Abstract
MicroRNAs (miRNAs) are short noncoding RNAs that regulate gene expression by posttranscriptional and epigenetic mechanisms and affect many cellular processes and disease states. Over 2,000 human mature miRNAs have been identified, and at least 60% of all human protein-coding genes are known to be regulated by miRNAs. MicroRNA biogenesis involves classical transcription regulation and processing by key ribonucleases, as well as other protein factors and epigenetic mechanisms. Diabetic nephropathy (DN), a severe microvascular complication frequently associated with diabetes mellitus, is a major cause of renal failure. Although several mechanisms of regulation of key renal genes implicated in DN pathogenesis have been identified, a greater understanding is needed to develop better treatment modalities. Recent studies show that miRNAs can be induced in renal cells in vivo and in vitro under diabetic conditions and promote the accumulation of extracellular matrix proteins related to fibrosis and glomerular dysfunction. In this review, we highlight the significance of the expression of miRNAs in various stages of DN and emerging approaches to exploit them as biomarkers for early detection or novel therapeutic targets to prevent progression of DN.
Keywords: microRNAs, diabetic nephropathy, transforming growth factor β1, signal transduction, biomarkers, therapeutics
Introduction
Debilitating microvascular complications, such as diabetic nephropathy (DN), are highly prevalent in both type-1 and type-2 diabetes.1, 2 About 50% of diabetic patients have end-stage renal disease (ESRD), requiring painful and costly dialysis.2 Unfortunately, these diabetic patients also have a higher risk of macrovascular complications.1-3 It is now widely accepted that improved glycemic control reduces the development and delays the progression of microvascular complications in both type-14, 5 and type-2 diabetes.6, 7 Obesity is associated with increased rates of not only type-2 diabetes but also DN.8 A recent study showed that weight loss in overweight or obese adults with type-2 diabetes has beneficial outcomes for renal functions.9 Although several classical biochemical and molecular mechanisms and pathways have been widely studied and implicated in the pathogenesis of DN, continued escalation in the incidence and prevalence of DN worldwide clearly indicates that more investigation is needed. In this review, we describe some new concepts related to the molecular mechanisms underlying DN, with special emphasis on microRNAs (miRNAs) that are emerging as important regulators of renal cell gene expression under diabetic conditions.
Mesangial expansion (related to hypertrophy) and accumulation of extracellular matrix (ECM) proteins, such as collagens (related to fibrosis), in the kidney glomerular and tubular compartments are major features of DN and are associated with renal failure in diabetes.10-13 Another key predictor of DN progression is a reduction in the number of podocytes per glomerulus, as well as podocyte effacement and apoptosis.12-15 In the initiation and progression of DN, several factors have been identified to work individually and cooperatively to promote the expression of genes that mediate these renal pathologies. These factors include hyperglycemia, advanced glycation end products (AGEs), growth factors, oxidant stress, inflammation via cytokines/chemokines, as well as the phenomenon of metabolic memory, which can be regulated by epigenetic mechanisms.10-17 In this review, we focus on emerging mechanisms, such as those involving miRNAs, by which growth factors promote the glomerular hypertrophy and ECM protein accumulation that are key features of the early stages of DN, because these mechanisms can be targeted for early intervention to more effectively prevent patients from progressing to renal failure and ESRD.10-13, 18-21
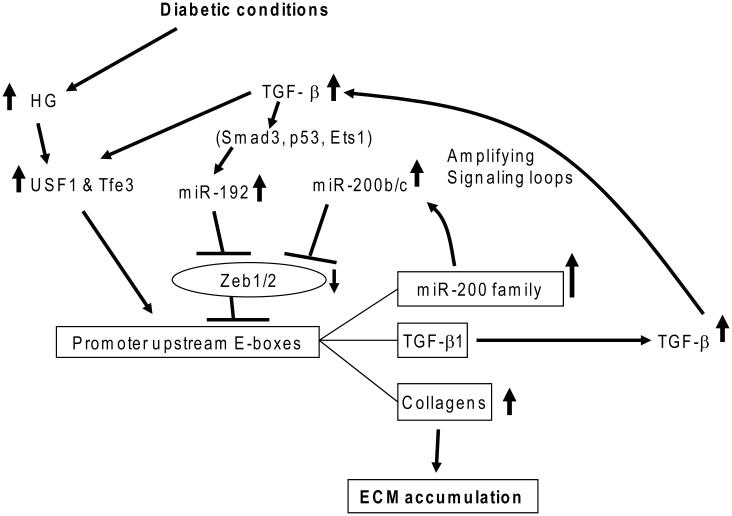
High-glucose (HG) and diabetic conditions have profound effects on all types of renal cells (such as podocytes, and mesangial, tubular, and endothelial cells) and on monocyte/macrophages (inducing their infiltration into the glomerulus). HG conditions also increase the formation of AGEs and production of growth factors, such as transforming growth factor-β1 (TGF-β1), angiotensin II, and platelet-derived growth factor (PDGF) in renal cells,13, 18, 19, 22, 23 via several well-described signaling mechanisms that promote renal hypertrophy, fibrosis, and inflammation. TGF-β1 is a major player in DN pathogenesis, mainly because of its potent profibrotic actions; its level is increased in most renal cells in diabetes.10,12,18,19 Evidence shows that upstream stimulatory factors (upstream transcription factor 1 (USF1)/USF2)) induced by HG conditions upregulate TGF-β1 in mesangial cells (MCs) through promoter E-box elements in Tgfb1 (Fig. 1).24, 25 TGF-β1 acts on renal cells via specific receptors, leading to the phosphorylation and nuclear translocation of Smad2/Smad3 transcription factors,26, 27 which bind to Smad-binding elements in the promoters of key TGF-β1 target genes to induce their expression.28, 29 In addition, mitogen-activated protein kinases, such as p38 and extracellular signal-related kinases (ERKs)30-32 and Akt kinases (through inhibition of Pten)33, 34 are also activated by TGF-β1. Besides HG and TGF-β1, other diabetogenic factors can also trigger such signaling events and gene regulation, and crosstalk between them can amplify and enhance the expression of pathological genes related to the progression of DN.13
Figure 1.
TGF-β1 induced by high-glucose conditions via upstream transcription factors (USFs) creates signaling circuits mediated by miRNAs. Diabetic conditions increase the expression of TGF-β1 initially through USFs that bind to E-boxes in the TGF-β1 promoter. Upregulated TGF-β1 induces miR192 (inhibitor of E-box repressors Zeb1/2) via mechanisms involving the actions of Smad3, Ets-1, and p53. MiR-200b/c are also upregulated due to the downregulation of Zeb1/2 by miR-192. MiR-200b/c targeting Zeb1/2 can lead to a further decrease of Zeb1/2 and thus further augments the expression of miR-200b/c, TGF-β1, Col1a2, and Col4a1 through a loss of repression, along with a gain of activation (via Tfe3 and/or USF1) at their E-boxes. These signaling loops accelerate TGF-β1 signaling and downstream ECM gene expression involved in the progression of chronic kidney diseases such as DN. Modified from Kato et al.25
Despite significant efforts in investigating the pathogenesis of DN, current therapies for DN do not consistently provide satisfactory prevention from progression to ESRD in many patients, implying that there may be additional unknown mediators and mechanisms that need to be explored. In particular, an in-depth examination of mechanisms mediating early events in DN, such as ECM protein accumulation and hypertrophy, could help identify targets whose inhibition could be a valuable approach to prevent further progression of DN. MicroRNAs present one such opportunity because recent studies demonstrate that, apart from coding genes, diabetogenic and growth factors can also induce noncoding RNAs (ncRNAs), such as miRNAs. These miRNAs are dysregulated in early DN and can promote the expression of ECM proteins and other genes associated with the initial stages of DN.
Here, we highlight the biogenesis and modes of action of miRNAs, their roles in regulating key genes associated with the pathology of early DN including participation in signaling circuits, mechanisms of their regulation (including epigenetics), and their potential to serve as noninvasive biomarkers for the early detection of DN, as well as for the prevention or treatment of DN.
MicroRNAs
MicroRNAs are naturally expressed, small ncRNAs (20–22 nucleotides) that regulate gene expression through posttranscriptional mechanisms. In general, miRNAs lower the expression of target genes by imperfect base pairing to the 3′-untranslated regions of target mRNAs via translational repression and/or mRNA degradation.35 More than 2,000 human mature miRNAs have now been identified, and at least 60% of all human protein-coding genes are known to be regulated by miRNAs.35 Therefore, miRNAs have tremendous potential to modulate the expression of numerous genes to change cellular and biochemical functions, potentially affecting the development or progression of various diseases. Since their discovery twenty years ago, miRNAs have been implicated in numerous biological processes, as well as in the pathology of several human diseases.
Biogenesis, processing, and functions of miRNAs in gene regulation
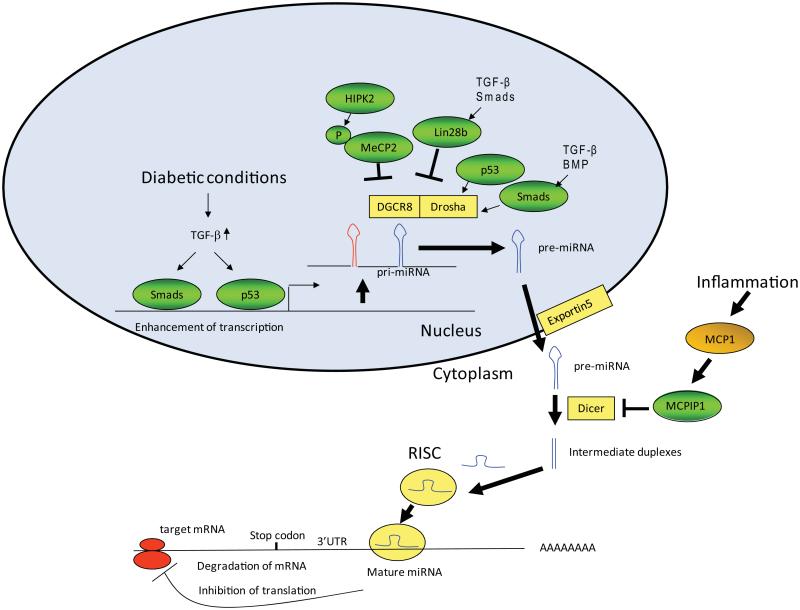
MicroRNA genes are quite often intergenic and transcribed independently, but they can also be transcribed along with a host gene that is either noncoding or protein coding. MicroRNAs are transcribed as primary transcripts (pri-miRNAs) in the nucleus by RNA polymerase II (Fig. 2). Pri-miRNAs are cleaved by the ribonuclease III Drosha in a microprocessor complex and processed to precursor miRNAs (pre-miRNAs; ~70 nucleotide stem-loop structures) in the nucleus (Fig. 2),36, 37 which are then translocated by exportin-5 to the cytoplasm where they are further processed to mature miRNAs by another ribonuclease, Dicer.36, 37 A mature miRNA duplex is recruited to an RNA-induced silencing complex (RISC), which includes the Argonaute (Ago) family proteins essential for miRNA-mediated gene silencing. Mature miRNAs interact with the 3′-untranslated region of their target mRNAs and induce translational repression or degradation in a RISC.36 Apart from these classical modes of miRNA biogenesis and processing, which are further influenced by various proteins within the miRNA microprocessor complex, other novel mechanisms have also been reported.16, 36, 37
Figure 2.
Biogenesis and actions of microRNAs. MicroRNAs (miRNAs) are initially transcribed in the nucleus as pri-miRNAs, which are processed into pre-miRNAs by the Drosha enzyme. Pre-miRNAs are further cleaved by Dicer to result in double-stranded miRNA duplexes. Drosha processing can be regulated by p53 or Smads (activated by TGF-β or BMPs). This process can also be inhibited via interactions of phosphorylated MeCP2 (by HIPK2) with DGCR8, a cofactor of Drosha. In addition, Dicer processing can be inhibited by MCPIP1, which is induced by MCP-1 (related to inflammation). Processing of let-7 family members is inhibited by Lin28b, which can be induced by TGF-β/Smad signaling. The miRNA duplexes are then unwound by the action of Dicer, and the mature miRNA guide strand is loaded into the RISC complex. Please refer to the main text for details. RISC, RNA-induced silencing complex; UTR, untranslated region. Modified from Kato et al.16
Modulators of miRNA biogenesis
Recent findings also show that miRNA biogenesis and target recognition can be modulated by additional mechanisms that involve specific RNA-binding proteins. Drosha-mediated processing was reported to be enhanced by Smads in response to TGF-β1 or bone morphogenetic proteins (BMPs),38,39 or by p5340 (Fig. 2). Biogenesis of the let-7 family of miRNAs, initially identified as tumor suppressors,41-43 is repressed by the RNA-binding protein Lin28,44, 45 which has two members: Lin28A, predominantly localized in the cytoplasm, and Lin28B, which is mainly in the nucleus due to the presence of nuclear localization signals. Lin28A and Lin28B have similar functions but different mechanisms for binding and inhibiting let-7 miRNAs.46 For instance, Lin28A mainly inhibits pre-let-7 processing by Dicer, and this enhances the degradation of oligouridylated pre-let-7 RNAs; Lin28B binds to pri-let-7 miRNAs in the nucleus and blocks processing by Drosha and DGCR8. The similar regulation of let-7 processing by the two Lin28 members is conserved in normal development in C. elegans.47 Because of the tumor-suppressor roles of let-7, the inhibitory activities of Lin28A and Lin28B on let-7 biogenesis can be translated to drug therapy.46 A recent report showed that homeodomain-interacting protein kinase 2 (HIPK2) induces phosphorylation of methyl CpG–binding protein 2 (MeCP2), which is known to act as a transcriptional repressor by binding to methylated cytosines in DNA, and phospho-MeCP2 binds to DGCR8 (another component of the microprocessor complex) to inhibit Drosha-mediated processing of pri-miRNAs (Fig. 2).48-50 This is noteworthy because HIPK2 was identified as a key regulator of kidney fibrosis,51 suggesting that the HIPK2-to-MeCP2 connection may also modulate the processing of key miRNAs involved in the early stages of DN, although more studies are necessary to examine this. Another modulator of Dicer action, monocyte chemoattractant protein-1–induced protein 1 (MCPIP1), was identified as a suppressor of miRNA biogenesis in response to inflammation (MCP1),52 which resulted in the inhibition of miRNA maturation (Fig. 2). Some pre-miRNAs have also been reported to be affected by adenosine deaminases, nuclear RNA-editing enzymes, which convert adenosine residues to inosine on double-stranded RNAs. This increases miRNA-mediated silencing of a target RNA through enhanced complex formation of the target RNA with Dicer.53 Methylation of 5′ monophosphate ends of miRNA precursors by BCDIN3D, an RNA methyltransferase, also inhibits miRNA processing. Enhanced miR-145 maturation by inhibition of BCDIN3D has been associated with breast cancer tumorigenesis.54 Interestingly, some miRNAs can also be processed in a Dicer-independent formation. For example, miR-451, a miRNA important in erythropoiesis, is trimmed by Ago2 slicer activity and matured without Dicer.55 In some instances, Dicer and Ago2 are destroyed by autophagy, resulting in reduced miRNA activity.56 Also, IRE1alpha RNase, an endoplasmic reticulum (ER) transmembrane kinase-endoribonuclease, induces rapid decay of certain miRNAs targeting caspase-2, which results in increased ER stress–induced cell death.57 Therefore, key modulators of miRNA biogenesis are emerging as additional players in controlling the expression of miRNAs, and thus the physiological and pathological functions of miRNAs. Together, it is clear that cellular levels of miRNAs can be modulated by various mechanisms.
Notably, the critical biological roles of miRNAs and their processing by Drosha and Dicer are exemplified by studies demonstrating severe cardiac and renal defects in mice deficient in these proteins.58-65 Importantly, such observations also indicate that these small miRNAs have tremendous potential to be exploited for uncovering new mechanisms underlying the progression of DN, as well as for developing novel biomarkers and therapeutic targets for this disease.
MicroRNAs in the early stage of DN and in signal-amplifying circuits
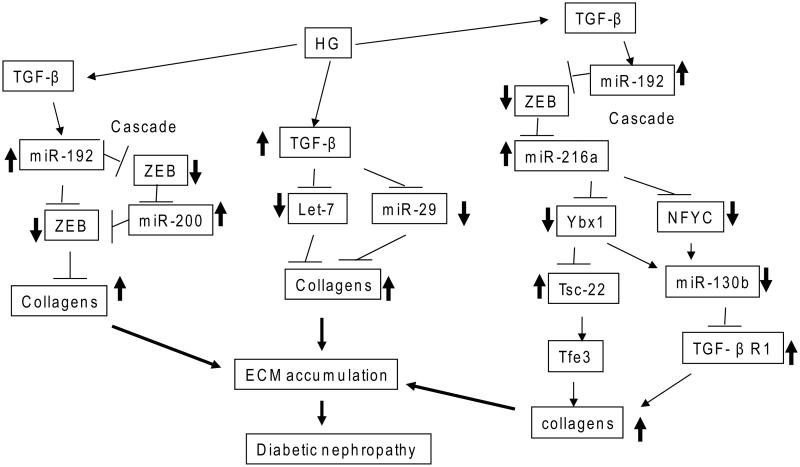
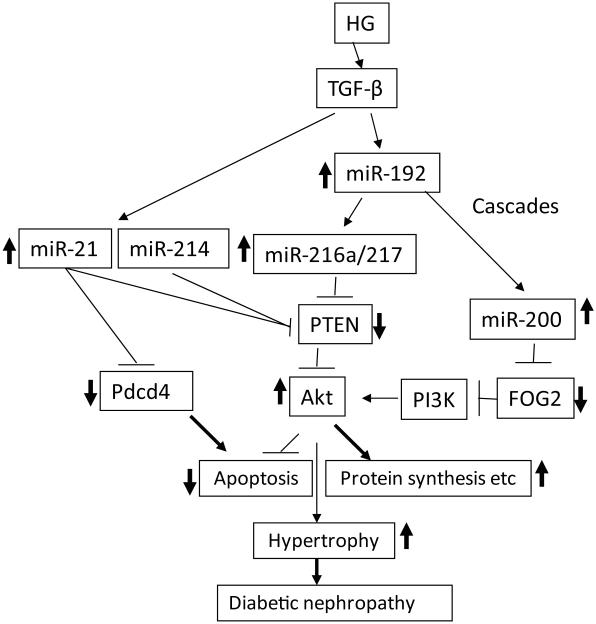
Strong evidence of the involvement of miRNAs in kidney dysfunction has come from observations of severe renal phenotypes in mice with podocyte-specific deletion of Dicer, phenotypes including proteinuria, podocyte foot process effacement and apoptosis, glomerulosclerosis, and tubulointerstitial fibrosis with renal failure.58,59,61 However, because this strategy inhibits many miRNAs, it did not clarify the functions of specific ones in kidney failure. In an early study, five miRNAs (miR-192, miR-194, miR-204, miR-215, and miR-216a) were identified to be enriched in the kidney compared to other organs, suggesting their potential function in the kidney.66 Another comprehensive study using rat kidney reported different miRNA expression patterns in the renal cortex and medulla, and suggested possible cell type– and tissue-specific functions.67 The earliest studies to examine the renal functions of miRNAs were focused on these five miRNAs because of their observed expression in the kidney. Higher expression of several miRNAs (miR-192, miR-200b/c, miR-216a, and miR-217) was detected in mouse MCs (MMCs) treated with TGF-β1 and in renal glomeruli from mouse models of diabetes depicting early stages of DN (streptozotocin-injected type-1 diabetic mice and type-2 diabetic db/db mice (a leptin receptor mutant)) compared to nondiabetic control mice.25,68-71 Several lines of evidence have shown that miR-192 upregulates the key fibrotic genes Col1a2 and Col4a1 related to early DN in MCs by targeting the E-box repressors Zeb1 and Zeb2 to relieve repression at the Col1a2 and Col4a1 promoters (Fig. 3). Interestingly, miR-192 upregulates other miRNAs in MCs, including miR-216a/miR-217 and miR-200b/c, also through targeting the E-box repressors Zeb1 and Zeb2 in their promoters, and this creates amplifying circuits to further augment collagen expression in DN (Fig. 3). Interestingly, both miR-216a and miR-217 activate Akt kinase by targeting Pten in MMCs treated with TGF-β1, enhancing cellular hypertrophy associated with Akt kinase activation (Fig. 4).69 MiR-200b/c can also upregulate collagen expression and the autoregulation of TGF-β1 in MMCs by inhibiting Zeb1 to relieve repression at the Tgfb1 promoter E-box elements (Figs. 1 and 3).25 Recent studies showed that miR-200b/c activates Akt by targeting FOG2, an inhibitor of phosphoinositide 3-kinase (PI3K) (Fig. 4).71 The in vivo involvement of miR-192 in the pathogenesis of DN was demonstrated with miR-192 gene knockout mice and mice treated with miR-192 inhibitor, both of which showed less severe phenotypes of DN (glomerular hypertrophy, fibrosis, and proteinuria) compared to wild-type mice.72, 73 In addition, mutual activation of miR-192 and p53 in an amplification loop augmented ECM gene expression and hypertrophy in response to TGF-β1 in MCs.72,74
Figure 3.
Extracellular matrix (ECM) protein accumulation by microRNA cascades in the early stages of DN. High-glucose (HG) conditions induce TGF-β1, which causes DN through increased fibrosis and ECM protein accumulation. Signaling cascades are depicted to delineate key intermediate black boxes in TGF-β1–induced signaling in DN. TGF-β1 increases ECM genes, such as collagens, through miR-192 and miR-200 and downregulation of their targets, E-box repressors (ZEB1/2). One cascade shown runs from miR-192 to miR-200 family members. In another, HG and TGF-β1 also induce collagens by inhibiting the expression of miR-29 family and let-7 family members that target several collagens. In a third cascade, miR-192 regulation of collagens is via the intermediate actions of miR-216a and miR-130b and their targets, including Ybx1, NFYC, and TGF-β1R1. These miRNA cascades can amplify the initial signal transduction events to accelerate chronic kidney diseases such as DN.
Figure 4.
MicroRNA cascades related to hypertrophy and apoptosis in the early stages of DN. High-glucose (HG) conditions induce TGF-β1, which causes DN through increased hypertrophy via activation of Akt kinase. TGF-β1 activates Akt kinase through the miR-216a/217/200 family, which target and downregulate PTEN or FOG2 and thereby promoter hypertrophy. HG and TGF-β1 also activate Akt by increasing miR-21 and miR-214 that target PTEN. Two cascades from miR-192 are depicted: to the miR-216a/217 cluster and to the miR-200 family. Increased protein synthesis and inhibition of apoptosis through Akt activation can lead to hypertrophy. Increased miR-21 can also affect apoptosis directly through its apoptosis-related targets, such as Pdcd4. These miRNA cascades can amplify the signaling events initiated by hyperglycemia and TGF-β1 and thereby promote chronic kidney diseases such as DN.
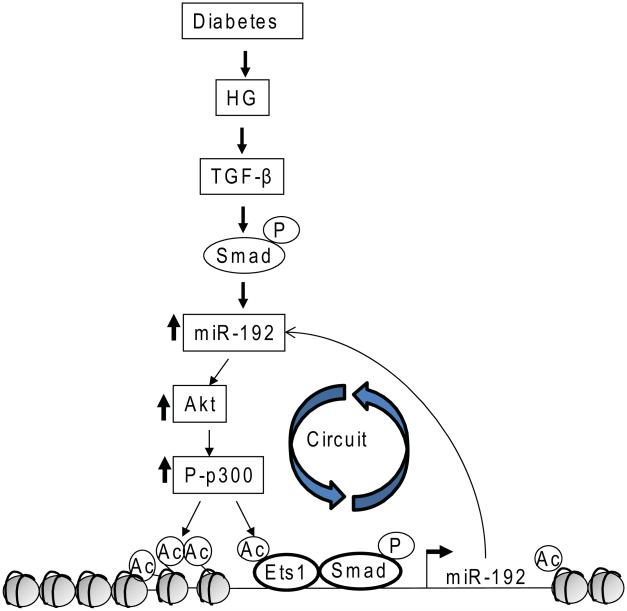
Detailed studies on the mechanism of miR-192 gene promoter regulation by TGF-β1 in MMCs revealed the involvement of not only Smads, but also Ets-1 transcription factors and novel chromatin remodeling mechanisms through histone acetylation mediated by the histone acetytransferase p300 and its phosphorylation/activation by Akt kinase (Fig. 5).75 Because miR-192 itself activates the Akt-p300 pathway, this illustrates another amplifying loop accelerating the signaling downstream of TGF-β1 (Fig. 5).74, 75 MiR-192 gene promoter regulation by Smads and p53, as well as by hepatocyte nuclear factor-1 (HNF-1), has been reported in tubular epithelial cells, which also showed renal cell–specific mechanisms.76, 77 Other studies also reported increased levels of these miRNAs (miR-192, miR-200 family, miR-216a, miR-217) in renal cells treated with HG or TGF-β1, or in animal models of kidney injury.76, 78-83 MiR-192 was increased in streptozotocin-injected mice fed with a high-fat diet in farnesoid X receptor gene knockout mice.79 MiR-192 and miR-200b were found to be higher in the glomeruli of diabetic db/db mice and in podocytes and renal microvascular endothelial cells treated with HG.80 Moreover, miR-192 and miR-215 were upregulated in MCs treated with TGF-β1 and in glomeruli from diabetic db/db mice, and were suggested to induce phenotype transition of MCs by targeting β-catenin–interacting protein-1 (CTNNB1).81 Together, studies with in vitro and in vivo mouse models of early DN have identified miR-192 as a master miRNA regulator in MCs treated with TGF-β1, and in the increased expression of ECM genes associated with the early stage of DN.25,68-72,74-76
Figure 5.
Epigenetic regulation of miRNA expression (with miR-192 as an example) and circuitry mediated by Akt/p300 signaling. Autoactivation of the miR-192 promoter by acetylation of chromatin histones and transcription factors (such as Ets-1) through p300. Activation of Akt is mediated by key miRNAs downstream of miR-192 that target and downregulate PTEN or FOG2 (Fig. 4). Continuous and uncontrolled activation of these signaling pathways in this circuitry may induce chronic kidney diseases such as DN.
In recent years, several other miRNAs have been reported to have functions related to DN. Among these, miR-21 has been studied extensively because many of its targets are both relevant to DN and related to Akt activation, hypertrophy, and apoptosis. MiR-21 was highly expressed in the renal cortex of OVE26 type-1 diabetic mice, and by targeting Pten it promotes Akt and mTOR activation, all factors associated with DN (Fig. 4).84 In db/db mice, miR-21 accelerated TGF-β1 signaling by targeting Smad7; thus, an inhibitor of miR-21 was evaluated as a therapeutic target in this model.85 MiR-21 was also shown to be upregulated in the KK-Ay mouse model of DN and contributed to renal fibrosis by targeting matrix metalloproteinase-9 and metalloproteinase inhibitor 1.86 Mir21−/− mice had reduced interstitial fibrosis in response to renal injury, while wild-type mice treated with anti-miR-21 oligonucleotides depicted reduced fibrosis.87 By contrast, miR-21 was also reported in one study to be downregulated in db/db mice, and its overexpression blocked MC proliferation.88 Moreover, more severe phenotypes including podocyte loss and albuminuria were observed in Mir21 knockout mice crossed with Tgfb1 transgenic mice, compared to that seen in Tgfb1 transgenic mice alone, suggesting that deletion of Mir21 is detrimental under these conditions, especially to podocytes, due to upregulation of the pro-apoptotic targets of miR-21.89 In other studies, miR-214 upregulated by TGF-β1 was found to activate Akt to reduce apoptosis in human monocyte THP1 cells treated with AGE products,90 and to promote renal fibrosis in a mouse model of unilateral ureteral obstruction (UUO)91 (Fig. 4). MiR-451 was downregulated in early DN in db/db mice, and this could induce hypertrophy by targeting Ywhaz, a protein required for activation of mitogen-activated protein kinase.92
Regarding ECM protein accumulation/fibrosis, studies demonstrate that downregulation of miR-29 family members, which directly target multiple collagens, can lead to increased collagen deposition in tubular cells, MCs, and podocytes treated with TGF-β93, 94 (Fig. 3). A profibrotic role for miR-192 and an antifibrotic role for miR-29 family members were reported even in other nondiabetic animal models of renal fibrosis.76,82,95 Downregulation of miR-29a by HG increased collagen type IV in human tubular HK-2 cells (Fig. 3).96 However, miR-29c was shown to be upregulated and to activate Rho kinase by targeting Spry-1, which induces ECM protein accumulation and podocyte apoptosis under similar diabetic conditions.80 A recent report revealed that linagliptin, a dipeptidyl peptidase-4 (DPP-4) inhibitor now used as an antidiabetic drug, can also confer renal protection and ameliorate fibrosis in a mouse model of DN by inducing miR-29, which targets DPP-4.97 MiR-377 was upregulated by HG or TGF-β1in MCs, and increased fibronectin expression and oxidant stress by targeting manganese superoxide dismutase and p21-activated kinase.78
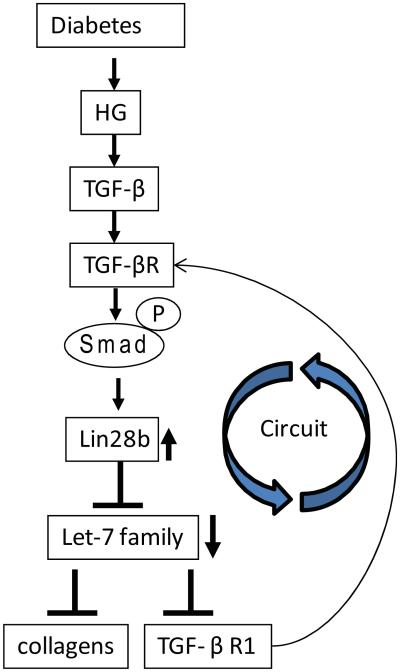
Let-7 family members were originally identified as tumor suppressors because these miRNAs were downregulated in several cancers.41-43 Increasing evidence shows that key let-7 members are also downregulated by TGF-β1 in renal cells, and this can promote fibrosis through upregulating its targets, TGF-β1 receptor type-1 (TGF-β1R1) and collagens (Fig. 3).98, 99 Moreover, examination of the mechanism of downregulation of let-7 family members revealed that downregulation can occur via Smad2/3-mediated upregulation of Lin28b (a negative regulator of let-7) in MMCs treated with HG or TGF-β1, as well as in glomeruli from streptozotocin (STZ) diabetic mice (Fig. 6).100 These studies have also identified Lin28 as a novel new target gene of TGF-β1. Because let-7 family members target TGF-β1R1, these data reveal yet another signal-amplifying loop leading to acceleration of DN74 (Fig. 6).
Figure 6.
Regulation of pro-fibrotic genes by diabetic conditions through Lin28-mediated inhibition of biogenesis of the let-7 family. TGF-β1 induced by diabetic conditions upregulates Lin28b through activation of Smad 2/3. Lin28b downregulates let-7 family miRNAs by inhibiting the processing of let-7. Decreased levels of let-7 members result in the upregulation of collagens (let-7 targets), leading to glomerular ECM protein accumulation and progression of DN, illustrating another circuit for signal perpetuation.
Signaling circuitry was also observed in increased TGF-β1–mediated actions and fibrosis by miR-433 through targeting antizyme inhibitor 1, which regulates polyamine synthesis.101 A recent report also showed that TGF-β1 can downregulate miR-130b, and that this induces the miR-130b target TGF-β1R1in MCs through mechanisms involving a cascade from miR-216a to Ybx1/NFYC (Fig. 3).102 MiR-135a was identified as upregulated in serum and renal tissue from patients with DN and db/db mice, and its levels were related to microalbuminuria and renal fibrosis.83 MiR-135a could promote MC proliferation and increase synthesis of ECM proteins by targeting transient receptor potential cation channel subfamily C member 1 (TRPC1). Decreases in the miR-30 family members were associated with accelerated DN due to upregulation of their target CTGF (connective tissue growth factor), another important profibrotic factor.103 MiR-22 was suggested to be a master regulator of BMP-7 and BMP-6, and to further increase TGF-β1 signaling in mouse kidney fibrosis models.104 MiR-93 was identified as a key miRNA downregulated in podocytes and renal microvascular endothelial cells treated with HG, as well as in glomeruli of diabetic db/db mice.105 Decreased miR-93 expression was shown to enhance angiogenesis via increases in its pro-angiogenic target, VEGF-A (vascular endothelial growth factor A).105
Oxidant stress has been implicated in the pathogenesis of DN, and several miRNAs that modulate oxidant stress have been identified. For example, miR-25 and miR-146a, which are downregulated in diabetic conditions, target Nox4, a key player in DN.106-110 Decreased miR-205 was associated with increased production of reactive oxygen species by targeting heme oxygenase and superoxide dismutase (SOD) in HK-2 tubular cells.111 The abovementioned miRNA cascades (miR-192, miR-216a, miR-217, and the miR-200 family) activate Akt and inhibit FoxO3a/SOD2 signaling in MCs.33, 69, 71 Aldose reductase downregulates miR-200a-3p and miR-141-3p, and regulates oxidant stress by targeting Keap1-Nrf2, TGF-β1/TGF-β2, and Zeb1/Zeb2 signaling in MCs and kidneys of diabetic mice.112 Thus, approaches to prevent renal oxidant stress and DN could involve inhibition of pro-oxidant miRNAs and enhancement of antioxidant miRNAs.16
Together, these findings clearly demonstrate that several miRNAs are dysregulated by diabetogenic factors in vitro and in in vivo models of early DN, a complex chronic disease with key features manifesting in the early versus late stages. Immense effort is being made to identify key biomarkers of early DN that could be translated to early intervention therapies aimed at preventing progression to renal failure. MiRNAs that are dysregulated by growth factors, HG, and related mediators in early DN could serve as valuable targets for early intervention.
MicroRNAs in the later stages of DN and renal cell–specific expression
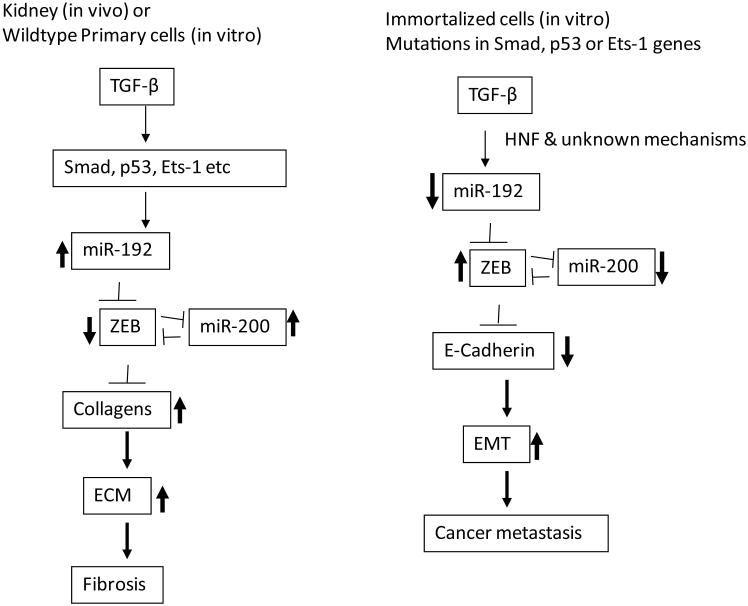
Several miRNAs, including miR-192, have reported to be downregulated in models of more severe or late-stage DN, such as diabetic ApoE-deficient (Apoe−/−) mice. MiR-192 was also reported in some studies of cultured proximal tubular epithelial cells treated with HG or TGF-β1, which was associated with increased fibrosis,113, 114 suggesting cell-specific effects. However, other reports show upregulation of miR-192 expression in tubular cells treated with TGF-β1 and increased fibrosis.76 These complexities could be due to cell type–specific effects of miRNAs and differences in the animal models studied. Tubular damage and necrosis/apoptosis are usually observed in the later stages of DN, which could lead to decreases in miRNA levels. However, cell-specific transcription factors could also dictate the actions of miRNAs. This is supported by data showing that key isoforms of hepatocyte nuclear factor (HNF)1 that mediate TGF-β1–induced downregulation of miR-192 in tubular cells are not present in MC.77 Because TGF-β1–induced miR-192 expression was abolished in MCs from p53−/− or Ets1−/− mice, the cell type–specific response of miR-192 to TGF-β1 can also be explained by the cellular status of p53 or Ets-1 (Fig. 7).72,75 Similarly, an increase in miR-192 in mouse models of kidney fibrosis was abolished in Smad3−/− mice.76 Although two reports showed an increase of miR-200b/c related to ECM protein accumulation and Akt activation in primary MMCs,25, 71 one report showed that miR-200a was downregulated in TGF-β1–treated proximal tubular epithelial cells, also related to fibrosis in which miR-200a targets TGF-β2.115 The miR-200 family has been well documented as regulators of the epithelial phenotype by targeting the transcriptional repressors of E-cadherin, Zeb1, and Zeb2,116 establishing an important link between miRNAs such as miR-200, TGF-β1 actions, and epithelial–mesenchymal transition (EMT), especially in cancer metastasis. On the other hand, increasing evidence shows that EMT may play a minor, or no, role in renal fibrosis and DN.117,118 Features of EMT are generally noticed mainly in immortalized cultured epithelial cell lines in vitro, but not well characterized in in vivo renal fibrosis in humans or animal models.119 Key differences are present in cancer models and immortalized cell lines relative to non-cancer models, with the former likely to have mutations in tumor-suppressor genes, such as p53 and oncogenes, which, along with cell type–specific transcription factors and epigenetic marks, can lead to differences in the cell-type miRNA responses to TGF-β1 (Fig. 7).
Figure 7.
Differential effects of miRNAs in kidney fibrosis and epithelial to mesenchymal transition (EMT) in cancer metastasis. In the early stage of DN, TGF-β1 induced by diabetic conditions upregulates miR-192 in mesangial and other renal cells through the activation of Smad 2/3, p53, or via Ets-1-mediated mechanisms. MiR-192 induces collagens by inhibiting E-box repressors (Zeb1/2) and also via increases in miR-200 family members. The miR-200 family also enhances collagen expression by targeting Zeb1/2 to amplify the signaling. On the other hand, in epithelial cancer cell lines or immortalized epithelial cell lines that have mutation in genes such as p53, Smads or Ets-1, TGF-β1 decreases miR-192. This also leads to decreases miR-200 family and E-Cadherin genes through E-box repressors (Zeb1/2) to induce EMT. Renal cell–specific transcription factors (such as HNF) can also be critical for cell-specific regulation of miRNAs in response to TGF-β1.
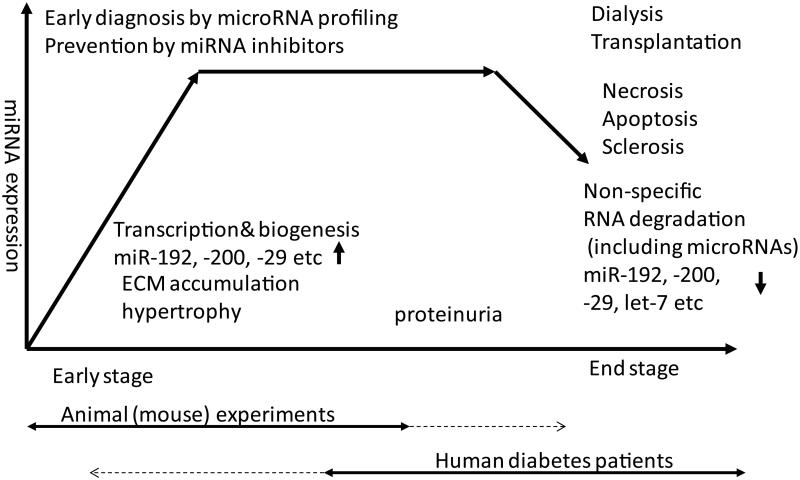
Discrepancies have also been noted in miRNA expression data from human DN patients versus animal models of DN (Fig. 8). Usually, renal biopsies used for studying miRNA expression are obtained from patients experiencing an ongoing progression (intermediate) or late stage of DN because of indications of more severe symptoms, including micro-and macro-albuminuria, that are less evident in most currently available animal models of DN.120 It is likely that levels of many RNAs (including coding and noncoding) are lower in samples obtained from such patients in late stages of DN, compared to those in intermediate or early DN as reported in a clinical study,113 due to tissue necrosis and apoptotic cells that would yield poor quality of RNA. Another confounding factor is the paucity of material from normal human kidneys for comparison. Thus, these data may lead to the interpretation that downregulation of certain renal miRNAs are associated with progression to end-stage renal failure. In the animal experiments, however, it is possible to monitor from the non-diabetic healthy stage to early- and even late-stage DN in some models. Therefore, certain miRNAs observed to be upregulated in animal models representing early DN may in fact present as being downregulated in samples from patients in later stages of DN because of poor quality of tissue or RNA. In fact, in one example, it was seen that inhibition of miR-192, which is upregulated in mouse models of early DN models13, 68, 75 but decreased in biopsies from patients in late stages of DN,113 can ameliorate DN in animal models.72,73 Therefore, while clinically challenging, it may be worthwhile to identify, target, and inhibit disease-inducing miRNAs in patients during the early stages of diabetes or DN (even before the development of overt symptoms) in order to prevent progression of renal dysfunction.
Figure 8.
DN stage–specific regulation of miRNAs: humans (~ late stage) versus animal models (~ early stage). Usually, renal biopsies are obtained from diabetic patients at the middle or late stage of DN because they present with micro- or macro-albuminuria, glomerular filtration rate (GFR) decline, or other clear symptoms of renal dysfunction. When compared, most RNAs (including coding and noncoding) are likely to depict lower expression in the end stage of DN relative to the intermediate stage. It might be interpreted that downregulation of RNA is associated with renal failure; however, decreases in RNA expression could be due to poor quality of RNAs in end-stage biopsies. In experimental models of DN, animals can be systematically monitored from the healthy to more advanced stages of renal disease, although most mouse models used to study miRNAs do not depict features of human DN. Therefore, some miRNAs found to be upregulated in the early stage of DN in animal experiments may appear to be downregulated in samples from humans who are in much later stages of DN, due to poor sample quality or non-comparable stages. Thus, miRNAs that are induced in early DN in animal models can be potentially assessed as therapeutic targets in humans because their inhibition might slow down progression to late- or end-stage disease even before clear symptoms are evident.
MicroRNAs as biomarkers for DN
Because miRNAs are stable and detectable in human biofluids, precise detection of miRNA profiles in biofluids is attractive in clinical translational research for biomarker development and diagnostics, as early diagnosis of DN can help effectively prevent the progression to renal failure. Several proteins, peptides, growth factors, and cytokines have been studied as known biomarkers of DN progression.121 Recently, miRNAs have become of great interest as sensitive, noninvasive, and precise stage-specific diagnostic biomarkers for DN, especially because of their stability in biofluids (such as urine and plasma) and in exosomes, and because of established techniques for reliable detection and quantification by sequencing, quantitative PCR, and microarrays.122-125 Several reports now demonstrate comprehensive profiles in patient urine, urinary sediment, and serum of miRNAs that could be correlated with specific stages of DN, fibrosis, and renal function decline (estimated by glomerular filtration rate, GFR).83, 126-135 In particular, exosomes in urine are an extremely valuable source for miRNA profiling in renal disorders because they originate from most renal cells.132 For example, miR-145 was reported to be enriched in urinary exosomes from type-1 diabetic patients showing microalbuminuria and in HG-treated MCs.136 MiRNAs that are dysregulated in renal tissues of animal models of DN or in patients (such as those discussed in above sections) could be evaluated in urine or serum samples at various times as candidate biomarkers of DN staging. 130 However, the caveat with clinical biomarker studies is the varied nature of patient cohorts, the number of patients per study, as well as the number of miRNAs estimated in each study and the unknown cellular source of the miRNAs. A comprehensive profiling of miRNAs was recently performed in urine samples obtained from patients with type-1 diabetes at different stages of DN that provided strong support for the use of miRNA profiles as molecular predictors of DN.137 Because miRNAs are stable even in paraffin-embedded sections, it may be possible to find new biomarkers from archived materials that have associated clinical information. MiRNAs that originate from dead or diseased cells may also be useful for detecting organ failure. On the other hand, since symptoms and molecular mechanisms of disease progression can vary from patient to patient, in the future patients may be segregated for personalized medicine and treatment on the basis of their individual miRNA profiles.74 As mentioned above, screening of key sets of miRNAs in patients who do not yet show symptoms of renal dysfunction can aid in detecting the onset of DN. Alternatively, miRNAs identified in animal models of early DN may be examined in patients to assess DN risk. Overall, although several challenges remain, it is clear that miRNAs are very attractive as simple and accurate biomarkers, particularly for kidney diseases such as DN.
MicroRNA-based therapy for DN
Despite numerous attempts to develop more effective drugs for the treatment of DN, not many treatments have reached clinical use. There are ongoing efforts, for example, to target TGF-β1 or to develop improved approaches for renin-angiotensin blockade.22,138,139 Clearly, new concepts should be investigated to design novel and better therapies.
Because several miRNAs have now been identified in the pathology of DN, these small molecules present new possibility for therapeutic intervention. Many attempts to downregulate or upregulate miRNAs using one of several delivery approaches in animal models of DN in vivo have been reported.13, 16, 132 Recent trends to control the expression of miRNA levels include the use of chemically-modified, stable, nuclease-resistant oligonucleotides (miRNA inhibitors and mimics), which could be developed for patient use in the future. Overexpression of miRNAs with sponges, or deletion of genomic regions of miRNAs in animal models, are useful as experimental models to examine the in vivo functions of key miRNAs, though they are not practical for use in humans. Anti-miRNAs modified with locked nucleic acid (LNA) have been broadly used to specifically inhibit particular miRNAs,69, 73, 140 including in some clinical trials.141, 142 Expression of miR-192, its downstream miRNAs (miR-216a, miR-217, and miR-200 family), and p53 were all effectively inhibited by LNA-modified anti-miR-192 in the renal cortex of normal and STZ-injected diabetic mice, and led to amelioration in DN symptoms.25, 69, 72, 73 The rates of DN in db/db mice were reduced by 2′-O-methyl antisense oligonucleotides targeting miR-29c.80 Furthermore, knockdown of miR-135a in diabetic kidneys in db/db mice restored levels of TRPC1 and reduced synthesis of ECM proteins.83 Other reports have shown MiR-21 inhibitors to be effective in treating animal models of renal failure, fibrosis, and diabetes.85, 87 In addition, approaches using antagomirs and agomirs have been tested and include the use of adeno-associated virus vectors and miRNA sponges,132 whereas bacteriophage MS2 virus-like particles have been evaluated for overexpression of downregulated miRNAs.143
Besides targeting miRNAs themselves, the host genes of miRNAs may also be evaluated as therapeutic targets. An early report showed that specific miRNAs (miR-216a and miR-217) were induced by TGF-β1 together with their host ncRNA RP23 in MCs,69 and that siRNA-mediated inhibition of RP23 mRNA could also reduce ECM gene expression. MiR-192 is co-regulated in MCs with its host ncRNA CJ241444, which is induced by TGF-β1 through CJ241444 gene promoter Smad-binding elements and epigenetic regulation via protein Ets-1 and histone acetylation.75 Another strategy is to target the upstream mechanisms controlling the expression/transcription of miRNA genes. For example, Mir192 is induced by TGF-β1 through promoter Smad-binding elements and epigenetic regulation via protein Ets-1 and histone acetylation, which can be activated by Akt.75 Therefore, miR-192 expression may be controlled by inhibitors of Akt, such as MK-2206 or histone acetyltransferases, and this may be effective to prevent DN. Interestingly, the mitotic inhibitor, paclitaxel (used in cancer chemotherapy) has been shown to downregulate miR-192 expression, resulting in attenuated fibrotic damage in the remnant kidney model.82 High-throughput screening of drugs that have potential to reduce miRNA expression may be another option.
Together, these findings provide evidence and hope that anti-miRNA therapies may become a reality for the treatment of human DN in the future. However, although miRNA-based therapies are being actively researched and clinical trials are already ongoing,141,142 there are still significant challenges and impediments before they can be developed as drugs for clinical use. Apart from optimizing stability, as well as organ-specific and cell-specific in vivo delivery, improved target specificity is needed to prevent off-target effects or toxicity to normal tissues and organs, as a given miRNA can have numerous targets.
Conclusions
In this review we discussed the emerging importance of the expression and functions of miRNAs in the pathology, diagnosis, and treatment of DN. We also highlighted the role of miRNAs in mediating the features of the early stages of DN, such glomerular hypertrophy and ECM accumulation. MiRNAs, such as miR-192 and miR-21, that are regulated abnormally in the early stage of DN and that mediate pathological phenotypes, could be useful for early diagnosis, treatment, or prevention of DN, as they could be targeted in patients even before the onset of symptoms of renal function decline. Because chemically-modified oligonucleotide inhibitors of some miRNAs have already been demonstrated to be effective in preventing features of DN in animal models, there is hope that such approaches will be clinically translatable to human DN. The field of ncRNA biology is growing exponentially and may help make this approach a reality. However, several challenges remain for translating animal studies to the clinic and evaluating therapies that combine miRNA targeting with currently available drugs for DN. Also needed are more standardized methods to monitor miRNA levels precisely in human biofluids, and to determine which miRNA(s) should be chosen for further targeting in patients on the basis of their DN staging and clinical profile.
Acknowledgements
The authors gratefully acknowledge funding from the National Institutes of Health (NIDDK and NHLBI), the Juvenile Diabetes Research Foundation, and the American Diabetes Association.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Fineberg D, Jandeleit-Dahm KA, Cooper ME. Diabetic nephropathy: diagnosis and treatment. Nat Rev Endocrinol. 2013;9:713–723. doi: 10.1038/nrendo.2013.184. [DOI] [PubMed] [Google Scholar]
- 2.Jones CA, et al. Epidemic of end-stage renal disease in people with diabetes in the United States population: do we know the cause? Kidney Int. 2005;67:1684–1691. doi: 10.1111/j.1523-1755.2005.00265.x. [DOI] [PubMed] [Google Scholar]
- 3.Sarnak MJ, et al. Kidney disease as a risk factor for development of cardiovascular disease: a statement from the American Heart Association Councils on Kidney in Cardiovascular Disease, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention. Circulation. 2003;108:2154–2169. doi: 10.1161/01.CIR.0000095676.90936.80. [DOI] [PubMed] [Google Scholar]
- 4.de Boer IH, et al. Intensive diabetes therapy and glomerular filtration rate in type 1 diabetes. N Engl J Med. 2011;365:2366–2376. doi: 10.1056/NEJMoa1111732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathan DM, et al. Diabetes control and complications trial/epidemiology of diabetes interventions and complications study at 30 years: advances and contributions. Diabetes. 2013;62:3976–3986. doi: 10.2337/db13-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colagiuri S, Cull CA, Holman RR. Are Lower Fasting Plasma Glucose Levels at Diagnosis of Type 2 Diabetes Associated With Improved Outcomes?: U. Prospective Diabetes Study 61. Diabetes care. 2002;25:1410–1417. doi: 10.2337/diacare.25.8.1410. [DOI] [PubMed] [Google Scholar]
- 7.Patel A, et al. Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. N Engl J Med. 2008;358:2560–2572. doi: 10.1056/NEJMoa0802987. [DOI] [PubMed] [Google Scholar]
- 8.Decleves AE, Sharma K. Obesity and kidney disease: differential effects of obesity on adipose tissue and kidney inflammation and fibrosis. Current opinion in nephrology and hypertension. 2015;24:28–36. doi: 10.1097/MNH.0000000000000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Look AHEAD Research Group Effect of a long-term behavioural weight loss intervention on nephropathy in overweight or obese adults with type 2 diabetes: a secondary analysis of the Look AHEAD randomised clinical trial. The Lancet Diabetes & Endocrinology. 2014;2:801–809. doi: 10.1016/S2213-8587(14)70156-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S, Jim B, Ziyadeh FN. Diabetic nephropathy and transforming growth factor-beta: transforming our view of glomerulosclerosis and fibrosis build-up. Seminars in nephrology. 2003;23:532–543. doi: 10.1053/s0270-9295(03)00132-3. [DOI] [PubMed] [Google Scholar]
- 11.Qian Y, et al. From fibrosis to sclerosis: mechanisms of glomerulosclerosis in diabetic nephropathy. Diabetes. 2008;57:1439–1445. doi: 10.2337/db08-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanwar YS, et al. A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annual review of pathology. 2011;6:395–423. doi: 10.1146/annurev.pathol.4.110807.092150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato M, Natarajan R. Diabetic nephropathy[mdash]emerging epigenetic mechanisms. Nat Rev Nephrol. 2014;10:517–530. doi: 10.1038/nrneph.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagtalunan ME, et al. Podocyte loss and progressive glomerular injury in type II diabetes. The Journal of clinical investigation. 1997;99:342–348. doi: 10.1172/JCI119163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer TW, Bennett PH, Nelson RG. Podocyte number predicts long-term urinary albumin excretion in Pima Indians with Type II diabetes and microalbuminuria. Diabetologia. 1999;42:1341–1344. doi: 10.1007/s001250051447. [DOI] [PubMed] [Google Scholar]
- 16.Kato M, Castro NE, Natarajan R. MicroRNAs: potential mediators and biomarkers of diabetic complications. Free Radic Biol Med. 2013;64:85–94. doi: 10.1016/j.freeradbiomed.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inagi R, Ishimoto Y, Nangaku M. Proteostasis in endoplasmic reticulum[mdash]new mechanisms in kidney disease. Nat Rev Nephrol. 2014;10:369–378. doi: 10.1038/nrneph.2014.67. [DOI] [PubMed] [Google Scholar]
- 18.Sharma K, Ziyadeh FN. Hyperglycemia and diabetic kidney disease. The case for transforming growth factor-beta as a key mediator. Diabetes. 1995;44:1139–1146. doi: 10.2337/diab.44.10.1139. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto T, et al. Expression of transforming growth factor beta is elevated in human and experimental diabetic nephropathy. Proc Natl Acad Sci U S A. 1993;90:1814–1818. doi: 10.1073/pnas.90.5.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato M, Park JT, Natarajan R. MicroRNAs and the glomerulus. Experimental cell research. 2012;318:993–1000. doi: 10.1016/j.yexcr.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 22.Ruggenenti P, Cravedi P, Remuzzi G. The RAAS in the pathogenesis and treatment of diabetic nephropathy. Nat Rev Nephrol. 2010;6:319–330. doi: 10.1038/nrneph.2010.58. [DOI] [PubMed] [Google Scholar]
- 23.Abboud HE. Role of platelet-derived growth factor in renal injury. Annual review of physiology. 1995;57:297–309. doi: 10.1146/annurev.ph.57.030195.001501. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y, et al. Role of upstream stimulatory factors in regulation of renal transforming growth factor-beta1. Diabetes. 2005;54:1976–1984. doi: 10.2337/diabetes.54.7.1976. [DOI] [PubMed] [Google Scholar]
- 25.Kato M, et al. A microRNA circuit mediates transforming growth factor-beta1 autoregulation in renal glomerular mesangial cells. Kidney Int. 2011;80:358–368. doi: 10.1038/ki.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, et al. Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 27.Roberts AB, McCune BK, Sporn MB. TGF-beta: regulation of extracellular matrix. Kidney Int. 1992;41:557–559. doi: 10.1038/ki.1992.81. [DOI] [PubMed] [Google Scholar]
- 28.Poncelet AC, Schnaper HW. Sp1 and Smad proteins cooperate to mediate transforming growth factor-beta 1-induced alpha 2(I) collagen expression in human glomerular mesangial cells. J Biol Chem. 2001;276:6983–6992. doi: 10.1074/jbc.M006442200. [DOI] [PubMed] [Google Scholar]
- 29.Tsuchida K, et al. Role of Smad4 on TGF-beta-induced extracellular matrix stimulation in mesangial cells. Kidney Int. 2003;63:2000–2009. doi: 10.1046/j.1523-1755.2003.00009.x. [DOI] [PubMed] [Google Scholar]
- 30.Kim YS, et al. Novel interactions between TGF-{beta}1 actions and the 12/15-lipoxygenase pathway in mesangial cells. J Am Soc Nephrol. 2005;16:352–362. doi: 10.1681/ASN.2004070568. [DOI] [PubMed] [Google Scholar]
- 31.Chin BY, et al. Stimulation of pro-alpha(1)(I) collagen by TGF-beta(1) in mesangial cells: role of the p38 MAPK pathway. Am J Physiol Renal Physiol. 2001;280:F495–504. doi: 10.1152/ajprenal.2001.280.3.F495. [DOI] [PubMed] [Google Scholar]
- 32.Hayashida T, et al. TGF-beta1 activates MAP kinase in human mesangial cells: a possible role in collagen expression. Kidney Int. 1999;56:1710–1720. doi: 10.1046/j.1523-1755.1999.00733.x. [DOI] [PubMed] [Google Scholar]
- 33.Kato M, et al. Role of the Akt/FoxO3a pathway in TGF-beta1-mediated mesangial cell dysfunction: a novel mechanism related to diabetic kidney disease. J Am Soc Nephrol. 2006;17:3325–3335. doi: 10.1681/ASN.2006070754. [DOI] [PubMed] [Google Scholar]
- 34.Mahimainathan L, et al. Mesangial cell hypertrophy by high glucose is mediated by downregulation of the tumor suppressor PTEN. Diabetes. 2006;55:2115–2125. doi: 10.2337/db05-1326. [DOI] [PubMed] [Google Scholar]
- 35.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 37.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature reviews. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 38.Davis BN, et al. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis BN, et al. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki HI, et al. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 41.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 42.Sampson VB, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer research. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 43.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature structural & molecular biology. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 46.Piskounova E, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruvkun G. The perfect storm of tiny RNAs. Nature medicine. 2008;14:1041–1045. doi: 10.1038/nm1008-1041. [DOI] [PubMed] [Google Scholar]
- 48.Cheng TL, et al. MeCP2 suppresses nuclear microRNA processing and dendritic growth by regulating the DGCR8/Drosha complex. Dev Cell. 2014;28:547–560. doi: 10.1016/j.devcel.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 49.Woo JS, Kim VN. MeCP2 caught moonlighting as a suppressor of MicroRNA processing. Dev Cell. 2014;28:477–478. doi: 10.1016/j.devcel.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 50.Bracaglia G, et al. Methyl-CpG-binding protein 2 is phosphorylated by homeodomain-interacting protein kinase 2 and contributes to apoptosis. EMBO reports. 2009;10:1327–1333. doi: 10.1038/embor.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin Y, et al. A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nature medicine. 2012;18:580–588. doi: 10.1038/nm.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki HI, et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 53.Ota H, et al. ADAR1 Forms a Complex with Dicer to Promote MicroRNA Processing and RNA-Induced Gene Silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xhemalce B, Robson SC, Kouzarides T. Human RNA Methyltransferase BCDIN3D Regulates MicroRNA Processing. Cell. 2012;151:278–288. doi: 10.1016/j.cell.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheloufi S, et al. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibbings D, et al. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Upton JP, et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harvey SJ, et al. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol. 2008;19:2150–2158. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho J, et al. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol. 2008;19:2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ho J, et al. The Pro-Apoptotic Protein Bim Is a MicroRNA Target in Kidney Progenitors. J Am Soc Nephrol. 2011;22:1053–1063. doi: 10.1681/ASN.2010080841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi S, et al. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol. 2008;19:2159–2169. doi: 10.1681/ASN.2008030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagalakshmi VK, et al. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int. 2011;79:317–330. doi: 10.1038/ki.2010.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhdanova O, et al. The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int. 2011;80:719–730. doi: 10.1038/ki.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 65.Chen JF, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Y, et al. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 2004;32:e188. doi: 10.1093/nar/gnh186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tian Z, et al. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome research. 2008;18:404–411. doi: 10.1101/gr.6587008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kato M, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kato M, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kato M, et al. Post-transcriptional up-regulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-{beta}-induced collagen expression in kidney cells. J Biol Chem. 2010;285:34004–34015. doi: 10.1074/jbc.M110.165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park JT, et al. FOG2 protein down-regulation by transforming growth factor-beta1-induced microRNA-200b/c leads to Akt kinase activation and glomerular mesangial hypertrophy related to diabetic nephropathy. J Biol Chem. 2013;288:22469–22480. doi: 10.1074/jbc.M113.453043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deshpande SD, et al. Transforming Growth Factor-beta-Induced Cross Talk Between p53 and a MicroRNA in the Pathogenesis of Diabetic Nephropathy. Diabetes. 2013;62:3151–3162. doi: 10.2337/db13-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Putta S, et al. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol. 2012;23:458–469. doi: 10.1681/ASN.2011050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kato M, Natarajan R. MicroRNA circuits in transforming growth factor-beta actions and diabetic nephropathy. Seminars in nephrology. 2012;32:253–260. doi: 10.1016/j.semnephrol.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kato M, et al. TGF-beta Induces Acetylation of Chromatin and of Ets-1 to Alleviate Repression of miR-192 in Diabetic Nephropathy. Sci Signal. 2013;6:ra43. doi: 10.1126/scisignal.2003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chung AC, et al. miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol. 2010;21:1317–1325. doi: 10.1681/ASN.2010020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jenkins RH, et al. Transforming growth factor beta1 represses proximal tubular cell microRNA-192 expression through decreased hepatocyte nuclear factor DNA binding. The Biochemical journal. 2012;443:407–416. doi: 10.1042/BJ20111861. [DOI] [PubMed] [Google Scholar]
- 78.Wang Q, et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. Faseb J. 2008;22:4126–4135. doi: 10.1096/fj.08-112326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang XX, et al. Diabetic Nephropathy is Accelerated by Farnesoid X Receptor Deficiency and Inhibited by Farnesoid X Receptor Activation in a Type 1 Diabetes Model. Diabetes. 2010;59:2916–2927. doi: 10.2337/db10-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Long J, et al. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem. 2011;286:11837–11848. doi: 10.1074/jbc.M110.194969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mu J, et al. Functional Implications of MicroRNA-215 in TGF-beta1-Induced Phenotypic Transition of Mesangial Cells by Targeting CTNNBIP1. PloS one. 2013;8:e58622. doi: 10.1371/journal.pone.0058622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun L, et al. Low-dose paclitaxel ameliorates fibrosis in the remnant kidney model by down-regulating miR-192. The Journal of pathology. 2011;225:364–377. doi: 10.1002/path.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.He F, et al. MiR-135a promotes renal fibrosis in diabetic nephropathy by regulating TRPC1. Diabetologia. 2014;57:1726–1736. doi: 10.1007/s00125-014-3282-0. [DOI] [PubMed] [Google Scholar]
- 84.Dey N, et al. MicroRNA-21 Orchestrates High Glucose-induced Signals to TOR Complex 1, Resulting in Renal Cell Pathology in Diabetes. J Biol Chem. 2011;286:25586–25603. doi: 10.1074/jbc.M110.208066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhong X, et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia. 2013;56:663–674. doi: 10.1007/s00125-012-2804-x. [DOI] [PubMed] [Google Scholar]
- 86.Wang J, et al. Effect of miR-21 on Renal Fibrosis by Regulating MMP-9 and TIMP1 in kk-ay Diabetic Nephropathy Mice. Cell Biochemistry and Biophysics. 2013;67:537–546. doi: 10.1007/s12013-013-9539-2. [DOI] [PubMed] [Google Scholar]
- 87.Chau BN, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Science translational medicine. 2012;4:121ra118. doi: 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Z, et al. MicroRNA-21 protects from mesangial cell proliferation induced by diabetic nephropathy in db/db mice. FEBS Lett. 2009;583:2009–2014. doi: 10.1016/j.febslet.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 89.Lai JY, et al. MicroRNA-21 in Glomerular Injury. J Am Soc Nephrol. 2014 doi: 10.1681/ASN.2013121274. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li L-M, et al. Role of MicroRNA-214–Targeting Phosphatase and Tensin Homolog in Advanced Glycation End Product-Induced Apoptosis Delay in Monocytes. The Journal of Immunology. 2011;186:2552–2560. doi: 10.4049/jimmunol.1001633. [DOI] [PubMed] [Google Scholar]
- 91.Denby L, et al. MicroRNA-214 Antagonism Protects against Renal Fibrosis. Journal of the American Society of Nephrology. 2014;25:65–80. doi: 10.1681/ASN.2013010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Z, et al. MicroRNA-451 regulates p38 MAPK signaling by targeting of Ywhaz and suppresses the mesangial hypertrophy in early diabetic nephropathy. FEBS Letters. 2012;586:20–26. doi: 10.1016/j.febslet.2011.07.042. [DOI] [PubMed] [Google Scholar]
- 93.Wang B, et al. Suppression of microRNA-29 Expression by TGF-beta1 Promotes Collagen Expression and Renal Fibrosis. J Am Soc Nephrol. 2012;23:252–265. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen HY, et al. MicroRNA-29b inhibits diabetic nephropathy in db/db mice. Mol Ther. 2014;22:842–853. doi: 10.1038/mt.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qin W, et al. TGF-beta/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephrol. 2011;22:1462–1474. doi: 10.1681/ASN.2010121308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Du B, et al. High glucose down-regulates miR-29a to increase collagen IV production in HK-2 cells. FEBS Letters. 584:811–816. doi: 10.1016/j.febslet.2009.12.053. [DOI] [PubMed] [Google Scholar]
- 97.Kanasaki K, et al. Linagliptin-Mediated DPP-4 Inhibition Ameliorates Kidney Fibrosis in Streptozotocin-Induced Diabetic Mice by Inhibiting Endothelial-to-Mesenchymal Transition in a Therapeutic Regimen. Diabetes. 2014;63:2120–2131. doi: 10.2337/db13-1029. [DOI] [PubMed] [Google Scholar]
- 98.Brennan EP, et al. Lipoxins attenuate renal fibrosis by inducing let-7c and suppressing TGFbetaR1. J Am Soc Nephrol. 2013;24:627–637. doi: 10.1681/ASN.2012060550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang B, et al. Transforming growth factor-beta1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int. 2014;85:352–361. doi: 10.1038/ki.2013.372. [DOI] [PubMed] [Google Scholar]
- 100.Park JT, et al. Repression of let-7 by Transforming Growth Factor-β1-induced Lin28 up-regulates collagen expression in glomerular mesangial cells under diabetic conditions. 2014 doi: 10.1152/ajprenal.00458.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li R, et al. The microRNA miR-433 promotes renal fibrosis by amplifying the TGF-beta/Smad3-Azin1 pathway. Kidney Int. 2013;84:1129–1144. doi: 10.1038/ki.2013.272. [DOI] [PubMed] [Google Scholar]
- 102.Castro NE, et al. Transforming Growth Factor beta1 (TGF-beta1) Enhances Expression of Profibrotic Genes through a Novel Signaling Cascade and MicroRNAs in Renal Mesangial Cells. J Biol Chem. 2014;289:29001–29013. doi: 10.1074/jbc.M114.600783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu J, et al. Downregulation of MicroRNA-30 Facilitates Podocyte Injury and Is Prevented by Glucocorticoids. Journal of the American Society of Nephrology. 2014;25:92–104. doi: 10.1681/ASN.2012111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Long J, et al. MicroRNA-22 Is a Master Regulator of Bone Morphogenetic Protein-7/6 Homeostasis in the Kidney. J Biol Chem. 2013;288:36202–36214. doi: 10.1074/jbc.M113.498634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Long J, et al. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J Biol Chem. 2010;285:23457–23465. doi: 10.1074/jbc.M110.136168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sedeek M, et al. Oxidative stress, Nox isoforms and complications of diabetes--potential targets for novel therapies. J Cardiovasc Transl Res. 2012;5:509–518. doi: 10.1007/s12265-012-9387-2. [DOI] [PubMed] [Google Scholar]
- 107.Babelova A, et al. Role of Nox4 in murine models of kidney disease. Free Radic Biol Med. 2012;53:842–853. doi: 10.1016/j.freeradbiomed.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 108.Zhu Y, Usui HK, Sharma K. Regulation of transforming growth factor beta in diabetic nephropathy: implications for treatment. Seminars in nephrology. 2007;27:153–160. doi: 10.1016/j.semnephrol.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fu Y, et al. Regulation of NADPH oxidase activity is associated with miRNA-25-mediated NOX4 expression in experimental diabetic nephropathy. Am J Nephrol. 2010;32:581–589. doi: 10.1159/000322105. [DOI] [PubMed] [Google Scholar]
- 110.Feng B, et al. miR-146a-Mediated extracellular matrix protein production in chronic diabetes complications. Diabetes. 2011;60:2975–2984. doi: 10.2337/db11-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Muratsu-Ikeda S, et al. Downregulation of miR-205 modulates cell susceptibility to oxidative and endoplasmic reticulum stresses in renal tubular cells. PloS one. 2012;7:e41462. doi: 10.1371/journal.pone.0041462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wei J, et al. Aldose reductase regulates miR-200a-3p/141-3p to coordinate Keap1–Nrf2, Tgfβ1/2, and Zeb1/2 signaling in renal mesangial cells and the renal cortex of diabetic mice. Free Radical Biology and Medicine. 2014;67:91–102. doi: 10.1016/j.freeradbiomed.2013.10.811. [DOI] [PubMed] [Google Scholar]
- 113.Krupa A, et al. Loss of MicroRNA-192 Promotes Fibrogenesis in Diabetic Nephropathy. J Am Soc Nephrol. 2010;21:438–447. doi: 10.1681/ASN.2009050530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang B, et al. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes. 2010;59:1794–1802. doi: 10.2337/db09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang B, et al. miR-200a Prevents renal fibrogenesis through repression of TGF-beta2 expression. Diabetes. 2011;60:280–287. doi: 10.2337/db10-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 117.Ren S, Duffield JS. Pericytes in kidney fibrosis. Current opinion in nephrology and hypertension. 2013;22:471–480. doi: 10.1097/MNH.0b013e328362485e. [DOI] [PubMed] [Google Scholar]
- 118.LeBleu VS, et al. Origin and function of myofibroblasts in kidney fibrosis. Nature medicine. 2013;19:1047–1053. doi: 10.1038/nm.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kriz W, Kaissling B, Le Hir M. Epithelial-mesenchymal transition (EMT) in kidney fibrosis: fact or fantasy? The Journal of clinical investigation. 2011;121:468–474. doi: 10.1172/JCI44595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Breyer MD, et al. Mouse models of diabetic nephropathy. J Am Soc Nephrol. 2005;16:27–45. doi: 10.1681/ASN.2004080648. [DOI] [PubMed] [Google Scholar]
- 121.Fassett RG, et al. Biomarkers in chronic kidney disease: a review. Kidney Int. 2011;80:806–821. doi: 10.1038/ki.2011.198. [DOI] [PubMed] [Google Scholar]
- 122.Farazi TA, et al. miRNAs in human cancer. The Journal of pathology. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.M. Fabbri. miRNAs as molecular biomarkers of cancer. Expert review of molecular diagnostics. 2010;10:435–444. doi: 10.1586/erm.10.27. [DOI] [PubMed] [Google Scholar]
- 124.Wang K, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tijsen AJ, et al. MiR423-5p as a circulating biomarker for heart failure. Circulation research. 2010;106:1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 126.Szeto CC, et al. Micro-RNA expression in the urinary sediment of patients with chronic kidney diseases. Disease markers. 2012;33:137–144. doi: 10.3233/DMA-2012-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Neal CS, et al. Circulating microRNA expression is reduced in chronic kidney disease. Nephrol Dial Transplant. 2011;26:3794–3802. doi: 10.1093/ndt/gfr485. [DOI] [PubMed] [Google Scholar]
- 128.Luk CC, et al. Urinary biomarkers for the prediction of reversibility in acute-on-chronic renal failure. Disease markers. 2013;34:179–185. doi: 10.3233/DMA-120959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang G, et al. Urinary sediment miRNA levels in adult nephrotic syndrome. Clinica chimica acta; international journal of clinical chemistry. 2013;418:5–11. doi: 10.1016/j.cca.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 130.Yang Y, et al. Urine miRNAs: potential biomarkers for monitoring progression of early stages of diabetic nephropathy. Medical hypotheses. 2013;81:274–278. doi: 10.1016/j.mehy.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ichii O, et al. Altered expression of microRNA miR-146a correlates with the development of chronic renal inflammation. Kidney Int. 2012;81:280–292. doi: 10.1038/ki.2011.345. [DOI] [PubMed] [Google Scholar]
- 132.DiStefano JK, Taila M, Alvarez ML. Emerging roles for miRNAs in the development, diagnosis, and treatment of diabetic nephropathy. Current diabetes reports. 2013;13:582–591. doi: 10.1007/s11892-013-0386-8. [DOI] [PubMed] [Google Scholar]
- 133.Cai X, et al. Serum microRNAs levels in primary focal segmental glomerulosclerosis. Pediatric nephrology (Berlin, Germany) 2013;28:1797–1801. doi: 10.1007/s00467-013-2434-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang Y, et al. Involvement of inflammation-related miR-155 and miR-146a in diabetic nephropathy: implications for glomerular endothelial injury. BMC nephrology. 2014;15:142. doi: 10.1186/1471-2369-15-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang G, et al. Urinary miR-21, miR-29, and miR-93: novel biomarkers of fibrosis. Am J Nephrol. 2012;36:412–418. doi: 10.1159/000343452. [DOI] [PubMed] [Google Scholar]
- 136.Barutta F, et al. Urinary Exosomal MicroRNAs in Incipient Diabetic Nephropathy. PloS one. 2013;8:e73798. doi: 10.1371/journal.pone.0073798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Argyropoulos C, et al. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PloS one. 2013;8:e54662. doi: 10.1371/journal.pone.0054662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ziyadeh FN, et al. Long-term prevention of renal insufficiency, excess matrix gene expression, and glomerular mesangial matrix expansion by treatment with monoclonal antitransforming growth factor-beta antibody in db/db diabetic mice. Proc Natl Acad Sci U S A. 2000;97:8015–8020. doi: 10.1073/pnas.120055097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Decleves AE, Sharma K. New pharmacological treatments for improving renal outcomes in diabetes. Nat Rev Nephrol. 2010;6:371–380. doi: 10.1038/nrneph.2010.57. [DOI] [PubMed] [Google Scholar]
- 140.Krutzfeldt J, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 141.van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 2014;6:851–864. doi: 10.15252/emmm.201100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J Cell Biol. 2012;199:407–412. doi: 10.1083/jcb.201208082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pan Y, et al. MS2 VLP-based delivery of microRNA-146a inhibits autoantibody production in lupus-prone mice. International journal of nanomedicine. 2012;7:5957–5967. doi: 10.2147/IJN.S37990. [DOI] [PMC free article] [PubMed] [Google Scholar]