the ancestral lineage of Oscines (fig. 3). Indeed, we observed that the five passerine birds studied carry a larger Tas2r repertoire compared with most other bird species (fig. 1). The varying coverage of genome sequences (Zhang et al. 2014) may affect gene identification, but it is not the case for these avian genomes because we identified a low number of Tas2rs from each avian genome, irrespective of the genome coverage. Our additional analysis did not detect a correlation between the fraction of partial genes and contig N50 length (ρ = 0.141, P = 0.374; supplementary fig. S6, Supplementary Material online), possibly because birds typically have fewer partial Tas2r genes (mean 0.6, median 0) and most birds (35 out of 48) have no partial Tas2rs (fig. 1). However, the small Tas2r repertoires in birds do not necessarily mean a reduced importance of bitter taste, which could be compensated for by either the recognition of more bitter compounds or the development of novel taste receptors. For example, all chicken and turkey Tas2r receptors were able to recognize a wide range of bitter chemicals (Behrens et al. 2014); the hummingbird repurposed the ancestral umami receptor to compensate for the loss of Tas1r2, which encodes a canonical sweet receptor (Zhao et al. 2003; Baldwin et al. 2014). Despite this, birds appear to have a less developed sense of bitter taste than mammals, as a higher number of Tas2rs allows the evolution of more specialized bitter taste receptors (Behrens et al. 2014).
Evolution of the narrow Tas2r gene repertoires in birds still reflects the changes in dietary preferences, with a positive correlation between the functional Tas2r gene number and abundance of potential toxins in the diet. Because herbivorous birds consume plant products that typically contain more toxins than animal tissues and insectivorous birds feed on insects that may release defensive secretions toxic to birds, both herbivorous and insectivorous birds are expected to require more Tas2rs than carnivorous birds eating noninsect animals. Our present findings appear to support the expectation that dietary toxins shaped the Tas2r gene repertoires in birds. In addition, we also observed some cases of discrepancies between the gene number and food habit. For example, three birds clearly have a diet consisting of potential toxins, yet have only 1 intact Tas2r and 3 in total (carmine bee-eater), and 2 intact Tas2rs and 3 in total (common cuckoo). These discrepancies may result from the narrowness of their diets, as proposed in vampire bats (Zhao et al. 2010; Hong and Zhao 2014). Future studies are needed to evaluate other ecological factors that are potentially involved.
Consistent with the observation across all of the vertebrates examined (Li and Zhang 2014), we observed a similar pattern in birds, a subgroup of vertebrates, suggesting that diet impacts Tas2r evolution at both large and small scales. It would be interesting to measure, at a smaller scale, the tuning properties of Tas2r receptors in populations or closely related species with variations in bitter taste ability. In contrast, a larger genome size cannot predict more Tas2rs in birds because all birds examined have similar genome sizes, ranging from 1.05 to 1.26 Gb (Zhang et al. 2014). However, other than diet, additional driving forces must be involved in shaping Tas2r diversity. For instance, all but one Tas2rs were pseudogenes in both toothed and baleen whales (Feng et al. 2014; Kishida et al. 2015), possibly driven by the high concentration of sodium in the ocean, the feeding behavior of swallowing food whole, and the dietary switch from plants to meat in ancient whales (Feng et al. 2014); while the two Tas2rs are intact in their outgroup species, both genes were pseudogenized in the common ancestor of all extant penguins, which may result from the extremely cold Antarctic (Zhao et al. 2015). All modern bird species lack teeth and swallow food without mastication (Meredith et al. 2014), and hence, this feeding behavior should not account for the Tas2r evolution in the case of bird species. In addition to diet selection, however, extraoral functions (e.g., in gastrointestinal tract or respiratory epithelium) (Wu et al. 2002; Finger et al. 2003) may also drive the evolution of bitter taste receptor genes in birds, a hypothesis that awaits future investigation.
Supplementary Material
Supplementary figures S1–S6, tables S1–S7, and data set S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank two anonymous referees and Wei Hong for helpful comments. This work was financially supported by National Natural Science Foundation of China (31300313) to H.Z.
Literature Cited
- Adler E, et al. 2000. A novel family of mammalian taste receptors. Cell 100:693–702. [DOI] [PubMed] [Google Scholar]
- Bachmanov AA, Beauchamp GK. 2007. Taste receptor genes. Annu Rev Nutr. 27:389–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin MW, et al. 2014. Evolution of sweet taste perception in hummingbirds by transformation of the ancestral umami receptor. Science 345:929–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M, Korsching SI, Meyerhof W. 2014. Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes. Mol Biol Evol. 31:3216–3227. [DOI] [PubMed] [Google Scholar]
- Berkhoudt H. 1985. Structure and function of avian taste recetors. In: Levy AS, McLelland J, editors. Form and function in birds III. New York: Academic press; p. 463–496. [Google Scholar]
- Blum M. 1981. Chemical defense of arthropods. New York: Academic Press. [Google Scholar]
- Chandrashekar J, et al. 2000. T2Rs function as bitter taste receptors. Cell 100:703–711. [DOI] [PubMed] [Google Scholar]
- Chen K, Durand M, Farach-Colton M. 2007. Notung: a program for dating gene duplications and optimizing gene family trees. J Comp Biol. 7:429–447. [DOI] [PubMed] [Google Scholar]
- Dalloul RA, et al. 2010. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 8:e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JK, et al. 2010. Evolution of a bitter taste receptor gene cluster in a New World sparrow. Genome Biol Evol. 2:358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong P, Holloway G, Brakefield PM, de Vos H. 1991. Chemical defence in ladybird beetles (Coccinellidae) II. Amount of reflex fluid, the alka-lois adaline and individual variation in defence in 2 spot ladybirds (Adalia bipunctata). Chemoecology 2:15–19. [Google Scholar]
- DeGolier TF, Mahoney SA, Duke GE. 1999. Relationships of avian cecal lengths to food habits, taxonomic position, and intestinal lengths. Condor 101:622–634. [Google Scholar]
- del Hoyo J, Elliott A, Sargatal J, Cabot J. 1992. Handbook of the birds of the world. Barcelona: Lynx Edicions. [Google Scholar]
- Dong D, Jones G, Zhang S. 2009. Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol Biol. 9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985a. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1985b. Phylogenies and the comparative method. Am Nat. 125:1–15. [Google Scholar]
- Feng P, Zheng JS, Rossiter SJ, Wang D, Zhao H. 2014. Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol Evol. 6:1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger TE, et al. 2003. Solitary chemoreceptor cells in the nasal cavity serve as sentinels of respiration. Proc Natl Acad Sci U S A. 100:8981–8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DO, Owens IP. 2004. The comparative method in conservation biology. Trends Ecol Evol. 19:391–398. [DOI] [PubMed] [Google Scholar]
- Ganchrow JR, Steiner JE, Bartana A. 1990. Behavioral reactions to gustatory stimuli in young chicks (Gallus gallus domesticus). Dev Psychobiol. 23:103–117. [DOI] [PubMed] [Google Scholar]
- Garica J, Hankins WG. 1975. The evolution of bitter and the acquisition of toxiphobia. In: Denton DA, Coghlan JP, editors. Olfaction and Taste V, Proceedings of the 5th International Symposium in Melbourne, Australia. New York: Academic Press; p. 39–45. [Google Scholar]
- Gilad Y, Wiebel V, Przeworski M, Lancet D, Paabo S. 2004. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glendinning JI. 1994. Is the bitter rejection response always adaptive? Physiol Behav. 56:1217–1227. [DOI] [PubMed] [Google Scholar]
- Go Y. 2006. Lineage-specific expansions and contractions of the bitter taste receptor gene repertoire in vertebrates. Mol Biol Evol. 23:964–972. [DOI] [PubMed] [Google Scholar]
- Green RE, et al. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346:1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Suzuki-Hashido N, Matsui A, Go Y. 2014. Frequent expansions of the bitter taste receptor gene repertoire during evolution of mammals in the Euarchontoglires clade. Mol Biol Evol. 31:2018–2031. [DOI] [PubMed] [Google Scholar]
- Hillier LW, et al. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716. [DOI] [PubMed] [Google Scholar]
- Hong W, Zhao H. 2014. Vampire bats exhibit evolutionary reduction of bitter taste receptor genes common to other bats. Proc Biol Sci. 281:20141079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke A, Arnason U. 1997. The complete mitochondrial genome of Alligator mississippiensis and the separation between recent archosauria (birds and crocodiles). Mol Biol Evol. 14:1266–1272. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida T, Thewissen J, Hayakawa T, Imai H, Agata K. 2015. Aquatic adaptation and the evolution of smell and taste in whales. Zool. Lett. 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhang J. 2014. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol Biol Evol. 31:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann B. 1996. Taste reception. Physiol Rev. 76:719–766. [DOI] [PubMed] [Google Scholar]
- Mason JR, Clark L. 2000. The chemical senses in birds. In: Whittow GC, editor. Sturkie’s avian physiology. 5th ed San Diego (CA): Academic press; p. 39–56. [Google Scholar]
- Matsunami H, Montmayeur JP, Buck LB. 2000. A family of candidate taste receptors in human and mouse. Nature 404:601–604. [DOI] [PubMed] [Google Scholar]
- Meredith RW, Zhang G, Gilbert MT, Jarvis ED, Springer MS. 2014. Evidence for a single loss of mineralized teeth in the common avian ancestor. Science 346:1254390. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York: Oxford University Press. [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 53:793–808. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Rouquier S, Blancher A, Giorgi D. 2000. The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates. Proc Natl Acad Sci U S A. 97:2870–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roura E, Baldwin MW, Klasing KC. 2013. The avian taste system: potential implications in poultry nutrition. Anim Feed Sci Technol. 180:1–9. [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- Sawyer S. 1989. Statistical tests for detecting gene conversion. Mol Biol Evol. 6:526–538. [DOI] [PubMed] [Google Scholar]
- Shi P, Zhang J. 2006. Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol Biol Evol. 23:292–300. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 6:175–182. [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Thomas SD, Zhang J. 2004. Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum Mol Genet. 13:2671–2678. [DOI] [PubMed] [Google Scholar]
- Warren WC, et al. 2010. The genome of a songbird. Nature 464:757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherston J, Percy JE. 1970. Arthropod defensive secretions. New York: Academic Press. [Google Scholar]
- Wilson MF. 1974. Avian community organization and habitat structure. Ecology 55:1017–1029. [Google Scholar]
- Wu SV, et al. 2002. Expression of bitter taste receptors of the T2R family in the gastrointestinal tract and enteroendocrine STC-1 cells. Proc Natl Acad Sci U S A. 99:2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. 1997. Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo Method. Mol Biol Evol. 14:717–724. [DOI] [PubMed] [Google Scholar]
- Yarmolinsky DA, Zuker CS, Ryba NJP. 2009. Common sense about taste: from mammals to insects. Cell 139:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GQ, et al. 2003. The receptors for mammalian sweet and umami taste. Cell 115:255–266. [DOI] [PubMed] [Google Scholar]
- Zhao H, Li J, Zhang J. 2015. Molecular evidence for the loss of three basic tastes in penguins. Curr Biol. 25:R141–R142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, et al. 2010. Evolution of the sweet taste receptor gene Tas1r2 in bats. Mol Biol Evol. 27:2642–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Dong D, Zhang S, Zhao H. 2009. Positive selection drives the evolution of bat bitter taste receptor genes. Biochem Genet. 47:207–215 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.