Abstract
Since their earliest days, humans have been struggling with infectious diseases. Caused by viruses, bacteria, protozoa, or even higher organisms like worms, these diseases depend critically on numerous intricate interactions between parasites and hosts, and while we have learned much about these interactions, many details are still obscure. It is evident that the combined host-parasite dynamics constitutes a complex system that involves components and processes at multiple scales of time, space, and biological organization. At one end of this hierarchy we know of individual molecules that play crucial roles for the survival of a parasite or for the response and survival of its host. At the other end, one realizes that the spread of infectious diseases by far exceeds specific locales and, due to today's easy travel of hosts carrying a multitude of organisms, can quickly reach global proportions.
The community of mathematical modelers has been addressing specific aspects of infectious diseases for a long time. Most of these efforts have focused on one or two select scales of a multi-level disease and used quite different computational approaches. This restriction to a molecular, physiological, or epidemiological level was prudent, as it has produced solid pillars of a foundation from which it might eventually be possible to launch comprehensive, multi-scale modeling efforts that make full use of the recent advances in biology and, in particular, the various high-throughput methodologies accompanying the emerging –omics revolution. This special issue contains contributions from biologists and modelers, most of whom presented and discussed their work at the workshop From within Host Dynamics to the Epidemiology of Infectious Disease, which was held at the Mathematical Biosciences Institute at Ohio State University in April 2014. These contributions highlight some of the forays into a deeper understanding of the dynamics between parasites and their hosts, and the consequences of this dynamics for the spread and treatment of infectious diseases.
Keywords: Epidemiology, Host-Parasite Interactions, Infectious Diseases, Malaria, Modeling, Systems Biology
Introduction
The need to cope with infectious diseases has always been a basic feature of the human condition, and in spite of the enormous advancements in modern medicine and hygiene, such diseases continue to be a scourge without equal. According to best estimates, malaria claimed about 600,000 deaths in 2013, among 200 million or more infected individuals in about 100 countries [1; 2]. Approximately 35 million individuals currently live with HIV/AIDS [3], about 9 million with tuberculosis [4], and 3 to 5 million with severe cases of influenza [5]. Almost 20 million Americans acquire sexually transmitted diseases each year, and infectious diseases are the leading cause of death among adults under the age of 60 [6]. In addition to the enormous pain and suffering caused by these diseases, the time spent for patient care and the economic costs due to lost work are enormous. In the United States alone, the costs of infectious diseases are about $120 billion per year, and antibiotic resistance in pathogenic bacteria incurs estimated costs of about $5 billion each year [7]. A pandemic flu outbreak in the U.S. is projected to have an economic impact of hundreds of billion dollars, even without accounting for disruptions to society and commerce [8].
These are staggering numbers that beg the question of how science may help alleviate the causes, symptoms, and consequences of infectious diseases. Of course, this question is neither new to biology and medicine, nor is it to mathematical modeling. However, it deserves to be given new attention, as the –omics revolution and the emerging field of systems biology are beginning to complement the traditional repertoire of biomedical methods and technologies with genuinely new tools and techniques that carry the potential of great progress. As with all innovations, these new tools have been applied at first to low-hanging fruit, but both fields, experimental –omics and computational systems biology, have matured to a point where one might legitimately ask whether their capabilities might be ready for a new, concerted attack on infectious diseases. This special issue, like the workshop where many of the materials described here were discussed [9], attempts to highlight some of the promising advances that are presently emerging toward this goal with respect to mathematical and computational methods. A particular case, which is in many ways representative and is addressed in several of the articles, as well as in this introduction, is malaria. Nevertheless, the same or similar scientific problems arise in many of the other infectious diseases, mutatis mutandis.
As recently as a few decades ago, mathematical modelers were frequently told that biology and medicine were too complicated for modeling. After all, diseases are complex, involving thousands of molecules and complicated interactions between pathogens, hosts, and, indeed, societies and their environments, whereas models at the time usually consisted of a handful of variables. Also, it was claimed that there was no way that a computer could ever mimic, let alone surpass, the mind of an experienced physician. At the time, the critique was justified to some degree, although it ignored, for instance, the great progress in our understanding of the spread of epidemics and of the drivers that allow a disease either to flourish or to perish. These models, often abstracted to the bare bones of disease processes and devoid of all possibly distracting details, established the fundamental structure of infectious diseases, their progression, timing, and ultimate outcome (for recent reviews, see, e.g., [10; 11]). Times have changed dramatically since the earlier epidemiological models, as computers now beat Grand Masters in chess, and biomedical information is increasing so rapidly that no physician can keep up even with a specialty subfield of medicine. As a case in point, a PubMed search for “immunology” reveals that about 100,000 papers related to the subject were published in 2013 and 2014, a number that corresponds to one new paper about every 10 minutes, day in, day out, without ceasing.
Research in recent years has brought forth plentiful new information that was obtained with methods of the traditional branches of biology and medicine, entirely novel experimental options afforded by the field of –omics, and incomparably greater computer and modeling power than just a few years ago. As a consequence, new types of questions and strategies pertaining to infectious diseases have come within the reach of computational modeling. Some of these strategies attempt to improve our generic understanding of diseases, while others address specific approaches toward specific diseases, as well as crisply targeted means of intervention at the molecular, physiological, societal, and global disease levels.
As specific case studies must involve the key particularities of the investigated disease, as well as crucial details regarding their pathogens and hosts, some of the new models have a drastically different appearance than the abstracted base models that preceded them in infectious disease research. New disease models may consist of hundreds of variables and parameters, which mandates a shift of their analytical focus from rigorous algebra and calculus to large-scale simulations of possibly important scenarios, exploratory Monte-Carlo simulations and, as some traditional biologists derogatorily used to say, “fishing expeditions” that have the goal of “seeing what's out there” and generating novel hypotheses based on it.
Thus, the field of computational disease research is in the midst of an exciting transition that permits, and indeed requires, both, pure mathematical modeling that targets the fundamental structures governing disease processes, as well as larger-scale computational approaches that pinpoint weaknesses in a pathogen's mode of attack, which might be exploited for manipulation, treatment, and possibly eradication of the disease. Adding to the excitement of the new opportunities is not only the fact that drastically new and much refined biological experiments are becoming possible, which were unthinkable just a decade ago, but also that biologists are increasingly eager to work with modelers toward a common goal. This eagerness is a tremendous asset to the field of computational systems biology, and its importance for the modeler arguably exceeds that of some of the new biological technologies, because the close interaction with subject area biologists is invaluable for the computational scientist when designing effective models. However, with the willingness to collaborate comes the challenge of truly effective communication between separate fields with different terminologies and expectations. This challenge will be revisited toward the end of this article.
As a paradigm for the challenges facing infectious disease modelers, the next section summarizes key aspects of malaria. Although much simplified, this summary will indicate how truly complex, multi-scaled, and multifaceted infectious diseases are.
Malaria as the Paradigm Infectious Disease
Malaria is a persistent and recurring infectious disease that is prevalent in close to 100 countries in Africa, Southeast Asia, and the Americas. The disease directly threatens about half of the world's population. All estimates regarding malaria naturally come with a large margin of error, and the 2014 World Malaria Report gives a range of 124 to 283 million infected individuals in 2013, most of whom were young children, and about 600,000 deaths. People of all ages lacking any or sufficient immunity are at high risk, and pregnant women comprise an especially vulnerable group that suffers severe consequences (reviewed in [2; 12]).
The disease is caused by five species of the apicomplexan parasitic protists in the genus Plasmodium: P. falciparum, P. vivax, P. malariae, P. ovale, and P. knowlesi. The two dominant parasite species affecting humans are P. falciparum, which is present worldwide, although most prevalent in Sub-Saharan Africa, and P. vivax, which is encountered primarily in Asia and South America, although it is a severe problem in at least 50 countries [13; 14]. In addition, Plasmodium knowlesi has gained increasing attention over the last ten years as a zoonotic parasite that naturally infects macaques in the forests of South East Asia but is making its way into human habitats, with thousands of cases of clinical illness on record and at least 16 deaths reported to date [15; 16; 17]. Modelling the transmission of each of these species, and accounting for frequently occurring ecological and epidemiological changes, is a major task that has been aided in recent years by novel strategies and tools using geographic information systems (GIS) and sophisticated spatial decision support systems (SDSS) [18].
The challenges in understanding the disease begin with the parasite's life cycle, which involves two hosts, namely female mosquitoes of the genus Anopheles and humans or non-human primates (NHPs) [19; 20], and a multitude of evolutionarily honed host-parasite interactions. Not all, but several other mammals, birds and reptiles can also be infected with Plasmodium parasites, but these species of Plasmodium are not infectious to humans [21; 22; 23].
Various intervention strategies, including the elimination of mosquito breeding sites, insecticide spraying, promotion of the use of protective insecticide-treated bed-nets, and improved treatments, have led to substantial reductions in the number of clinical malaria cases over the past 5 to 10 years [1; 2; 12; 24]. However, effective coverage with such interventions is still limited on a global scale, has many logistical challenges, and is not necessarily sustainable. Pharmaceutical treatments are confronted with the parasite's ability to become resistant to their modes of action; in essence, by evolving to survive in the presence of these drugs. As a result, drug resistance remains a looming global concern that prohibits the ensured effective treatment of parasitized individuals. Indeed, this issue must be continually addressed in the context of today's global malaria elimination and eradication goals and strategies, with new drug options and combination therapies being brought to the forefront [25; 26; 27]. Moreover, for malaria elimination strategies to succeed, both symptomatic individuals and asymptomatic carriers must be considered, which imposes diagnostic and treatment challenges [28; 29; 30]. Mass drug administration modelling and interdisciplinary debates have become necessary to address the utility and ethical benefits and constraints of drug treatment policies and protocols [31; 32; 33].
Malaria is a systemic illness that disturbs the normal functioning of the blood in its main roles of delivering oxygen (red blood cells) and fighting infectious agents (white blood cells), and subsequently other tissues and organs including the brain, lungs, kidneys, spleen and bone marrow [34]. Clinical symptoms attributed to malaria include fevers, chills, nausea, headache, vomiting, and muscle pain. Anemia and respiratory distress are common results, and, in the most severe cases, neurological involvement can lead to coma, and multi-organ failure can result in death [34; 35; 36; 37].
The enormous complexity of the disease is due to numerous factors, from differing disease transmission characteristics in different geographical environments, to the 60 or so existing species of Anopheles mosquitoes with different susceptibilities and efficiencies as vectors, to the manner in which the immune system responds to each species and the many diverse strains of the invading pathogen. Individual and population genetics and gene expression patterns come into play [38; 39; 40; 41; 42; 43; 44; 45; 46; 47]. The parasite undergoes continuous growth and development, throughout the different stages of its life cycle. With the chance of mutations, these may be associated with the parasite becoming resistant to drugs [48; 49; 50; 51]. The transfer and propagation of offspring within and between its hosts is requisite for its survival, and modeling efforts are of paramount importance to project and understand individual and population-based parasite loads, transmission and disease characteristics, and treatment regimens, as well as to devise new means for intervention [52; 53].
When an infected female Anopheles mosquito bites an individual, the asexual sporozoite forms of the parasite enter the human skin, blood stream and lymphatics. Innate immune responses are activated, which at present are largely unexplored [54]. Surviving sporozoites migrate to the liver, where they take temporary refuge in hepatocytes [55]. There, one sporozoite can multiply within a week to 10 days to form tens of thousands of distinctly different asexual parasite forms, called merozoites. The incubation and expansion period in the liver is asymptomatic, until the matured merozoites leave the liver in bulk and enter the bloodstream, where they invade red blood cells (RBCs) [56; 57; 58]; it is also not unusual for as many as 2 or 3 merozoites to invade, co-exist and develop in the same host RBC. The invasion of RBCs is an intricate process that can be dependent or restricted by the age or phenotype of the RBCs [47; 59] and involves a cascade of host-parasite interactions, which are different for each species [47; 59; 60; 61; 62] and many of which are ill characterized or unknown. Within the RBCs, the parasites take over the cells’ normal physiology [63; 64; 65]. They asexually replicate for about 24 to 72 hours, depending on the species, which results in eight to 32 newly formed merozoites per each infected erythrocyte. Upon maturation, the parasites are released from the infected RBCs and immediately begin the process of infecting other available RBCs in circulation and possibly in some tissue compartments; again, many of the specific details are yet to be discovered [30; 66; 67; 68]. All clinical symptoms of malaria occur as the parasites are being released from the host RBCs along with the cell's contents at this mature stage of development, which include remnant membranous materials, proteins, lipids, nucleic acids, and various unknown metabolites. Some merozoites alternatively transform inside the RBCs to form male or female gametocytes, the sexual stages of the parasite, which can infect mosquitoes when these feed on their primate hosts [69; 70].
After repeated infections over time, the human host typically acquires anti-parasite and anti-disease immunity. Even so, individuals may have recurring asymptomatic infections with parasitemia. Children gain immunological protection, but it is incomplete and not always long-lasting. Thus, adults remain susceptible to infection, and women can become especially vulnerable to malaria complications during pregnancy [71; 72; 73; 74]. These are all factors to be considered for the development, licensing and eventual use of vaccines [75]. Furthermore, for certain species (P. vivax and P. ovale), the parasite, in the form of hypnozoites, may remain dormant in the liver for weeks, months, or over a year, only to emerge at a later time and initiate a new disease cycle [60; 76; 77; 78; 79; 80].
The human or non-human primate host responds to the pathogen assault in numerous ways, and the scope and precise details of the immune response from the time the parasite enters its host remain largely unexplored and still pose many open questions (e.g., see [20; 36; 74; 81; 82; 83; 84; 85; 86; 87; 88]). Because the parasite is threatened by the immune system, it has evolved escape strategies, including the intriguing use of antigenic variation mechanisms [89; 90; 91]. Most notably, P. falciparum and P. knowlesi have large multigene families, respectively, called the var and SICAvar gene families with about 60 (var) and 100 (SICAvar) members, that encode related variant proteins with different antigenic phenotypes [92; 93; 94; 95]. This topic is discussed again further below in relation to modeling within and between host-infection dynamics.
In a simplistic view, the mechanism behind this immune evasion strategy is the following: As soon as the parasite has invaded an RBC, it begins to produce specific variant proteins that become expressed at the outer surface of the RBC. Once antibodies are produced against them, the parasite switches its variant antigen gene expression profile, such that different variant proteins are produced by the parasite and positioned at the surface of the infected host RBC membrane. By expressing different parasite-encoded antigens on the outer surface of its host cell, which the immune system does not yet recognize, the parasite gains extra time to replicate without being destroyed by the antibody mediated host immune response. In P. falciparum, the variant proteins are especially ‘sticky’ and cause the infected RBCs to become sequestered in postcapillary venules [96; 97]. These proteins have multiple domains that interact with various endothelial receptors in the vasculature, causing the infected RBCs to adhere to the walls of the venules and obstruct the normal blood flow in these vessels. This adhesion can lead to secondary effects including hypoxia, local inflammation and, possibly cerebral malaria, when the parasite becomes sequestered in the venules of the brain. High parasitemias and associated immunological or other factors arguably may also lead to the observed pathological consequences [36; 98]. It is furthermore intriguing to note that specific signals may work in concert to trigger gametocytogenesis, in essence telling the parasite to ‘jump ship’ [70; 99; 100]. Additionally, extracellular vesicles may play a role in cell-cell communication, immunity, pathogenesis, and gametocytogenesis. This field is just beginning to be investigated in malaria research [101; 102; 103].
The human or primate host responds to an infection not only through the immune system, but also with changes in the blood-forming system, which responds to the moderate to severe anemia that tends to accompany malaria [84; 104; 105]. One might intuitively expect that the loss of RBCs is due to the invasion by parasites and the subsequent bursting of these host cells. While loss of infected RBCs is certainly a factor, ten to forty times as many RBCs, infected or not, are actually cleared by macrophages in a complicated cascade of events that involve a number of molecular factors and anti-erythrocytic antibodies [106]. As a consequence, the RBC count decreases precipitously, thereby causing anemia. Under more typical circumstances of anemia, the spleen senses that the oxygen level in the blood becomes abnormally low, and the body produces the hormone erythropoietin, which stimulates the formation of new blood cells in the bone marrow. This blood cell differentiation process in the bone marrow is a complex and highly regulated system, which becomes dysregulated during malaria, thereby greatly aggravating the disease. Modelling of erythropoiesis during malaria is discussed elsewhere in this special issue [107].
This short description will have rendered it evident that malaria, just like other infectious diseases, is a multi-scale, systemic disease. Survival of both the host and the parasite requires the development of complex molecular relationships, with numerous interactions and intimate host-pathogen interdependence. The disease course affects the physiological systems of both hosts and the parasite. The normal physiology of the hosts is continuously perturbed as the parasite grows, multiplies and advances from one tissue to another, but in humans and non-human primates (NHPs) it is collectively robust enough to tolerate most instances of the disease. Severe disease and death occur in a comparatively small number of cases, and there is much to be learned about what prevents those individuals from overcoming the onslaught of the invading parasite. In addition to physiological aspects, geographical, environmental, socioeconomic, political and cultural factors affecting human health and mobility come into play, as well as transmission cycles and spatial distribution patterns of the Anopheles mosquitoes.
Modeling From Within-Host to Between-Host Dynamics
In contrast to vector-transmitted diseases like malaria, the circulation of many pathogens is often the result of “contagious hosts” shedding infectious agents into the environment. The subsequent pathogen uptake and colonization of suitable new hosts is the fundamental driver of epidemics. Due to a host's immune response, exposure to low levels of a pathogen may not result in infection. Furthermore, pathogens may be unable to find a suitable host before degradation. Therefore, the onset of an infection requires the direct or indirect (vector-driven) uptake of a minimum dose of infectious agents, such as viruses, bacteria, protozoa, or helminths. The minimum infectious dose that colonizes a host with 50% probability of success is known as ID50. It has a very large range of variation, from a minimum of a single infectious agent for, e.g., Coxiella burnetii, to more than 2x1010 for Gardnerella vaginalis [108]. Once the host has been colonized above the minimum infectious dose, the pathogen's virulence and the host-pathogen interactions determine disease course and environmental shedding, which in turn determine the dynamics of epidemics. The virulence of a pathogen responds to evolutionary forces shaped by host type and availability, environmental factors that influence between-host survival, route of intake, within-host refuges and substrates, and the host's immune response.
Under a microscope, pathogens of the same species and stage of development often look very similar. However, modern methods of genomics demonstrate very clearly that microdiversity is extensive among strains of the vast majority of pathogens. This diversity is revealed through two mechanisms: (1) Simultaneous Infection: An infection can simultaneously include several distinct pathogen genomes even of the same species and, of course, from multiple species. As a consequence, the rates of transition between hosts are not constant, but functions of the ecological competition between multiple strains and the immune system response. (2) Antigenic diversity and variation: Antigenic diversity, defined as antigenic differences between pathogens in a population, and antigenic variation, defined as the ability of a pathogen to change antigens presented to the immune system during an infection, are central to a pathogen's ability to infect previously exposed hosts and to maintain a long-term infection in the face of the host's immune response. Immune evasion facilitated by this variability is a critical factor in the dynamics of pathogen growth, and therefore, transmission. Antigenic variation has been successfully modeled in some cases [109; 110; 111; 112; 113; 114; 115].
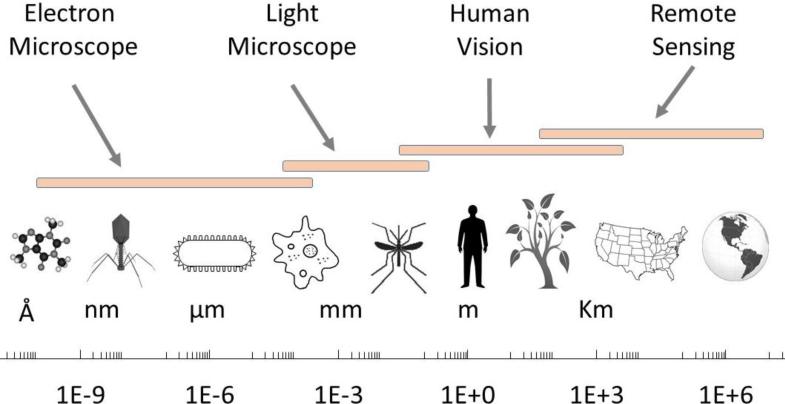
Thus, pathogens in circulation are not uniform. Instead, they constitute a complex multivariate distribution defined by the expression of different genetic, pathogenic and population dynamic traits. Similarly, the circulation of pathogens with different genotypes is multifactorial and can depend heavily on human movement dynamics, and, in some situations, vector availability and competence. Together, these anthropological, ecological, molecular, and immunological factors are fundamental drivers in the transmission of infectious disease, and their correct characterization, requires a comprehensive interdisciplinary multi-scale modeling approach. Figure 1 shows the full scale involved in many epidemiological processes, from molecules to continents.
Figure 1.
Infectious disease processes span many scales. They start with the molecular mechanisms of disease and depend on complex dispersal dynamics over scales ranging from local to intercontinental.
At the largest scale of infectious diseases, uncounted modeling studies have addressed the transmission of infections throughout populations (e.g., see [10; 11]). The traditional epidemiological approach for this purpose consists of compartmentalizing hosts into susceptible, exposed, infected, recovered (SEIR), and/or vectors into susceptible, exposed, and infected (SEI); many variations of this paradigm have been proposed over the past century. This family of models can be traced back to Sir Ronald Ross' 1916-1917 phenomenological models to study malaria. In the late 1890s, Ross had demonstrated the life-cycle of the malaria parasite in the mosquito, and in the early 1900s he published a series of papers using mathematical models to study the transmission of malaria [116; 117; 118]. In particular, Ross developed a model that explains the relationship between the number of mosquitoes and incidence of malaria in humans. This simple model was expanded by the classic work of Macdonald [119], who introduced an epidemiological compartment for exposed mosquitoes, and explicitly considered the time (~10 days) that Plasmodium spends growing and multiplying inside the mosquito before it harbors infectious sporozoite forms of the parasite in its salivary glands.
Anderson and May [120] added to Macdonald's model the ~21 days that pass before a person exposed to P. falciparum becomes infectious with gametocytes in the blood. Many models have been developed that expand Ross' model by considering factors, such as age-related differential susceptibility to malaria in human populations [120; 121], acquired immunity [122], and spatial and genetic heterogeneity of the host and parasite [123; 124].
Compartmentalized models, initiated with the study of malaria, have been successfully used to study many other disease transmission dynamics, thanks to the seminal work of Kermack and McKendrick [125]. For relatively large populations and under the assumption of free movement of all individuals, these models permit the investigation of the speed of infection and recovery, persistence of the disease, and a global assessment of which parameters in this system drive the dynamics and ultimate outcome of the disease. Indeed, these models have been the dominating paradigm in epidemiology for the past 90 years, and proved to be effective in characterizing important aspects of epidemics [126].
The distinction between susceptible and infected hosts is the foundation of these compartment models. However, in the real world this distinction might not be clear due to imperfect diagnosis methods, asymptomaticity, and variable rates of infectivity and transmission. In the particular case of malaria, instances have been reported where more than 90% of the exposed individuals were likely infected with chronic asymptomatic malaria [127], thus providing a well-distributed reservoir for the parasite. Similar situations of asymptomatic malaria still occur worldwide [128]. It has been reported that asymptomaticity is produced in some cases by specific strains [129], and it is conjectured that it plays a major role in transmission [130]. Under-reporting of asymptomatically infected individuals in public health information systems is prevalent, and since these sources are later used to calibrate epidemiological models, a situation emerges in which data are frequently insufficient to parameterize compartment models, which in turn might not be well defined when most individuals are infected.
Some previous work based on compartment models has considered the interplay between within-host and between-host dynamics. For example, Schweitzer and Anderson [131] proposed that a low level of pathogens might result in immunity, whereas a larger exposure might overwhelm the immune system, leading to transmission; in the case of malaria, this could mean a switch to the production and circulation of more gametocyte-infected RBCs. Gupta et al. [132] introduced a model in which the host population is divided into susceptible and infected, and the pathogen population is comprised of a number of strains; in this framework the rate of infection depends on the total pathogen population for each strain. Dushoff [133] proposed a model in which the rate of disease is proportional to the force of infection, that is, the rate at which uninfected individuals acquire an infectious disease. In this model, the probability of reaching different infected states depends not only on the state of the individual but also on the level of the disease in the population. Anderson [134] explicitly mentioned the possibility of quantifying changes in parasite abundance by means of within- and between-host models that had been calibrated with molecular data. Particularly striking is his observation that “the area of melding within-host and between-host parasite models is one of much interest and requires further development,” which still holds true today. Reluga et al. [135] showed that within-host dynamics of immunity may have important consequences for population-level dynamics, when coupled to non-monotone effects in the immune response to infection. More recently, Johnston et al. [69] were able to calculate the basic reproductive number (R0) linking within-host dynamics and transmission.
Obviously, the assumptions behind the original model of Kermack and McKendrick [125] were quite simplistic, but an incredible flourishing of this type of approach over the past ninety years has explored uncounted variations and twists, many of which make these models much more realistic. One particular focus has been the heterogeneity of the physical space within which the disease spreads and which leads to different, yet mutually dependent infection scenarios within different “patches” that are, however, functionally connected through the movements of hosts, pathogens, or both. Examples of this type of modeling analysis are presented in some of the papers in this special issue.
At the physiological scale, numerous mathematical models have focused on organ systems. With respect to the heart, considerable progress has been made, and modern models effectively mimic the electrical and physiological features of healthy and diseased hearts (e.g., [136; 137]). Some of these models are detailed enough to connect heart failure to genomic features, through a continuous chain of causes and effects. Similarly, strong efforts have been devoted to the physiology and pathology of the liver [138] [139]. For the context of infectious diseases, these models of organ systems are good indicators of what is achievable in terms of comprehensiveness and realism, but they may not be as important as models of the immune system, which are intimately associated with infections, but so far are vastly underdeveloped.
Molecular events are probably supported most comprehensively by experimental data, and a plethora of models has been focusing on gene regulatory, metabolic, and signaling systems. With respect to infectious diseases, the challenge at this level is that minute details may become very important, thereby requiring new models at many a juncture. For instance, the clinical and biological characteristics of a first malaria infection differ substantially from those of a second infection even with the same parasite strain. Infections strongly depend on particular parasite species, even within the same family, and, while exhibiting many similarities, often cause considerable differences in outcome among individuals and between human and non-human primate hosts. They are also greatly affected by the host's health history. Thus, while models for the spread of an infection can possibly afford to ignore these differences, a host's response systems are multifold, distributed, and adaptive to different parasites, parasite strains, and even antigenic variations within the same strain over time.
At all levels of disease modeling, the field has made enormous progress. At the same time, it is quite evident that most models so far have been focusing on a single level of organization, thereby creating distinct silos with their own questions and methods. At the highest levels, models have helped explain the modes with which infectious diseases spread, what the dominant transmission pathways are, how non-homogeneous environments affect the persistence or intrinsic disappearance of a disease, and how intervention can affect the course of a disease. At the physiological and molecular levels, models have focused on specific aspects of specific diseases, trying to understand their drivers and hoping to identify most likely points of successful intervention. Also at these levels, a few models have attempted to capture the dynamics of interactions between hosts and parasites, and co-infections with different pathogens. Other models have provided support for investigations of the evolution of diseases and of issues of co-evolution and fitness of pathogens and hosts, while yet other models have focused on generic questions of personalized health and disease [140].
Spatial Aspects of Infectious Disease: Modeling the Macro-World
Challenges abound in modeling spatial aspects of disease agent, disease vector, and disease host dynamics, particularly in the context of malaria and other vector-borne diseases (e.g., see [141; 142]). Especially important for disease transmission are spatial distributions and movement patterns for the agents, vectors and hosts. These patterns have long been recognized as important in understanding species interactions and ecological communities in spatial ecology (e.g., see [143; 144] and references therein). Consequently, it is frequently useful to regard disease agents, vectors and hosts as mobile, interacting, structured ecological species. In so doing, complications arise due to various disparities between vectors and hosts such as: (i) Life spans of vectors and agents are typically much shorter than those of hosts; (ii) Vectors may have a much more restrictive natural spatial range of dispersal than do hosts; (iii) Locally, agents and vectors may be far more numerous than hosts; (iv) There may be vast differences in body sizes between hosts and vectors; and (v) Vectors and hosts may utilize very different movement modes and primary sensory cues; e.g., hosts such as humans principally rely on visual cues to navigate the environment, whereas vectors such as mosquitoes move primarily in response to chemical concentrations. These disparities are further exacerbated by two kinds of heterogeneity: (i) spatio-temporal heterogeneity in the landscape that hosts and vectors inhabit, including such features as topography and terrain, elevation, hydrology, and climate; and (ii) heterogeneity in the range of behaviors of hosts and vectors.
Consequently, the long-standing paradigm in epidemiology [125] of using spatially homogeneous systems of ODE's such as SEIR models, which are often useful in understanding disease systems at a local level, is limited in its utility when modeling larger scale spatial effects of disease transmission. Indeed, it is the case that these disparities in components of agent-vector-host systems frequently necessitate the use of hybrid models vis- à-vis time, space and an appropriate organizational framework. Here, the term organizational framework is meant to refer to systems where one could, for example, think of hosts as individuals and think of vectors as population densities.
Spatial mathematical models in ecology, roughly speaking, fall into two classes. The first is spatially explicit, wherein spatial location is explicitly tracked. Analytic models of this type include reaction-diffusion-advection [143], integro-difference [145; 146], and integro-differential equation systems. Such models allow the tracking of population densities over spatio-temporally heterogeneous landscapes of various sizes and geographical configurations and admit a broad range of local and non-local species movement. The other class is spatially implicit. Here, locations are treated as homogeneous patches and idealized as points with some notion of connectivity among them, but without explicit reference to relative position. Spatially implicit models include metapopulation models which are widely used in the context of large potentially fragmented landscapes (e.g., see [147] and references therein) and oftentimes patch island models based on discrete diffusion. Such models may incorporate parameters that reflect the distance between patches. In some cases involving disease dynamics, discrete diffusion models can be spatially explicit (for instance, when disease transmission involves city-to-city movement). All these model types [143] can incorporate a notion of persistence of a population or ecological system over a spatial landscape in terms of a numerical threshold value akin to the notion of the basic reproduction number. A comparison of the information that can be gleaned from different spatial modeling formats when transitioning from small to large-scale environments can be found in [148].
Frequently, it is of interest to understand disease dynamics on small, intermediate and large spatial scales, and simultaneously to consider nonlinear feedback loops in the transmission process. Regarding scale transitions, it is important to consider various issues. First, natural movement scales of vectors frequently will be local. However, it is often the case that disease agents and vectors can be transported vast distances either by natural forces such as wind or by host movement (such as migrating birds in the case of West Nile virus) or via man-made devices, such as planes, trains, automobiles and boats. As a consequence, one needs not only to track populations or population densities of vectors and/or hosts at diverse particular localities, but also the network of spatio-temporal pathways linking them. Here multi-patch spatially implicit models play an important role (see, e.g., [149; 150] and references therein). Going forward, we will also need to combine “micro-scale” models with an explicit spatial representation of vector densities with spatially implicit or explicit “macro-scale” rules for connecting the local models such that each level informs the other. Second, the scale of observation affects when it is appropriate to track individuals, population densities, or some other continuous variable. Another somewhat subtle but significant consideration regards the identity of individuals [151] as they move from locale to locale. When vectors or hosts move from one locale to another, they may, on the one hand, blend into a comparable cohort in the new location; that is, they may be thought of as immigrants. On the other hand, they might retain their identities as merely visitors in the new location if they move back and forth between different locales on a regular basis.
Additionally, complicated feedback loops are created by various factors at play in the system. For example, agents (such as pathogens) may exhibit genetic diversity and may experience evolutionary changes or mutations on time scales that are very short relative to life spans of host organisms. An important category of such mutations is that of agents acquiring resistance to therapeutic drugs or vectors acquiring resistance to chemical controls. Secondly, there may be spatially heterogeneous distributions of patterns of resistance which then interact with movements of host populations. Moreover, host activity patterns (particularly in the case of humans) and disease transmission can influence each other and, again, there may be spatial patterns to such activities; for example, life styles in urban areas may be very different from rural areas. This idea has been captured in the epidemiological literature through the notion of activity spaces (e.g., [152]). Finally, all ecological processes are subject to aspects of global change.
Model Design in the –Omics World
During the decades preceding the recent revolution in high-throughput biology, a mathematical model was typically designed based on a diagram of a biological phenomenon or system. This diagram had been constructed by biologists or by the modeler from available biological information. Given the computational and informational limitations of the times, the resulting model structure typically contained between a few and maybe a dozen variables, as well as a corresponding number of interactions among them. Once the model structure was established, an attempt was made to convert kinetic, molecular, ecological and other pertinent information into specific parameter values and other settings of the model. In most cases, the available information was incomplete, requiring complementary assumptions based on biological or mathematical rationale. With all parameter values and assumed settings specified, the model was diagnosed with respect to stability, sensitivities, dynamic repertoires, and reasonableness. Subsequently, refinements and amendments were introduced to reconcile inconsistencies between data and model results. Due to the paucity of data, validation of the model was often not truly possible.
The –omics revolution has changed the overall situation from data-poor to data-rich [153] and ignited the field of systems biology. Instead of scrambling for the value of a needed kinetic parameter, many a modeler now faces the opposite situation of data overload. In fact, many experiments now generate so many raw data that it is difficult to extract information from them, especially if the noise level is high. It is obvious that the tried and true strategies of model design are critically affected by this situation. New modeling tools need to be developed and applied to the raw data in order to reduce their complexity and denoise them, before dynamic models in the traditional style can be designed from –omics data. These new models are of a different nature and fall within the overlap area of statistics, artificial intelligence, and bioinformatics. While discussed in some detail below, one paper in this special issue expands upon these topics [154].
Generically, the modeling process for –omics data could have the following structure. In the first phase, the large-scale data (“BigData”) are preprocessed. In the case of infectious diseases, these data do not necessarily come from molecular and experimental systems biology, but could also be individualized health data, epidemiological information, or public health records. In the majority of cases, though, they are expected to derive from experimental datasets, such as genomics, proteomics, metabolomics, lipidomics, glycomics, immune profiling, or signaling data.
If feasible, statistical tests are employed to determine summary metrics, trends, and risk factors, as well as general diagnostics of the computed results. These steps may be executable with standard methods of statistics, but given the enormity of some modern datasets, they are more likely to require advanced machine learning techniques. These newer techniques discern information from noise, establish complex, often nonlinear correlations or patterns among some of the measured variables, and infer associative or causal network structures. In particular, they can identify clusters of variables that behave similarly with respect to certain features or aspects. For instance, a cluster analysis might sort genes by their expression patterns in response to some stimulus or in a direct comparison between healthy and diseased cells. Correlations or clusters that can be determined with such methods yield two important streams of information. On the one hand, they allow a grouping of variables and possibly a reduction in complexity, due to the fact that it might be justified to substitute all members of the same groupings with one representative per group. On the other hand, the results from a machine learning analysis may reveal patterns within the data that had never been seen before and that can be converted into hypotheses, which might be testable in the biological laboratory or with more refined dynamic models. Such patterns could consist of yet unknown relationships between the components of a system or even of fully connected networks that suggest hierarchies among the variables. In less favorable cases it may also be possible that the machine learning analysis reveals that the degree of noise and uncertainty is so high that a mechanistic modeling analysis, or even a valid interpretation of the data, is likely not feasible.
Ideally, the computational preprocessing “cleans” the raw data, via denoising and the statistically valid elimination of outliers, and suggests different types of hypotheses. For instance, a metabolomics analysis might indicate systematic changes in the profiles of certain metabolites. The superposition of these metabolites on pathways in a database like KEGG [155] or MetaCyc [156] might furthermore suggest which metabolic systems are affected. Again under ideal conditions, corresponding data regarding the genome and proteome would reflect these changes. In most realistic cases, the supporting data will not be as comprehensive or they will not fit together quite so neatly, but they can nevertheless be explored with a dynamic model, once a hypothesis has been formed. Furthermore, such a model is able to incorporate external information that might be relevant. For example, the database BRENDA [157] might contain values or ranges of critical kinetic parameters and identify inhibitors and other modulators that could be used to design and instantiate a model of the pathway suggested by the machine learning analysis. In most practical cases, the transition from statistical and machine learning analyses to the design of mechanistic models is not automatic, and much curation and intensive investigator efforts are needed to support the data pipeline between an –omics experiment and a valid dynamic model. It is to be expected that this pipeline will be the target of much investigation in the future.
The models suggested by –omics data, combined with more traditional information, may be of many different varieties. Most obvious and prevalent are models of gene regulatory systems and metabolic pathways, but new methods like mass cytometry [158] also make it possible to analyze signaling systems in a single-cell granularity that was considered impossible just a few years ago. Similarly, appropriate data can form the basis for specialized model designs, for instance, supporting investigations of the roles and dynamics of different types of immune cells, cytokines and chemokines in the host response to infectious agents [159], or of alterations in the hematopoietic system of the host that are effected by a disease like malaria.
No matter how straightforward or complicated is the translation of raw data into a dynamic model, the result, if it can be validated, can be very valuable. Namely, instead of looking at noisy snapshots in the form of raw data, one has achieved an integration of often heterogeneous data into a computational construct that permits numerous simple, fast, and cheap explorations. In contrast to new laboratory experiments, these computational model analyses can easily screen thousands of scenarios that represent slightly changed conditions, combinations of perturbations, and responses to interventions, and facilitate the personalization of health and disease models [160]. Valid models may also be scanned for particularly sensitive chokepoints that could become the targets of pharmaceutical treatments or for the optimization of combination treatments.
Scale-Bridging Modeling Approaches
The historical emergence of silos with different classes of models is understandable but also raises the question of whether models for different aspects should intercalate and how “silo-bridging” model integration could possibly be accomplished. Clearly, infectious diseases are driven by processes that occur at distinct spatial and temporal levels ranging from molecular events and host-pathogen interactions to global epidemiology and public health. Yet, the multitudinous processes at all these scales depend on each other, and one should ask whether it might be feasible, or even desirable, to create all-comprehensive “supermodels” that encompass the different levels of biological organization. Has the time come to declare such models a hallmark goal for the foreseeable future of the systems biology of infectious diseases? Otherwise, if we do not concatenate models functionally by integrating them into superstructures, how can a model at one scale effectively inform a model at another scale? Will it ultimately be sufficient to restrict each model to one or two levels and to develop off-line bridging mechanisms with which one model might inform another? Time will tell.
At present, these questions are almost moot as the development of such scale-bridging, all-comprehensive models is not feasible and realistic models integrating over all scales are out of reach for numerous technical reasons. For instance, it is clear that not every molecular process can (or should) be retained in a physiological organ model, let alone a whole-host or epidemiological model. A traditional strategy for addressing the multi-scale challenge is the separation of time scales, which assumes that very slow processes are essentially constant at a fast scale and that very fast processes are essentially always in steady state at a slow time scale. The consequence of these assumptions is that some of the differential equations in a dynamical model are reduced to algebraic equations or even to singular parameter values. It is unclear whether or to what degree this strategy is feasible and optimal in the case of infectious diseases.
A related question is whether the mathematical structures of a multi-scale model are necessarily specific for a single level or scale or whether it might be possible to reuse models from other levels. For instance, would it be possible to represent the interactions between neutrophils and bacteria as a predator-prey model? Is it useful to employ “canonical” models whose structures are independent of scale [161; 162; 163; 164; 165; 166]?
Finally, the recent literature has documented intense discussions about biological design and operating principles [167]. Is it reasonable and feasible to search for general biological or mathematical principles that drive infectious diseases? These questions cannot be answered satisfactorily at present, but they might be a foundation for launching or redirecting strategies of computational modeling in infectious disease research.
So far, little has been done regarding multi-scale integration, but some articles in this special issue discuss some of the challenges and potential strategies forward. They provide examples of how modeling in the era of systems biology and –omics research may help close the gaps between organizational scales in biology.
The within-host link between the molecular and cellular realms is bridged by data analysis of –omics technologies, which inform molecular quantities at specific points in time. Yin et al. [154] present a novel analysis pipeline that addresses thousands of measurements originating from a few observations, dimensionality reduction, and Bayesian inference. The result is a reduced set of meaningful molecular variables that can inform mathematical models involving molecular and cellular processes.
In some cases, the within-host dynamics of infection can be a beneficial process for the host. Jacobsen and Pilyugin [168] demonstrate this fact with the analysis of tumor therapy based on fusogenic oncolytic viruses. The infection targets in their study are cancer cells. Long-term tumor radius is shown to decrease with increasing values of viral burst size, while the effect of the rate of fusion on tumor growth is demonstrated to be non-monotonic.
A within-host viral infection with virus-to-cell and cell-to-cell transmission can be used to characterize HBV, HIV, or HTLV-1 infections. Yang et al. [169] study this problem with a model that accounts for three distributed delays. The first describes the intracellular latency for the virus-to-cell infection, while the second delay represents the intracellular latency for the cell-to-cell infection, and the third delay describes the time period for viruses to penetrate into cells and infected cells to release new virions. This model determines a globally asymptotically stable equilibrium that is consistent with chronic infection.
In the case of vector-borne diseases, the epidemiological dynamics is significantly affected by the movement of the host, which, in the case of humans, has become increasingly complex in the age of globalization. Cosner [170] presents several alternatives to studying host movement. Space can be considered either explicitly (Eulerian models) in the form of variation of population density over time, or implicitly (Lagrangian models) by characterizing time budgets and movement patterns. Both approaches pose open questions and offer different types of insight.
SIR models (and their variations) have been extensively studied for almost one hundred years. However, many questions persist. Schwartz et al. [171] study the average number of secondary influenza virus infections, which does not match observations, when it is estimated via maximum likelihood estimation (MLE). By contrast, when the fitting is done with least-squares estimation (LSE), the calculation of secondary infections performs better. The reason is that MLE and LSE coincide when the variables form a multi-variate normal distribution; however, if the distribution is not normal, the Gauss-Markov theorem establishes that LSE is the most efficient unbiased estimator.
Pathogen virulence is subject to evolutionary pressure, as studied by Feng et al. [172]. This study of the evolution of virulence at the population level indicates that disease prevalence at the positive stable equilibrium is increasing as a function of the within-host or between-host reproduction numbers. Thus, the computation of threshold conditions for disease control should incorporate explicitly the links between within-host and between-host dynamics. This presents a problem, though. The canonical approach to link slow and fast processes—that is molecular and epidemiological time scales—is to use a globally stable equilibrium of the fast variables as a constant for the slow variables. However, because there could be multiple attractors, it is necessary to account for the dynamical behaviors of the fast system in detail.
Host-pathogen systems can be heterogeneous in multiple ways. An example of this is the study of a two-pathogen vector borne disease. White et al. [173] present an analysis of the dynamics of the lone star tick (Amblyomma americanum), and the pathogens it can carry (Rickettsia parkeri and Rickettsia amblyommii), which cause rickettsiosis and possibly typhus, both endemic and epidemic, Rocky Mountain spotted fever, and Rickettsialpox. A numerical exploration shows the importance of transmission of the pathogen from a vector to another when they feed in close proximity. This co-feeding mechanism, in conjunction with slow host recovery rates, can suffice to sustain an endemic status of the disease.
Eukaryotic parasites have life cycles that are orders of magnitude more complex than those of viruses and bacteria, and their mathematical characterization is correspondingly more challenging. Yan et al. [174] present a coupled system of transport PDEs and ODEs to capture the disease dynamics of malaria. The PDEs are formulated to take into considerations the variation in the age of RBCs and the parasites, along with interactions with the innate and adaptive immune system. This combined system is capable of characterizing the evolution of a first infection (which is possibly fatal), re-infections (coexistence of the host and the pathogen), and the end of chronic infection (clearance of the parasite by the immune system).
Diseases like malaria directly affect the blood system and cause severe anemia. A detailed understanding of the host process of erythropoiesis (i.e., the creation of new RBCs) is a necessary preliminary step before attempting to characterize hemodynamics of disease. Fonseca and Voit [107] compare ordinary differential equations (ODE), delay differential equations (DDE), and discrete recursive equations (DRE) as base frameworks for modeling the process of erythropoiesis. The technique found to be most suitable for this purpose uses DREs with age classes.
The within-host dynamics of viral infection can be studied in animal models. Allen and Schwartz [175] present the dynamics of equine infectious anemia, which is caused by a retrovirus that infects horses and ponies. This virus comes in two strains: susceptible to clearance by the immune system, and resistant to the immune system. The system shows an array of behaviors including coexistence of the two strains, a resistant strain equilibrium, and a virus clearance equilibrium. The transitions depend on the proportion of cell-to-cell transmission versus free virus transmission.
Epidemiological dynamics is driven by the uptake of pathogens by suitable hosts, as described earlier in this article. Hence, the study of environmental pathogen shedding is central to the link between within-host dynamics and between-host transmission. Barfield et al. [176] study how the rate of shedding is likely to evolve, and what factors permit coexistence of alternative shedding rates in a pathogen population. A trade-off develops according to which within-host competition favors clones with low shedding rates while between-host competition benefits clones with higher shedding rates.
The study of dispersal of species over landscapes requires the proper consideration of space heterogeneities. Gutierrez et al. [177] explore the use of bivariate splines to produce a unique solution of nonlinear PDEs over irregular domains. The within-host dynamics can be taken into account to determine environmental pathogen shedding, which in turn determines the dynamics of interaction between pathogens, vectors and hosts. Such formulation requires very complicated segmentations of the spatial domain for each species considered. The characterization of the dynamics in this setting can only be undertaken with numerical methods. It is in this context that bivariate splines offer a reliable alternative to quantify dispersal.
Communication and Education
As recently as one or two decades ago, true collaborations between biologists and physicians on the one hand and mathematicians and computer scientists on the other were rare and mostly limited to questions of data analysis. This situation has changed incredibly for the better, in part triggered by the absolute necessity of computational support for analyses of the wealth of modern large-scale, high-throughput datasets, where experience and common sense are quickly overwhelmed by the sheer size and complexity of the data. Now, forward-looking biomedical scientists readily ask for computational support, and modelers find themselves in a new world of enormous opportunity, unprecedented data largess, and daunting technical challenges.
While it might initially be acceptable if a modeler disappears with a biologist's data for a while and then returns with an appropriate analysis and interpretation, as it used to be commonplace, the truly successful collaboration of the future will require that both sides learn from each other much more than what was typically expected in the past. The primary reason is that the enormous complexity of scientific problems that can be addressed with today's tools requires substantive brainstorming early on, before experiments are designed, and even with respect to asking the right scientific questions (for a detailed example, see Chapter 11 of [178]). This type of brainstorming is only beneficial if each side has a good impression of what the other side is capable of accomplishing, and gaining such an impression, in turn, requires a common language that permits effective communication. The vernacular is oftentimes not suited for this purpose, as most common words are only loosely defined and technical terminology and jargon are often unknown. Thus, few biologists would be able to explain what a bifurcation analysis is or does, and many mathematicians’ eyes would glaze over during a technical discussion about the details of optimal experimental conditions for an investigation in molecular biology.
Anthropologists and ethnographers are very familiar with this issue, because they have studied it intensely in other contexts [179]. As a paradigmatic situation, imagine traders from Xi'an and from Tehran meeting along the Silk Road in Tashkent. Both are intrigued by the other's wares and both are motivated and interested in trading or bartering. However, their different languages are a natural hindrance that somehow must be overcome. A translator could be a solution, but both sides would probably feel that they could incur the risk of the translator's favoritism for the other side.
Biology, mathematics, and computer science face the same dilemma. As an anthropologist may say, the three parties enter a trading zone or “agora,” where practitioners from different disciplines enter via their particular on-ramps to study a “boundary object” [180; 181]. In the present context, this boundary object could be an infectious disease, about which the different parties know rather different details. The practitioners on all sides have genuine strengths, knowledge, different experiences and backgrounds, and a disposition toward complex problem solving, but they are bound to face language barriers in the agora. Such barriers are initially addressed with an impromptu and highly unstable “Pidgin” language that is provisional, dynamic, and full of neologisms, vaguely defined terminologies, initial code switching, and new categorization schemes, that are summarily termed “basilectalization.” Driven by the desire for scientific trade and potentially beneficial collaboration, the initially immature gibberish steadily morphs into a fully defined and refined “Creole” language, which subsequently becomes the means of communication in the agora. Thus, the practitioners retain their “native” language, but over time become bidialectical or bilingual [182]. One can see this unfolding process very clearly in initial scientific meetings between a biologist and a mathematician or computer scientist. After all, what is a biologist to do with a statement such as “all eigenvalues have negative real parts,” which a mathematician immediately interprets as a confirmation of system stability at a fixed point.
In addition to new terminologies in the emerging, common Creole, the two sides need to develop a basic understanding of what is important to the other side, what are their metrics of success, and what drives their progress (N. Nersessian, pers. comm.). Without this awareness, a biologist might not understand why an impact factor or an h-index is of no particular concern to a mathematician, and a mathematician might not appreciate the slew of control experiments that may be needed to affirm a new finding.
It is quite evident that the new type of collaborative systems biology critically depends on suitable language skills, possibly in a new “Systems Biology Creole,” which are to be acquired through new ways of education and learning and require substantial cognitive flexibility of all involved [183; 184]. Such a bidialectical education of integrative thinkers does not strive to “homogenize” them but to achieve transdisciplinarity through carefully nurtured communication channels, a process which demands interdisciplinary knowledge as well as substantial cognitive flexibility. In the case of infectious diseases, communication is even more complicated than presented here, as not only biologists, mathematicians, and computer scientists are involved in comprehensive investigations, but also physicians, epidemiologists, public health officials, sociologists, and practitioners of many other disciplines. It is not to be expected that the practitioners from all these areas will fluidly converse about all aspects of infectious disease models. Nonetheless, it would be desirable if all sides developed a “feel” for the systemic nature of infectious diseases and their complex and highly cyclical and adaptive systems of causes and effects [185]. Some of these transdisciplinary challenges have been taken on in recent years by centers of excellence and large systems biology consortia including, specifically in the context of infectious diseases, the Malaria Host-Pathogen Interaction Center [186].
Discussion and Conclusions
Whether an infectious disease or some other biological phenomenon is the target of a computational analysis, good modeling starts with a trio of ingredients: a set of focused questions, sufficient data, and a suitable modeling framework [178]. These ingredients are of course not independent of each other but, in fact, determine each other's specifications. In the context of infectious diseases, it is immediately evident that very different data and very different modeling frameworks are needed, because the questions surrounding infectious diseases are just so different. If one sorts them by scale, diseases like malaria, SARS, or MERS leave little doubt that some important questions concern a global scale, which naturally is accompanied by a corresponding temporal scale of months or years. If a disease remains more regional, the spatial and temporal scales are somewhat reduced, and the key questions may shift to individuals, actual disease transmission mechanisms and the heterogeneity or “patchiness” of the particular environment. At the level of individual suffering, the physiological scale is at the center of modeling, with processes at the rate of minutes to days. Direct host-parasite interactions occur at the cellular level, within and between cells, and if one studies the responses of the host to an invading species, the models focus increasingly on molecular events, and the associated scales become even smaller and faster.
The wide span of scales implies directly that models of infectious diseases can have rather different goals. For academicians, the overriding goal will always be to understand the inner workings of the systems that drive the disease. These systems are different for the various levels of biological organization, and truly comprehending one or all of them would be a significant achievement. Given the amount of available information, models at the molecular level may currently have the best chance of revealing detailed mechanisms, and understanding the molecular disease processes could provide the means to identifying bottlenecks or chokepoints within the system. Such points are very sensitive to alterations and could therefore be valuable drug targets or sites of other potential interventions that could facilitate the disruption of the chain of events leading from an infection event to disease. At the other end of the spectrum, the goal of infectious disease models may be a better grasp of the epidemiology of the disease and its drivers. Here, the unit for modeling is typically a subpopulation or maybe the individual. To be of practical interest to epidemiologists and public health professionals, the abovementioned models focusing on the molecular level would have to be expanded to questions of how the molecular events within hosts translate into disease outbreaks, transmission, spread, and possible eradication. Yet a different goal of an infectious disease model could be a disease simulator that would combine molecular and physiological levels [187]. Like a flight simulator, which trains pilots to practice routine tasks as well as the avoidance of catastrophes, a disease simulator would mimic all aspects of a disease within a person so well that it could be used to train medical students in the art of diagnosis and treatment. Certainly, an effective disease simulator would be very welcome.
No matter what the ultimate goal of the modeling analysis, once the specific focus has been selected, it is necessary to assess the types of data that are available or could become available within the foreseeable future. Simultaneously considering research questions and data typically provides clues regarding the most appropriate modeling framework. The history of disease modeling provides many illustrations of such choices.
The quantitative analysis of the range of dynamics spanning aspects from within-host dynamics to the epidemiology of infectious disease demands the integration of phenomena that usually is studied in isolation. The mathematical modeling of within and between living system interactions faces obstacles of increasing complexity as the level of detail of molecular data increases. Until very recently, the amount of information that could be collected regarding a single organism for the purpose of producing predictive models was manageable by a single investigator or a single lab. While small, focused efforts are and will remain in place, it is increasingly common to have access to comprehensive and heterogeneous data sets produced by interdisciplinary cooperative teams. However, the integration of multiple data sources to characterize a living system remains an unsolved problem given the heterogeneity of information (transcriptomic, metabolomic, genomic, lipidomic, proteomic, clinical, geographic, epidemiological, sociological data), the difference in scale with respect to observations (molecular, cellular, physiological, epidemiological), and the different inherent quality and certainty of observations. The tools, skills and approaches needed to probe these systems require a transdisciplinary framework and the development of novel modeling tools. Arguably for the first time, we are positioned to study the full range of interactions involved in infectious diseases. The integration of all scales has not yet occurred, but it is a clear goal for the field.
Acknowledgments
This project was funded, in part, by Federal funds from the U.S. National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract # HHSN272201200031C, and the International Centers for Excellence in Malaria Research - Center for non-Amazonian regions of Latin America (CLAIM), NIH's NIAID cooperative agreement U19AI089702. The workshop From within Host Dynamics to the Epidemiology of Infectious Disease, where some of this material was presented, was held at the Mathematical Biosciences Institute at Ohio State University in April 2014 with support from the National Science Foundation, grant DMS-0931642.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.WHO Malaria. 2013 http://www.who.int/malaria/media/world_malaria_report_2013/en/
- 2.WHO World Malaria Report. 2014 www.who.int/malaria/publications/world_malaria_report_2014/en/
- 3.AIDS AIDS: Global Statistics. 2014 https://www.aids.gov/hiv-aids-basics/hiv-aids-101/global-statistics/
- 4.WHO Tuberculosis. 2015 http://www.who.int/mediacentre/factsheets/fs104/en/
- 5.WHO Influenza. 2014 http://www.who.int/topics/influenza/en/
- 6.CDC, 2013 Sexually Transmitted Disease Surveillance. 2014 http://www.cdc.gov/std/stats13/default.htm.
- 7.Lasker Infectious Diseases. 2014 http://www.laskerfoundation.org/media/pdf/factsheet15infectiousdiseases.pdf.
- 8.Moffatt M. The economic cost of a flu pandemic. 2015 http://economics.about.com/od/healthcareeconomics/a/flu_pandemic.htm.
- 9.MBI From within Host Dynamics to the Epidemiology of Infectious Disease. 2014 http://mbi.osu.edu/event/?id=715.
- 10.Clayton D, Hills M. Statistical Models in Epidemiology. Oxford University Press; Oxford, UK: 2013. [Google Scholar]
- 11.Feng Z. Applications of Epidemiological Models to Public Health Policymaking. World Scientific Publishing; Singapore: 2014. [Google Scholar]
- 12.Alonso PL, Tanner M. Public health challenges and prospects for malaria control and elimination. Nat Med. 2013;19:150–5. doi: 10.1038/nm.3077. [DOI] [PubMed] [Google Scholar]
- 13.Howes RE, Dewi M, Piel FB, Monteiro WM, Battle KE, Messina JP, Sakuntabhai A, Satyagraha AW, Williams TN, Baird JK, Hay SI. Spatial distribution of G6PD deficiency variants across malaria-endemic regions. Malar J. 2013;12:418. doi: 10.1186/1475-2875-12-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moyes CL, Henry AJ, Golding N, Huang Z, Singh B, Baird JK, Newton PN, Huffman M, Duda KA, Drakeley CJ, Elyazar IR, Anstey NM, Chen Q, Zommers Z, Bhatt S, Gething PW, Hay SI. Defining the geographical range of the Plasmodium knowlesi reservoir. PLoS Negl Trop Dis. 2014;8:e2780. doi: 10.1371/journal.pntd.0002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galinski MR, Barnwell JW. Monkey malaria kills four humans. Trends Parasitol. 2009;25:200–4. doi: 10.1016/j.pt.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–24. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 17.William T, Jelip J, Menon J, Anderios F, Mohammad R, Awang Mohammad TA, Grigg MJ, Yeo TW, Anstey NM, Barber BE. Changing epidemiology of malaria in Sabah, Malaysia: increasing incidence of Plasmodium knowlesi. Malar J. 2014;13:390. doi: 10.1186/1475-2875-13-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly GC, Tanner M, Vallely A, Clements A. Malaria elimination: moving forward with spatial decision support systems. Trends Parasitol. 2012;28:297–304. doi: 10.1016/j.pt.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Coatney GR, Collins WE, Warren M, Contacos PG. The Primate Malarias (original book published in 1971), Division of Parasitic Diseases. Centers for Disease Control and Prevention; Atlanta, GA, USA: 1971. 2003. p. 366. [Google Scholar]
- 20.Galinski MR, Barnwell JW. Non-human Primate Models for Human Malaria Research, Nonhuman Primates in Biomedical Research: Diseases. Elsevier, Inc.; 2012. pp. 299–323. [Google Scholar]
- 21.Ayala SC, D'Alessandro A, Mackenzie R, Angel D. Hemoparasite infections in 830 wild animals from the eastern Llanos of Colombia. J Parasitol. 1973;59:52–9. [PubMed] [Google Scholar]
- 22.Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl Acad Sci U S A. 1994;91:11373–7. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayakawa T, Tachibana S, Hikosaka K, Arisue N, Matsui A, Horii T, Tanabe K. Age of the last common ancestor of extant Plasmodium parasite lineages. Gene. 2012;502:36–9. doi: 10.1016/j.gene.2012.04.037. [DOI] [PubMed] [Google Scholar]
- 24.Hall BF, Fauci AS. Malaria control, elimination, and eradication: the role of the evolving biomedical research agenda. J Infect Dis. 2009;200:1639–43. doi: 10.1086/646611. [DOI] [PubMed] [Google Scholar]
- 25.Sibley CH. Understanding drug resistance in malaria parasites: basic science for public health. Mol Biochem Parasitol. 2014;195:107–14. doi: 10.1016/j.molbiopara.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Sinha S, Medhi B, Sehgal R. Challenges of drug-resistant malaria. Parasite. 2014;21:61. doi: 10.1051/parasite/2014059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visser BJ, van Vugt M, Grobusch MP. Malaria: an update on current chemotherapy. Expert Opin Pharmacother. 2014;15:2219–54. doi: 10.1517/14656566.2014.944499. [DOI] [PubMed] [Google Scholar]
- 28.Chen N, LaCrue AN, Teuscher F, Waters NC, Gatton ML, Kyle DE, Cheng Q. Fatty acid synthesis and pyruvate metabolism pathways remain active in dihydroartemisinin-induced dormant ring stages of Plasmodium falciparum. Antimicrob Agents Chemother. 2014;58:4773–81. doi: 10.1128/AAC.02647-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Q, Cunningham J, Gatton ML. Systematic Review of Sub-microscopic P. vivax Infections: Prevalence and Determining Factors. PLoS Negl Trop Dis. 2015;9:e3413. doi: 10.1371/journal.pntd.0003413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopes SC, Albrecht L, Carvalho BO, Siqueira AM, Thomson-Luque R, Nogueira PA, Fernandez-Becerra C, Del Portillo HA, Russell BM, Renia L, Lacerda MV, Costa FT. Paucity of Plasmodium vivax mature schizonts in peripheral blood is associated with their increased cytoadhesive potential. J Infect Dis. 2014;209:1403–7. doi: 10.1093/infdis/jiu018. [DOI] [PubMed] [Google Scholar]
- 31.Kondrashin A, Baranova AM, Ashley EA, Recht J, White NJ, Sergiev VP. Mass primaquine treatment to eliminate vivax malaria: lessons from the past. Malar J. 2014;13:51. doi: 10.1186/1475-2875-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maude RJ, Nguon C, Dondorp AM, White LJ, White NJ. The diminishing returns of atovaquone-proguanil for elimination of Plasmodium falciparum malaria: modelling mass drug administration and treatment. Malar J. 2014;13:380. doi: 10.1186/1475-2875-13-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirot E, Skarbinski J, Sinclair D, Kachur SP, Slutsker L, Hwang J. Mass drug administration for malaria. Cochrane Database Syst Rev. 2013;12:CD008846. doi: 10.1002/14651858.CD008846.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu OA, Dondorp AM. Malaria. Lancet. 2014;383:723–35. doi: 10.1016/S0140-6736(13)60024-0. [DOI] [PubMed] [Google Scholar]
- 35.Baird JK. Pernicious and threatening Plasmodium vivax as reality. Am J Trop Med Hyg. 2014;91:1–2. doi: 10.4269/ajtmh.14-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cunnington AJ, Riley EM, Walther M. Stuck in a rut? Reconsidering the role of parasite sequestration in severe malaria syndromes. Trends Parasitol. 2013;29:585–92. doi: 10.1016/j.pt.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price RN, Douglas NM, Anstey NM. New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr Opin Infect Dis. 2009;22:430–5. doi: 10.1097/QCO.0b013e32832f14c1. [DOI] [PubMed] [Google Scholar]
- 38.Ali IM, Evehe MS, Netongo PM, Atogho-Tiedeu B, Akindeh-Nji M, Ngora H, Domkam IK, Diakite M, Baldip K, Ranford-Cartwright L, Mimche PN, Lamb T, Mbacham WF. Host candidate gene polymorphisms and associated clearance of P. falciparum amodiaquine and fansidar resistance mutants in children less than 5 years in Cameroon. Pathog Glob Health. 2014;108:323–33. doi: 10.1179/2047773214Y.0000000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alout H, Yameogo B, Djogbenou LS, Chandre F, Dabire RK, Corbel V, Cohuet A. Interplay between Plasmodium infection and resistance to insecticides in vector mosquitoes. J Infect Dis. 2014;210:1464–70. doi: 10.1093/infdis/jiu276. [DOI] [PubMed] [Google Scholar]
- 40.Nair S, Nkhoma SC, Serre D, Zimmerman PA, Gorena K, Daniel BJ, Nosten F, Anderson TJ, Cheeseman IH. Single-cell genomics for dissection of complex malaria infections. Genome Res. 2014;24:1028–38. doi: 10.1101/gr.168286.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nwakanma DC, Duffy CW, Amambua-Ngwa A, Oriero EC, Bojang KA, Pinder M, Drakeley CJ, Sutherland CJ, Milligan PJ, Macinnis B, Kwiatkowski DP, Clark TG, Greenwood BM, Conway DJ. Changes in malaria parasite drug resistance in an endemic population over a 25-year period with resulting genomic evidence of selection. J Infect Dis. 2014;209:1126–35. doi: 10.1093/infdis/jit618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ocholla H, Preston MD, Mipando M, Jensen AT, Campino S, MacInnis B, Alcock D, Terlouw A, Zongo I, Oudraogo JB, Djimde AA, Assefa S, Doumbo OK, Borrmann S, Nzila A, Marsh K, Fairhurst RM, Nosten F, Anderson TJ, Kwiatkowski DP, Craig A, Clark TG, Montgomery J. Whole-genome scans provide evidence of adaptive evolution in Malawian Plasmodium falciparum isolates. J Infect Dis. 2014;210:1991–2000. doi: 10.1093/infdis/jiu349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preston MD, Assefa SA, Ocholla H, Sutherland CJ, Borrmann S, Nzila A, Michon P, Hien TT, Bousema T, Drakeley CJ, Zongo I, Ouedraogo JB, Djimde AA, Doumbo OK, Nosten F, Fairhurst RM, Conway DJ, Roper C, Clark TG. PlasmoView: a web-based resource to visualise global Plasmodium falciparum genomic variation. J Infect Dis. 2014;209:1808–15. doi: 10.1093/infdis/jit812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preston MD, Campino S, Assefa SA, Echeverry DF, Ocholla H, Amambua-Ngwa A, Stewart LB, Conway DJ, Borrmann S, Michon P, Zongo I, Ouedraogo JB, Djimde AA, Doumbo OK, Nosten F, Pain A, Bousema T, Drakeley CJ, Fairhurst RM, Sutherland CJ, Roper C, Clark TG. A barcode of organellar genome polymorphisms identifies the geographic origin of Plasmodium falciparum strains. Nat Commun. 2014;5:4052. doi: 10.1038/ncomms5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor SM, Fairhurst RM. Malaria parasites and red cell variants: when a house is not a home. Curr Opin Hematol. 2014;21:193–200. doi: 10.1097/MOH.0000000000000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamagishi J, Natori A, Tolba ME, Mongan AE, Sugimoto C, Katayama T, Kawashima S, Makalowski W, Maeda R, Eshita Y, Tuda J, Suzuki Y. Interactive transcriptome analysis of malaria patients and infecting Plasmodium falciparum. Genome Res. 2014;24:1433–44. doi: 10.1101/gr.158980.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmerman PA, Ferreira MU, Howes RE, Mercereau-Puijalon O. Red blood cell polymorphism and susceptibility to Plasmodium vivax. Adv Parasitol. 2013;81:27–76. doi: 10.1016/B978-0-12-407826-0.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Menard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Menard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–5. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Menard D, Fidock DA. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2014 doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wendler JP, Okombo J, Amato R, Miotto O, Kiara SM, Mwai L, Pole L, O'Brien J, Manske M, Alcock D, Drury E, Sanders M, Oyola SO, Malangone C, Jyothi D, Miles A, Rockett KA, MacInnis BL, Marsh K, Bejon P, Nzila A, Kwiatkowski DP. A genome wide association study of Plasmodium falciparum susceptibility to 22 antimalarial drugs in Kenya. PLoS One. 2014;9:e96486. doi: 10.1371/journal.pone.0096486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winzeler EA, Manary MJ. Drug resistance genomics of the antimalarial drug artemisinin. Genome Biol. 2014;15:544. doi: 10.1186/s13059-014-0544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel K, Simpson JA, Batty KT, Zaloumis S, Kirkpatrick CM. Modelling the time course of antimalarial parasite killing: a tour of animal and human models, translation and challenges. Br J Clin Pharmacol. 2015;79:97–107. doi: 10.1111/bcp.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor AR, Flegg JA, Nsobya SL, Yeka A, Kamya MR, Rosenthal PJ, Dorsey G, Sibley CH, Guerin PJ, Holmes CC. Estimation of malaria haplotype and genotype frequencies: a statistical approach to overcome the challenge associated with multiclonal infections. Malar J. 2014;13:102. doi: 10.1186/1475-2875-13-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liehl P, Meireles P, Albuquerque IS, Pinkevych M, Baptista F, Mota MM, Davenport MP, Prudencio M. Innate immunity induced by Plasmodium liver infection inhibits malaria reinfections. Infect Immun. 2015 doi: 10.1128/IAI.02796-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frevert U, Nacer A. Immunobiology of Plasmodium in liver and brain. Parasite Immunol. 2013;35:267–82. doi: 10.1111/pim.12039. [DOI] [PubMed] [Google Scholar]
- 56.Baer K, Klotz C, Kappe SH, Schnieder T, Frevert U. Release of hepatic Plasmodium yoelii merozoites into the pulmonary microvasculature. PLoS Pathog. 2007;3:e171. doi: 10.1371/journal.ppat.0030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graewe S, Rankin KE, Lehmann C, Deschermeier C, Hecht L, Froehlke U, Stanway RR, Heussler V. Hostile takeover by Plasmodium: reorganization of parasite and host cell membranes during liver stage egress. PLoS Pathog. 2011;7:e1002224. doi: 10.1371/journal.ppat.1002224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thiberge S, Blazquez S, Baldacci P, Renaud O, Shorte S, Menard R, Amino R. In vivo imaging of malaria parasites in the murine liver. Nat Protoc. 2007;2:1811–8. doi: 10.1038/nprot.2007.257. [DOI] [PubMed] [Google Scholar]
- 59.Clark MA, Goheen MM, Spidale NA, Kasthuri RS, Fulford A, Cerami C. RBC barcoding allows for the study of erythrocyte population dynamics and P. falciparum merozoite invasion. PLoS One. 2014;9:e101041. doi: 10.1371/journal.pone.0101041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galinski MR, Meyer EV, Barnwell JW. Plasmodium vivax: modern strategies to study a persistent parasite's life cycle. Adv Parasitol. 2013;81:1–26. doi: 10.1016/B978-0-12-407826-0.00001-1. [DOI] [PubMed] [Google Scholar]
- 61.Harvey KL, Gilson PR, Crabb BS. A model for the progression of receptor-ligand interactions during erythrocyte invasion by Plasmodium falciparum. Int. J. Parasitol. 2012;42:567–573. doi: 10.1016/j.ijpara.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 62.Kerlin DH, Gatton ML. Preferential invasion by Plasmodium merozoites and the self-regulation of parasite burden. PLoS One. 2013;8:e57434. doi: 10.1371/journal.pone.0057434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aikawa M, Miller LH, Rabbege J. Caveola--vesicle complexes in the plasmalemma of erythrocytes infected by Plasmodium vivax and P cynomolgi. Unique structures related to Schuffner's dots. Am J Pathol. 1975;79:285–300. [PMC free article] [PubMed] [Google Scholar]
- 64.Akinyi S, Hanssen E, Meyer EV, Jiang J, Korir CC, Singh B, Lapp S, Barnwell JW, Tilley L, Galinski MR. A 95 kDa protein of Plasmodium vivax and P. cynomolgi visualized by three-dimensional tomography in the caveola-vesicle complexes (Schuffner's dots) of infected erythrocytes is a member of the PHIST family. Mol Microbiol. 2012;84:816–31. doi: 10.1111/j.1365-2958.2012.08060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirk K, Lehane AM. Membrane transport in the malaria parasite and its host erythrocyte. Biochem J. 2014;457:1–18. doi: 10.1042/BJ20131007. [DOI] [PubMed] [Google Scholar]
- 66.Carmona-Fonseca J, Arango E, Maestre A. Placental malaria in Colombia: histopathologic findings in Plasmodium vivax and P. falciparum infections. Am J Trop Med Hyg. 2013;88:1093–101. doi: 10.4269/ajtmh.12-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aguilar R, Magallon-Tejada A, Achtman AH, Moraleda C, Joice R, Cistero P, Li Wai Suen CS, Nhabomba A, Macete E, Mueller I, Marti M, Alonso PL, Menendez C, Schofield L, Mayor A. Molecular evidence for the localization of Plasmodium falciparum immature gametocytes in bone marrow. Blood. 2014;123:959–66. doi: 10.1182/blood-2013-08-520767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joice R, Nilsson SK, Montgomery J, Dankwa S, Egan E, Morahan B, Seydel KB, Bertuccini L, Alano P, Williamson KC, Duraisingh MT, Taylor TE, Milner DA, Marti M. Plasmodium falciparum transmission stages accumulate in the human bone marrow. Sci Transl Med. 2014;6:244re5. doi: 10.1126/scitranslmed.3008882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnston GL, Smith DL, Fidock DA. Malaria's missing number: calculating the human component of R0 by a within-host mechanistic model of Plasmodium falciparum infection and transmission. PLoS Comput Biol. 2013;9:e1003025. doi: 10.1371/journal.pcbi.1003025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alano P. The sound of sexual commitment breaks the silencing of malaria parasites. Trends Parasitol. 2014;30:509–10. doi: 10.1016/j.pt.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Ataide R, Mayor A, Rogerson SJ. Malaria, primigravidae, and antibodies: knowledge gained and future perspectives. Trends Parasitol. 2014;30:85–94. doi: 10.1016/j.pt.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 72.Takem EN, D'Alessandro U. Malaria in pregnancy. Mediterr J Hematol Infect Dis. 2013;5:e2013010. doi: 10.4084/MJHID.2013.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thanapongpichat S, McGready R, Luxemburger C, Day NP, White NJ, Nosten F, Snounou G, Imwong M. Microsatellite genotyping of Plasmodium vivax infections and their relapses in pregnant and non-pregnant patients on the Thai-Myanmar border. Malar J. 2013;12:275. doi: 10.1186/1475-2875-12-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walker PG, Griffin JT, Cairns M, Rogerson SJ, van Eijk AM, ter Kuile F, Ghani AC. A model of parity-dependent immunity to placental malaria. Nat Commun. 2013;4:1609. doi: 10.1038/ncomms2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Corradin G, Engers H. Malaria Vaccine Development: Over 40 Years of Trials and Tribulations. 2014 www.futuremedicine.com/doi/book/10.2217/9781780844411.
- 76.Joyner CJ, Barnwell J, Galinski MR. No more monkeying around: Primate malaria model systems are key to understanding plasmodium vivax liver-stage biology, hypnozoites, and Rrelapses. Front. Microbiol. 2015;6:145. doi: 10.3389/fmicb.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.White MT, Karl S, Battle KE, Hay SI, Mueller I, Ghani AC. Modelling the contribution of the hypnozoite reservoir to Plasmodium vivax transmission. Elife. 2014;3 doi: 10.7554/eLife.04692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Betuela I, Rosanas-Urgell A, Kiniboro B, Stanisic DI, Samol L, de Lazzari E, Del Portillo HA, Siba P, Alonso PL, Bassat Q, Mueller I. Relapses contribute significantly to the risk of Plasmodium vivax infection and disease in Papua New Guinean children 1-5 years of age. J Infect Dis. 2012;206:1771–80. doi: 10.1093/infdis/jis580. [DOI] [PubMed] [Google Scholar]
- 79.Krotoski WA, Collins WE, Bray RS, Garnham PC, Cogswell FB, Gwadz RW, Killick-Kendrick R, Wolf R, Sinden R, Koontz LC, Stanfill PS. Demonstration of hypnozoites in sporozoite-transmitted Plasmodium vivax infection. Am J Trop Med Hyg. 1982;31:1291–3. doi: 10.4269/ajtmh.1982.31.1291. [DOI] [PubMed] [Google Scholar]
- 80.Reyes-Sandoval A, Bachmann MF. Plasmodium vivax malaria vaccines: why are we where we are? Hum Vaccin Immunother. 2013;9:2558–65. doi: 10.4161/hv.26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chua CL, Brown G, Hamilton JA, Rogerson S, Boeuf P. Monocytes and macrophages in malaria: protection or pathology? Trends Parasitol. 2013;29:26–34. doi: 10.1016/j.pt.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 82.Doolan DL, Dobano C, Baird JK. Acquired immunity to malaria. Clin Microbiol Rev. 2009;22:13–36. doi: 10.1128/CMR.00025-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ioannidis LJ, Nie CQ, Hansen DS. The role of chemokines in severe malaria: more than meets the eye. Parasitology. 2014;141:602–13. doi: 10.1017/S0031182013001984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moreno A, Cabrera-Mora M, Garcia A, Orkin J, Strobert E, Barnwell JW, Galinski MR. Plasmodium coatneyi in rhesus macaques replicates the multisystemic dysfunction of severe malaria in humans. Infect Immun. 2013;81:1889–904. doi: 10.1128/IAI.00027-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Praba-Egge AD, Montenegro S, Cogswell FB, Hopper T, James MA. Cytokine responses during acute simian Plasmodium cynomolgi and Plasmodium knowlesi infections. Am J Trop Med Hyg. 2002;67:586–96. doi: 10.4269/ajtmh.2002.67.586. [DOI] [PubMed] [Google Scholar]
- 86.Teo A, Hasang W, Randall LM, Feng G, Bell L, Unger H, Langer C, Beeson JG, Siba PM, Mueller I, Molyneux ME, Brown GV, Rogerson SJ. Decreasing malaria prevalence and its potential consequences for immunity in pregnant women. J Infect Dis. 2014;210:1444–55. doi: 10.1093/infdis/jiu264. [DOI] [PubMed] [Google Scholar]
- 87.Viriyavejakul P, Khachonsaksumet V, Punsawad C. Liver changes in severe Plasmodium falciparum malaria: histopathology, apoptosis and nuclear factor kappa B expression. Malar J. 2014;13:106. doi: 10.1186/1475-2875-13-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wunderlich F, Al-Quraishy S, Dkhil MA. Liver-inherent immune system: its role in blood-stage malaria. Front Microbiol. 2014;5:559. doi: 10.3389/fmicb.2014.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blyuss KB. The effects of symmetry on the dynamics of antigenic variation. J Math Biol. 2013;66:115–37. doi: 10.1007/s00285-012-0508-y. [DOI] [PubMed] [Google Scholar]
- 90.Guizetti J, Scherf A. Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell Microbiol. 2013;15:718–26. doi: 10.1111/cmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Warimwe GM, Recker M, Kiragu EW, Buckee CO, Wambua J, Musyoki JN, Marsh K, Bull PC. Plasmodium falciparum var gene expression homogeneity as a marker of the host-parasite relationship under different levels of naturally acquired immunity to malaria. PLoS One. 2013;8:e70467. doi: 10.1371/journal.pone.0070467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Galinski MR, Corredor V. Variant antigen expression in malaria infections: posttranscriptional gene silencing, virulence and severe pathology. Mol Biochem Parasitol. 2004;134:17–25. doi: 10.1016/j.molbiopara.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 93.Korir CC, Galinski MR. Proteomic studies of Plasmodium knowlesi SICA variant antigens demonstrate their relationship with P. falciparum EMP1. Infect Genet Evol. 2006;6:75–9. doi: 10.1016/j.meegid.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 94.Lapp SA, Korir-Morrison C, Jiang J, Bai Y, Corredor V, Galinski MR. Spleen-dependent regulation of antigenic variation in malaria parasites: Plasmodium knowlesi SICAvar expression profiles in splenic and asplenic hosts. PLoS One. 2013;8:e78014. doi: 10.1371/journal.pone.0078014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Voss TS, Bozdech Z, Bartfai R. Epigenetic memory takes center stage in the survival strategy of malaria parasites. Curr Opin Microbiol. 2014;20:88–95. doi: 10.1016/j.mib.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 96.Avril M, Brazier AJ, Melcher M, Sampath S, Smith JD. DC8 and DC13 var Genes Associated with Severe Malaria Bind Avidly to Diverse Endothelial Cells. PLoS Pathog. 2013;9:e1003430. doi: 10.1371/journal.ppat.1003430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith JD, Rowe JA, Higgins MK, Lavstsen T. Malaria's Deadly Grip: Cytoadhesion of Plasmodium falciparum Infected Erythrocytes. Cell Microbiol. 2013 doi: 10.1111/cmi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abdi AI, Fegan G, Muthui M, Kiragu E, Musyoki JN, Opiyo M, Marsh K, Warimwe GM, Bull PC. Plasmodium falciparum antigenic variation: relationships between widespread endothelial activation, parasite PfEMP1 expression and severe malaria. BMC Infect Dis. 2014;14:170. doi: 10.1186/1471-2334-14-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brancucci NM, Bertschi NL, Zhu L, Niederwieser I, Chin WH, Wampfler R, Freymond C, Rottmann M, Felger I, Bozdech Z, Voss TS. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe. 2014;16:165–76. doi: 10.1016/j.chom.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 100.Coleman BI, Skillman KM, Jiang RH, Childs LM, Altenhofen LM, Ganter M, Leung Y, Goldowitz I, Kafsack BF, Marti M, Llinas M, Buckee CO, Duraisingh MT. A Plasmodium falciparum histone deacetylase regulates antigenic variation and gametocyte conversion. Cell Host Microbe. 2014;16:177–86. doi: 10.1016/j.chom.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mantel PY, Hoang AN, Goldowitz I, Potashnikova D, Hamza B, Vorobjev I, Ghiran I, Toner M, Irimia D, Ivanov AR, Barteneva N, Marti M. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe. 2013;13:521–34. doi: 10.1016/j.chom.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marcilla A, Martin-Jaular L, Trelis M, de Menezes-Neto A, Osuna A, Bernal D, Fernandez-Becerra C, Almeida IC, Del Portillo HA. Extracellular vesicles in parasitic diseases. J Extracell Vesicles. 2014;3:25040. doi: 10.3402/jev.v3.25040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Regev-Rudzki N, Wilson DW, Carvalho TG, Sisquella X, Coleman BM, Rug M, Bursac D, Angrisano F, Gee M, Hill AF, Baum J, Cowman AF. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell. 2013;153:1120–33. doi: 10.1016/j.cell.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 104.Douglas NM, Anstey NM, Buffet PA, Poespoprodjo JR, Yeo TW, White NJ, Price RN. The anaemia of Plasmodium vivax malaria. Malar J. 2012;11:135. doi: 10.1186/1475-2875-11-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Perkins DJ, Were T, Davenport GC, Kempaiah P, Hittner JB, Ong'echa JM. Severe malarial anemia: innate immunity and pathogenesis. Int J Biol Sci. 2011;7:1427–42. doi: 10.7150/ijbs.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jakeman GN, Saul A, Hogarth WL, Collins WE. Anaemia of acute malaria infections in non-immune patients primarily results from destruction of uninfected erythrocytes. Parasitology. 1999;119:127–133. doi: 10.1017/s0031182099004564. [DOI] [PubMed] [Google Scholar]
- 107.Fonseca LL, Voit EO. Comparison of mathematical frameworks for modeling erythropoiesis in the context of malaria infection. Math Biosci this volume. 2015 doi: 10.1016/j.mbs.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gama JA, Abby SS, Vieira-Silva S, Dionisio F, Rocha EP. Immune subversion and quorum-sensing shape the variation in infectious dose among bacterial pathogens. PLoS pathogens. 2012;8:e1002503. doi: 10.1371/journal.ppat.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nowak MA, May RM. Mathematical biology of HIV infections: antigenic variation and diversity threshold. Mathematical Biosciences. 1991;106:1–21. doi: 10.1016/0025-5564(91)90037-j. [DOI] [PubMed] [Google Scholar]
- 110.Agur Z, Abiri D, Van der Ploeg L. Ordered appearance of antigenic variants of African trypanosomes explained in a mathematical model based on a stochastic switch process and immune-selection against putative switch intermediates. Proceedings of the National Academy of Sciences. 1989;86:9626–9630. doi: 10.1073/pnas.86.23.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Recker M, Buckee CO, Serazin A, Kyes S, Pinches R, Christodoulou Z, Springer AL, Gupta S, Newbold CI. Antigenic Variation in Plasmodium falciparum Malaria Involves a Highly Structured Switching Pattern. PLoS Pathog. 2011;7:e1001306. doi: 10.1371/journal.ppat.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gupta S, Trenholme K, Anderson R, Day K. Antigenic diversity and the transmission dynamics of Plasmodium falciparum. Science. 1994;263:961–963. doi: 10.1126/science.8310293. [DOI] [PubMed] [Google Scholar]
- 113.Arnot DE, Jensen AT. Antigenic Variation and the Genetics and Epigenetics of the PfEMP1 Erythrocyte Surface Antigens in Plasmodium falciparum Malaria. Adv Appl Microbiol. 2011;74:77–96. doi: 10.1016/B978-0-12-387022-3.00007-0. [DOI] [PubMed] [Google Scholar]
- 114.Eckhoff P. P. falciparum Infection Durations and Infectiousness Are Shaped by Antigenic Variation and Innate and Adaptive Host Immunity in a Mathematical Model. PLoS ONE. 2012;7:e44950. doi: 10.1371/journal.pone.0044950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ross R. An application of the theory of probabilities to the study of a priori pathometry. Part I. Proceedings of the Royal Society A: Mathematical, Physical and Engineering Sciences. 1916;92:204–230. [Google Scholar]
- 117.Ross R, Hudson H. An application of the theory of probabilities to the study of a priori pathometry. Part III. Proceedings of the Royal Society A: Mathematical, Physical and Engineering Sciences. 1917;93:225–240. [Google Scholar]
- 118.Ross R, Hudson H. An application of the theory of probabilities to the study of a priori pathometry. Part II. Proceedings of the Royal Society A: Mathematical, Physical and Engineering Sciences. 1917;93:212–225. [Google Scholar]
- 119.Macdonald G. The epidemiology and control of malaria. Oxford University Pres; London: 1957. [Google Scholar]
- 120.Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. OUP Oxford: 1992. [Google Scholar]
- 121.Dietz K. Mathematical models for transmission and control of malaria, Principles and Practice of Malariology. Churchill Livingston, Edinburgh: 1988. pp. 1091–1133. [Google Scholar]
- 122.Filipe JAN, Riley EM, Drakeley CJ, Sutherland CJ, Ghani AC. Determination of the Processes Driving the Acquisition of Immunity to Malaria Using a Mathematical Transmission Model. PLoS Comput Biol. 2007;3:e255. doi: 10.1371/journal.pcbi.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rodriguez D, Torres-Sorando L. Models of infectious diseases in spatially heterogeneous environments. Bull Math Biol. 2001;63:547–571. doi: 10.1006/bulm.2001.0231. [DOI] [PubMed] [Google Scholar]
- 124.Smith DL, Dushoff J, McKenzie FE. The risk of a mosquito-borne infectionin a heterogeneous environment. PLoS Biol. 2004;2:e368. doi: 10.1371/journal.pbio.0020368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kermack W, McKendrick AG. A Contribution to the Mathematical Theory of Epidemics. Proceedings of the Royal Society of London. Series A, Containing Papers of a Mathematical and Physical Character. 1927;115:700–721. [Google Scholar]
- 126.Brauer F, Castillo-Chávez C. Mathematical models in population biology and epidemiology. Springer; 2012. [Google Scholar]
- 127.Bottius E, Guanzirolli A, Trape J.-F.c., Rogier C, Konate L, Druilhe P. Malaria: even more chronic in nature than previously thought; evidence for subpatent parasitaemia detectable by the polymerase chain reaction. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1996;90:15–19. doi: 10.1016/s0035-9203(96)90463-0. [DOI] [PubMed] [Google Scholar]
- 128.Laishram DD, Sutton PL, Nanda N, Sharma VL, Sobti RC, Carlton JM, Joshi H. The complexities of malaria disease manifestations with a focus on asymptomatic malaria. Malar J. 2012;11:29. doi: 10.1186/1475-2875-11-29. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Farnert A, Snounou G, Rooth I, Bjorkman A. Daily dynamics of Plasmodium falciparum subpopulation in asymptomatic children in a holoendemic area. Am J Trop Med Hyg. 1997;56:538–547. doi: 10.4269/ajtmh.1997.56.538. [DOI] [PubMed] [Google Scholar]
- 130.da Silva-Nunes M, Moreno M, Conn J, Gamboa D, Abelesb S, Vinetz JM, Ferreira M. Amazonian malaria: Asymptomatic human reservoirs, diagnostic challenges, environmentally driven changes in mosquito vector populations, and the mandate for sustainable control strategies. Acta Tropica. 2012;121:281–291. doi: 10.1016/j.actatropica.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schweitzer A, Anderson R. Dynamic interaction between CD4+ T cells and parasitic helminths: mathematical models of heterogeneity in outcome. Parasitology. 1992;105:513–522. doi: 10.1017/s0031182000074692. [DOI] [PubMed] [Google Scholar]
- 132.Gupta S, Maiden MC, Feavers IM, Nee S, May RM, Anderson RM. The maintenance of strain structure in populations of recombining infectious agents. Nature Medicine. 1996;2:437–442. doi: 10.1038/nm0496-437. [DOI] [PubMed] [Google Scholar]
- 133.Dushoff J. Incorporating immunological ideas in epidemiological models. Journal of theoretical biology. 1996;180:181–187. doi: 10.1006/jtbi.1996.0094. [DOI] [PubMed] [Google Scholar]
- 134.Anderson RM. Complex dynamic behaviours in the interaction between parasite populations and the host's immune system. International Journal of Parasitology. 1998;28:551–566. doi: 10.1016/s0020-7519(97)00207-5. [DOI] [PubMed] [Google Scholar]
- 135.Reluga TC, Medlock J, Perelson AS. Backward bifurcations and multiple equilibria in epidemic models with structured immunity. Journal of theoretical biology. 2008;252:155–165. doi: 10.1016/j.jtbi.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nickerson D, Nash MP, Nielsen PMF, Smith NP, Hunter PJ. Computational multiscale modeling in the physiome project: modeling cardiac electromechanics. IBM J. Res. Developm. 2006;50:617–630. [Google Scholar]
- 137.Physiome, The Physiome Project 2015 http://physiomeproject.org/
- 138.Virtual-Liver 2015 http://www.virtual-liver.de/ [Google Scholar]
- 139.Holzhütter HG, Drasdo D, Preusser T, Lippert J, Henney AM. The virtual liver: a multidisciplinary, multilevel challenge for systems biology. Wiley Interdiscip Rev Syst Biol Med. 2012;4:221–35. doi: 10.1002/wsbm.1158. [DOI] [PubMed] [Google Scholar]
- 140.Voit EO. A systems-theoretical framework for health and disease: inflammation and preconditioning from an abstract modeling point of view. Math. Biosci. 2009;217:11–8. doi: 10.1016/j.mbs.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Childs LM, Abuelezam NN, Dye C, Gupta S, Murray MB, Williams BG, Buckee CO. Modelling challenges in context: lessons from malaria, HIV, and tuberculosis. Epidemics. 2015 doi: 10.1016/j.epidem.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Riley S, Eames K, Isham V, Mollison D, Trapman P. Five challenges for spatial epidemic models. Epidemics. 2014 doi: 10.1016/j.epidem.2014.07.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cantrell RS, Cosner C. Spatial Ecology via Reaction-Diffusion Equations. John Wiley & Sons; 2003. [Google Scholar]
- 144.Murray JD. Mathematical Biology. II Spatial Models and Biomedical Applications {Interdisciplinary Applied Mathematics V. 18} Springer-Verlag New York Incorporated; 2001. [Google Scholar]
- 145.Kot M, Lewis MA, van den Driessche P. Dispersal data and the spread of invading organisms. Ecology. 1996;77:2027–2042. [Google Scholar]
- 146.Kot M, Schaffer WM. Discrete-time growth-dispersal models. Math. Biosci. 1986;80:109–136. [Google Scholar]
- 147.Ovaskainen O, Hanski I. Spatially structured metapopulation models: Global and local assessment of metapopulation capacity. Theor. Pop. Biol. 2001;60:281–302. doi: 10.1006/tpbi.2001.1548. [DOI] [PubMed] [Google Scholar]
- 148.Cantrell RS, Cosner C, Fagan WF. The implications of model formulation when transitioning from spatial to landscape ecology. Math. Biosci. Eng. 2012;9:27–60. doi: 10.3934/mbe.2012.9.27. [DOI] [PubMed] [Google Scholar]
- 149.Gao D, Ruan S. A multipatch malaria model with logistic growth populations. SIAM J. Appl. Math. 2012;72:819–841. doi: 10.1137/110850761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao D, Ruan S. A multipatch malaria model with logistic growth populations. SIAM journal on applied mathematics. 2012;72:819–841. doi: 10.1137/110850761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cosner C, Beier JC, Cantrell RS, Impoinvil D, Kapitanski L, Potts MD, Troyo A, Ruan S. The effects of human movement on the persistence of vector-borne diseases. J. Theor. Biol. 2009;258:550–60. doi: 10.1016/j.jtbi.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stoddard ST, Morrison AC, Vazquez-Prokopec GM, Soldan VP, Kochel TJ, Kitron U, Elder JP, Scott TW. The role of human movement in the transmission of vector-borne pathogens. PLoS Neglected Tropical Diseases. 2009;3:e481. doi: 10.1371/journal.pntd.0000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Voit EO. Models-of-data and models-of-processes in the post-genomic era. Math. Biosci. 2002;180:263–274. doi: 10.1016/s0025-5564(02)00115-3. [DOI] [PubMed] [Google Scholar]
- 154.Yin W, Kissinger JC, Moreno A, Galinski MR, Styczynski MP. From genome-scale data to models of infectious disease: a Bayesian network-based strategy to drive model development. Mathematical Biosciences. 2015 doi: 10.1016/j.mbs.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.KEGG 2015 http://www.genome.jp/kegg/
- 156.MetaCyc 2015 MetaCyc.org.
- 157.BRENDA 2015 http://www.brenda-enzymes.org/
- 158.Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, Nolan GP. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotech. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mihret A, Abebe M. Cytokines and chemokines as biomarkers of tuberculosis. J. Mycobac. Dis. 2013;3 [Google Scholar]
- 160.Voit EO, Brigham KL. The role of systems biology in predictive health and personalized medicine. The Open Path. J. 2008;2:68–70. [Google Scholar]
- 161.Voit EO, editor. S-System Approach to Understanding Complexity. Van Nostrand Reinhold, NY: 1991. Canonical Nonlinear Modeling. [Google Scholar]
- 162.Voit EO, Savageau MA. Equivalence between S-systems and Volterra-systems. Mathem. Biosci. 1986;78:47–55. [Google Scholar]
- 163.Voit EO. Biochemical Systems Theory: A review. Int. Scholarly Res. Network (ISRN – Biomathematics) Article. 2013;897658:1–53. [Google Scholar]
- 164.Savageau MA. Allometric morphogenesis of complex systems: Derivation of the basic equations from first principles. Proc Natl Acad Sci U S A. 1979;76:6023–5. doi: 10.1073/pnas.76.12.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Peschel M, Mende W. The Predator-Prey Model: Do we Live in a Volterra World? Akademie-Verlag; Berlin: 1986. [Google Scholar]
- 166.Voit EO. Canonical modeling: A review of concepts with emphasis on environmental health. Environ Health Perspect. 2000;108(Suppl 5):895–909. doi: 10.1289/ehp.00108s5895. [DOI] [PubMed] [Google Scholar]
- 167.Alves R, Sorribas A. Special issue on biological design principles. Math Biosci. 2011;231:1–2. doi: 10.1016/j.mbs.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 168.Jacobsen K, Pilyugin SS. Analysis of a mathematical model for tumor therapy with a fusogenic oncolytic virus. Mathematical Biosciences. 2015 doi: 10.1016/j.mbs.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 169.Yang Y, Zou L, Ruan S. Global Dynamics of a Delayed Within-host Viral Infection Model with both Virus-to-cell and Cell-to-cell Transmissions. Mathematical Biosciences. 2015 doi: 10.1016/j.mbs.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 170.Cosner C. Models for the effects of host movement in vector-borne disease systems. Mathematical Biosciences. 2015 doi: 10.1016/j.mbs.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 171.Schwartz EJ, Choi B, Rempala GA. Estimating Epidemic Parameters: Application to H1N1 Pandemic Data. Mathematical Biosciences. 2015 doi: 10.1016/j.mbs.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 172.Feng Z, Cen X, Zhao Y, Velasco-Hernandez JX. Coupled within-host and between-host dynamics and evolution of virulence. Mathematical Biosciences. 2015 doi: 10.1016/j.mbs.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 173.White A, Schaefer E, Wright C, Gaff H. Dynamics of two pathogens in a single tick population. Mathematical Biosciences. 2015 [Google Scholar]
- 174.Yan Y, Adam B, Moreno A, Galinski M, Kissinger JC, Gutierrez JB. Mathematical model of within-host interaction between a Plasmodium parasite and the immune system. Mathematical Biosciences. 2015 doi: 10.1016/j.mbs.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 175.Allen LJS, Schwartz EJ. Free-virus and cell-to-cell transmission in models of equine infectious anemia virus infection. Mathematical Biosciences. 2015 doi: 10.1016/j.mbs.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 176.Barfield M, Orive ME, Holt RD. The role of pathogen shedding in linking within- and between-host pathogen dynamics. Mathematical Biosciences. 2015 doi: 10.1016/j.mbs.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gutierrez JB, Lai M-J, Slavov G. Bivariate spline solution of time dependent nonlinear PDE for a population density over irregular domains. Mathematical Biosciences. 2015 doi: 10.1016/j.mbs.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 178.Voit EO. A First Course in Systems Biology. Garland Science; New York, NY: 2012. [Google Scholar]
- 179.Osbeck LM, Nersessian NJ, Malone KR, Newstetter WC. Science as Psychology: Sense-Making and Identity in Science Practice. Cambridge University Press; Cambridge, U.K.: 2010. [Google Scholar]
- 180.Galison PL. Image and Logic: A Material Culture of Microphysics. U. Chicago Press; 1997. [Google Scholar]
- 181.Star SL, Griesemer JR. Institutional ecology, ‘translations’ and boundary objects: amateurs and professionals in Berkeley's Museum of Vertebrate Zoology. Social Studies of Science. 1989;19:387–420. [Google Scholar]
- 182.Savageau MA. The challenge of reconstruction. New Biol. 1991;3:101–2. [PubMed] [Google Scholar]
- 183.Nat.Res.Council . Beyond Productivity: Information, Technology, Innovation, and Creativity. Nat. Acad. Press; 2003. [Google Scholar]
- 184.Spiro RJ. Cognitive Flexibility Theory Advanced Knowledge Structures in Ill-structured Domains. University of Illinois Press; 1988. [Google Scholar]
- 185.Voit EO, Newstetter WC, Kemp ML. A feel for systems. Mol. Syst. Biol. 2012;8:609. doi: 10.1038/msb.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.MaHPIC, Malaria Host-Pathogen Interaction Center 2012 www.systemsbiology.emory.edu/
- 187.Voit EO, Qi Z, Kikuchi S. Mesoscopic models of neurotransmission as intermediates between disease simulators and tools for discovering design principles. Pharmacopsychiatry. 2012;45(Suppl 1):S22–30. doi: 10.1055/s-0032-1304653. [DOI] [PubMed] [Google Scholar]