Abstract
This review discusses the challenges facing research in ‘functional glycomics’ and the novel technologies that are being developed to advance the field. The structural complexity of glycans and glycoconjugates makes studies of both their structures and recognition difficult. However, these intricate structures can be captured from their natural sources, isolated and fluorescently-tagged for detailed structural analysis and for presentation on glycan microarrays for functional recognition by glycan-binding proteins. These advances in glycan preparation and manipulation enable the streamlining of functional glycomics studies and will help to propel the field forward in studying natural, biologically relevant glycans.
Keywords: Functional Glycomics, glycans, glycan microarray, shotgun glycomics, natural glycans
Introduction
Functional glycomics is the systematic study of the structure and function of glycans and has been recognized as an important, emerging field in biomedical research [1–5]. The functions of glycans have been identified in many biological pathways, often through their interactions and recognition by proteins and/or microorganisms. Despite its well-recognized importance, glycomics has lagged far behind genomics and proteomics, which are the systematic studies of DNA and proteins, respectively. This lag is mostly due to the unique structural complexity of glycans, creating unique technical challenges to their characterization. The overall structures of DNA and proteins are linear with a limited number of building blocks, and the biosynthesis of DNA and proteins are template-driven, in which the genetic codes can be transcribed and translated linearly. High throughput sequencing and automatic synthesis are readily available for DNA and peptides. Modern proteomics can carry out high throughput sequencing of proteins either through correlating with genomic data or through de novo sequencing. Recombinant proteins can be expressed readily in various systems, and even total protein microarrays are now being generated, and several such arrays also include post-translational modifications of proteins [6], albeit now without the major types of protein glycosylation. Overall, from small amounts of biological specimens, structural information on DNA and proteins can be readily obtained; and from virtual structural information, DNA and proteins can be readily obtained by amplification techniques for experimental functional studies. This free flow of in silico information from and to actual biomolecules is, however, not available yet for glycans.
By contrast, structural and functional analyses of glycans and glycoconjugates are much more complicated (Fig. 1) than that for peptide/protein and DNA [7–10]. Glycans are composed of multiple monosaccharide blocks connected into complex branched structures. The complete structural data needs to include composition, sequence, branching position, anomeric configuration, and possible modifications such as acetylation, sulfation, phosphorylation, etc. [11,1,12]. Although these multi-layer structural data can be elucidated using techniques including mass spectrometry (MS), chromatography, and nuclear magnetic resonance (NMR), the process is often time consuming and requires special expertise that is only available in specialized laboratories. Similarly, the synthesis of complex, branched glycans requires sophisticated approaches and is often an insurmountable task and limits production to expert laboratories [13]. Automatic synthesis is promising but is still far from being widely adapted, due to the technical challenges and hindrances to accessing the expensive building blocks [14,15]. In addition, the functional recognition of such glycans by glycan-binding proteins (GBPs) is also difficult to explore. Overall, the lack of free flow between glycan and structural information hinders studies in functional glycomics from advancing as fast as genomics and proteomics.
Figure 1.
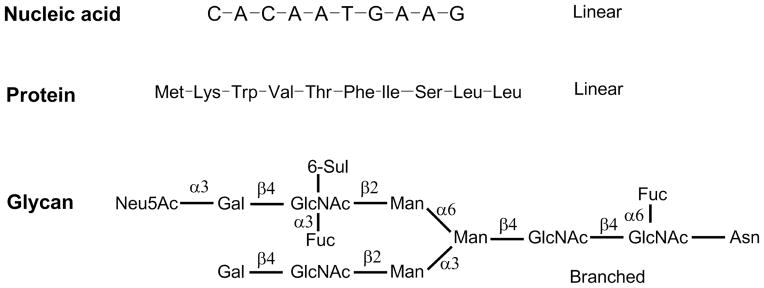
The structural complexity of glycans compared to nucleic acids and proteins/peptides.
Nevertheless, glycoscience is rapidly advancing and the systematic study of functional glycomics has become possible for addressing the urgent need for biomedical studies. This is at least partly due to great technological advancements in MS, high performance liquid chromatography (HPLC), and development of synthetic and natural glycan microarrays. The glycan microarray [3,16–22] has become one of the most important technological breakthroughs in functional glycomics. A glycan microarray is defined as the presentation of a large number of glycan structures immobilized on a single chip or surface, which can be interrogated with GBPs or microorganisms for the study of protein-glycan interactions. This technology evolved from ELISA-based studies of neoglycoconjugates, glycoproteins, and glycolipids, as well as overlay techniques using thin layer chromatography [23]. In the past decades, glycan microarrays have quickly become the natural choice for the screening of binding specificities of GBPs [24]. The success of a glycan microarray is dictated by the size, diversity, and biological relevance of the library of glycans that is used to prepare the microarray, and to the lesser degree, by the appropriate presentation of the glycans. Therefore the most urgent task is to establish large, diverse, and biologically relevant glycan libraries.
The two major routes to build glycan libraries are through chemical/enzymatic synthesis and through isolation of natural glycans from biological sources [25,24,26–29]. While these two routes are very different, they can be used in a complementary fashion to enrich the glycan libraries. Synthesis can be used to generate relatively large amounts of defined glycan structures, however, it is often technically too challenging for a large number of glycans to be made, especially for non-specialized laboratories. Therefore, the target glycan structures for synthesis are chosen because of known or speculated biological activity. This information, however, is often obtained through natural material. Natural sources of glycans are readily available, and many are potentially abundant for functional study. This is especially true for glycan microarray preparation, as glycans at nanomole levels are sufficient for the preparation of glycan microarrays. Natural glycans are extremely diverse and inherently biologically relevant. Based on our understanding of the urgent needs of the field and the current technological readiness, we have rationalized that natural glycans are the most imminent and relevant source for enriching our glycan repertoire [5], advancing glycan microarray and functional glycomics. In this review article, we will discuss several aspects in the utilization of natural glycans for functional glycomics.
Release of glycans from natural glycoconjugates for functional study
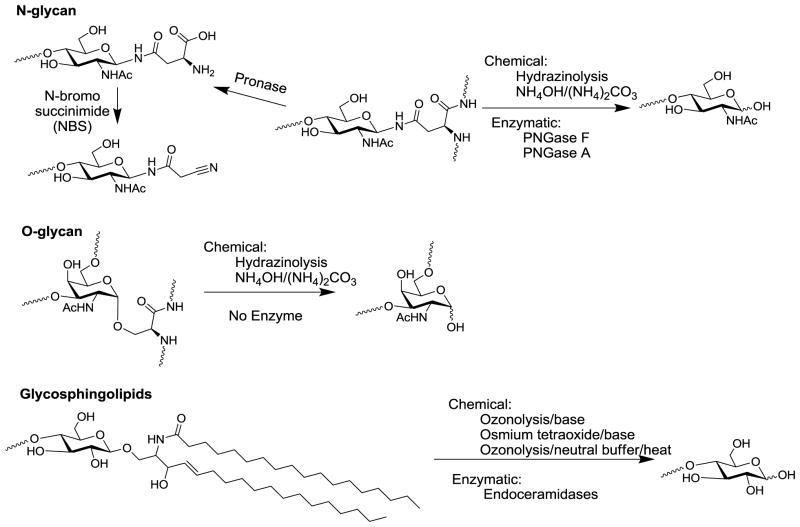
Free glycans exist in biological material, such as in human milk, while other natural glycans exist as glycoconjugates, linked to other biomolecules such as glycoproteins, glycolipids, proteoglycans, and GPI-anchored proteins [1]. To analyze the glycan structures and functions without interference from the non-glycan moieties, researchers often release glycans from the glycoconjugates for structural/functional glycomic analysis. Since the linkages of glycans to aglycons of each class of glycoconjugates are different, they often require specific releasing methods, as summarized in Fig. 2.
Figure 2.
Common methods to release N- and O-glycans from glycoproteins and glycosphingolipid (GSL) glycans.
Preparation of N-glycans for functional glycomics
N-glycans are by far the most studied class of glycans, probably due to the availability of a collection of tools for their release from glycoproteins, and the common structural motif of high mannose-, hybrid- and complex-type N-glycans in mammals [30,31]. A number of endoglycosidases [32,33] and N-glycanases [34–36] have been discovered that cleave the chitobiose glycosidic bond or the asparagine-glycan amide linkage respectively. PNGase F has been widely used as an efficient tool to release most intact N-glycans from glycoproteins, except N-glycans with core α3-fucosylation [37] existing in plants and lower animals such as insects. PNGase F is extremely useful for the release and analysis of mammalian N-glycans. However, the enzyme is often too expensive when large scale N-glycan preparation is needed. PNGase A is an N-glycanase that can release core α3-fucosylated N-glycans but it is even much more expensive than PNGase F. Chemical release of N-glycans requires very harsh conditions, such as hydrazinolysis at >90°C [38,39]. The harsh condition and toxic reagent make this an undesirable method. N-glycans can also be released by long treatment of ammonia/ammonium carbonate [40], which is primarily an O-glycan release method for microscale samples. However, the condition is still fairly harsh for many labile modification groups, such as O-acetyl group, and peeling reactions can occur [41].
We envisioned that new methods to release N-glycans are needed to avoid expensive enzymes, toxic reagents, and harsh reaction conditions to prevent loss of structural integrities. As one approach, we explored exhaustive Pronase digestion of glycoproteins. The resulting glycopeptides/glycoamino acids generated can be used in the preparation of glycan microarrays. To facilitate the separation of these glycopeptides/glycoamino acids by HPLC, they are tagged with a cleavable Fmoc group and rendered fluorescent [26]. Although this strategy can be used for glycoproteins with relatively simple glycan profiles, it becomes impractical due to the complexity in the heterogeneous peptide moiety generated by inevitably incomplete protease digestion. The problem was recently solved by the introduction of a mild oxidative reagent, N-bromosuccinimide (NBS) [42]. Pronase digestion of glycoproteins generates a mixture glycoamino acids and glycopeptides with short peptide moieties. A simple treatment of this mixture by NBS trimmed the small aglycon moieties to a nitrile. This nitrile can be specifically, fluorescently-tagged to generate glycan derivatives whose glycan profiles, as evidenced by MALDI-TOF-MS, are very similar to that generated by PNGase F digestion. This approach is mild, inexpensive, and can be applied to large amounts of tissues/organs for the preparation of large numbers/amounts of glycans for glycan microarrays.
Preparation of O-glycans for glycan microarray
In comparison to N-glycans, O-glycans are less studied, largely due to the lack of good tools for the release of intact reducing O-glycans. There is currently no O-glycanase with broad-spectrum activity available; the only available “O-glycanase” (endo-α-N-acetylgalactosaminidase) works only on O-glycans with a short core 1 or core 3 disaccharide structure [43]. Chemically, the traditional release of O-glycans has been through reductive elimination, in which O-glycans released by β-elimination with sodium hydroxide (NaOH) are immediately reduced by sodium borohydride (NaBH4) to prevent further peeling reactions [44,45]. Recently, two groups have developed a new methodology to release/tag O-glycans using 1-phenyl-3-methyl-5-pyrazolone (PMP) [46,47] under basic conditions for direct MS analysis. Although very useful for O-glycan structural profiling, these methods also destroy the reducing end, preventing further functional tagging and solid phase immobilization of the released glycans. Therefore these methods in their current form are not useful for glycan microarray preparation and other applications. Other non-reducing release methods for O-glycans are available with significant drawbacks. Hydrazine has been used to release O-glycans at a lower temperature than for N-glycans [48,49]. Other methods utilize ethylamine [50], ammonium hydroxide/ammonium carbonate [40] or carbamate [51] to release O-glycans without reduction of the reducing end. The protection of the reducing end is proposed to occur through the formation of glycosylamine carbamate. These methods, however, still use very strong basic conditions and present significant “peeling” side products [41]. Even if the reducing end is preserved, the next step to immobilize these glycans often requires ring-opening tagging, which significantly affects the structural integrities of the O-glycans due to their relatively smaller size compared to N-glycans. The reducing end of glycans is often critical to their recognition by antibodies and other molecules [52]. For O-glycans the reducing end GalNAc often serves as a branching point for O-glycans. Overall, better general methods that are not based on β-elimination are needed. Ideally, the new methods should also preserve the reducing end linkage. While regenerating N-linkage is relatively simple, regenerating an α-O-linkage of a natural O-glycan terminated with GalNAc continues to be challenging.
Preparation of glycans from glycosphingolipids (GSLs) for glycan microarray
Glycosphingolipids are amphipathic glycoconjugates that can be directly immobilized onto hydrophobic surfaces such as nitrocellulose slides utilizing its strongly hydrophobic lipid moiety. On the other hand, it is often of interest to remove the lipid moiety for easier handling of glycans and also comparison with other classes of glycans on the same microarray slides. Ceramidases [53–56] are available to remove ceramides and release free reducing glycans, however, these enzymes are expensive and often specific to certain GSL structures. Traditionally, the chemical release of free glycans from GSLs took advantage of the C=C double bond in the sphingosine moiety, which can be oxidized by ozonolysis [57] or osmium tetraoxide [58]. Base-catalyzed β-elimination releases free glycans from oxidized GSLs. To avoid the potentially detrimental base treatment, we have developed two routes to utilize the GSLs for microarray preparation through covalent immobilization. One route is direct fluorescent tagging of the ozonized GSLs [27], which preserves a larger component of the lipid moiety and inserts a fluorescent tag. The products can either be immobilized covalently on N-hydroxysuccinimide (NHS) ester or epoxy slides, or non-covalently on nitrocellulose slides. In another route, we found that gentle heating of the ozonized GSLs under neutral pH efficiently release free reducing glycans [59], apparently not through a β-elimination mechanism. These methods could be used for the preparation of glycan microarray from GSLs.
Other classes of glycans for microarray
Although natural GAG oligomers have been used for glycan microarray analysis, there is no systematic study of this topic [60,61]. The challenge lies presumably in the high heterogeneity of the sulfation patterns of the GAG chains, making the large scale separation of GAG glycans an intimidating task. Nevertheless, with the great advances in MS and HPLC analysis, it is certainly a plausible way to utilize natural GAG glycans to prepare comprehensive glycan microarrays for functional study of many known or unknown GAG-binding proteins.
To our knowledge, the utilization of natural GPI-anchor glycans for microarray preparation is not well studied, presumably due to the lack of available glycan diversity.
Functional and fluorescent tagging of glycans for preparation of glycan microarray
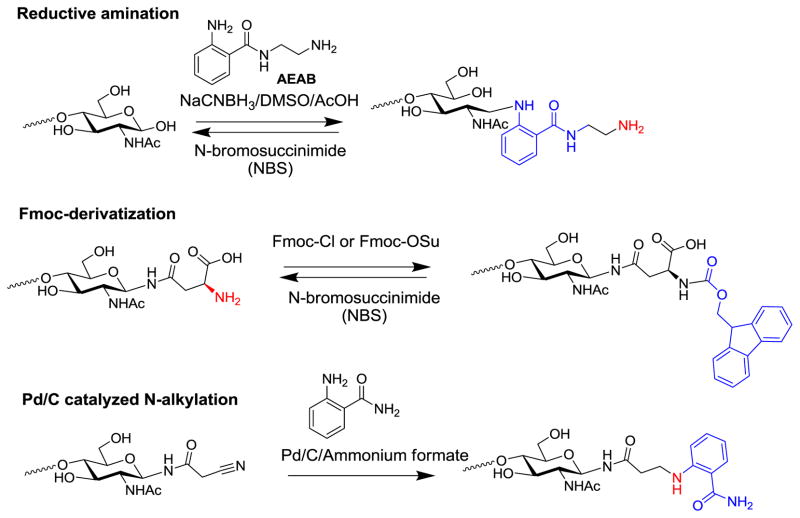
Glycans usually exist as a heterogeneous mixture in any biological sample. Often the mixtures of glycans need to be separated, either for analysis or for preparation of pure glycans. The absence of an exploitable chromophore in glycans creates a challenge for the monitoring glycans during chromatographic separation. On the other hand, the free reducing end has very limited reactivity and is not amenable to reaction with common surface activation chemistry for microarray slides. Therefore it is often necessary to install a functional group such as amino group for the efficient solid phase immobilization. Overall, bifunctional fluorescent tagging has become an important derivatization of natural glycans for their utilization in the preparation of glycan microarrays (Fig. 3).
Figure 3.
Several methods for fluorescently tagging glycans/glycoamino acids for glycan microarray preparation.
Fluorescent bifunctional tagging by reductive amination
Reductive amination of free reducing glycans with fluorescent amines has long been used for the HPLC profiling of glycans, such as 2-aminopyridine (2-AP) [62], 2-aminobenzamide (2-AB) [63], 2-aminobenzoic acid or anthranilic acid (2-AA) [64]. These small fluorescent amines can be efficiently installed onto the reducing end of glycans, resulting in an open-ring structure that greatly improves the sensitivity of HPLC detection. However, after conjugation with glycans, these fluorescent tags lack a functional group for efficient solid phase immobilization or covalent derivatization. As a homobifunctional tag, 2,6-diaminopyridine (DAP) was introduced to address this problem [65]. With an aromatic amino group, glycan-DAP conjugates can be immobilized onto activated surfaces for microarray preparation, especially on epoxy coated slides [66]. Interestingly, glycan conjugates with AA and 2-aminobenzamide (AB) can also be immobilized onto epoxy coated slides, although with a lower yield, via reactivity of the secondary amine [67].
To efficiently immobilize precious natural glycans, we developed a novel heterobifunctional tag, 2-amino-N-(2-aminoethyl)benzamide (AEAB), which contains an arylamine and an alkylamine in the same molecule [68]. Upon appropriate pH control, the aromatic amine conjugates with the glycan reducing end exclusively by reductive amination, leaving the alkylamine unmodified for efficient solid phase immobilization onto both NHS and epoxy activated glass slides. To prevent the ring opening at the reducing end, we have also developed a procedure to prepare glycan AEAB conjugates with intact reducing end ring structure. Another versatile linker, p-nitrophenyl anthranilate (PNPA), can also be conjugated to the reducing end of glycans through reductive amination [69]. This reaction introduces an activated ester that can react with amines to form amides. Interestingly, although glycan-PNPA conjugates are not fluorescent, they are rendered fluorescent after reaction with amines. Therefore, glycan-PNPA conjugates can be easily conjugated to proteins/peptides or converted to glycan-AEAB conjugates for microarray printing.
Besides sensitive detection of glycans by fluorescence, the installation of a fluorescent tag to the reducing end through reductive amination often increases the sensitivity of MS analysis compared to non-derivatized glycans. However, these tags generate structural complexity during permethylation, which is considered a necessary step for detailed sequencing by tandem MS.
Reversibility of fluorescent-tags for glycans
Because tags at the reducing end of glycans can complicate structural analyses by MS methods, we developed a facile and mild method to remove tags of reductively aminated glycans, which regenerate free reducing glycans amenable to permethylation and MS analysis [70]. Treatment of fluorescently-tagged glycans with N-bromosuccinimide (NBS) efficiently releases the tag and regenerates free reducing glycans. This method can be applied to tagged glycans with either a HexNAc or a hexose end. The method is efficient toward all types of tags installed through reductive amination, including 2-AP, 2-AB, 2-AA, and AEAB. Thus, potential problems in structural analysis of fluorescently-tagged glycans due to the introduction of the fluorescent tag have been resolved.
N-Fluorenylmethyloxycarbonyl (Fmoc) as a cleavable fluorescent tag
Fmoc group is a widely used protecting group for hydroxyl or amino groups in organic chemistry, particularly in solid phase peptide synthesis because of the easy installation and removal. For a glycan or glycoconjugate with a free amino group, Fmoc can be easily installed in aqueous solution. The fluorescent Fmoc group can greatly enhance the detection and quantitation of microscale glycans during chromatographic separation. It can also serve as an affinity tag due to its hydrophobicity. After simple removal, the free amino group can be regenerated for solid phase immobilization such as in microarray printing. It has been used to print glycoamino acids [26] generated by digestion with the mixed microbial proteases in Pronase and glycosylamines generated from free reducing glycans [71].
Natural glycan microarrays
With efficient glycan release from various classes of natural glycoconjugates and fluorescent/functional tagging, natural glycans can be readily separated and immobilized on microarray slides. Modern microarray printing requires only microgram levels of glycans, making microarray preparation plausible even for small amounts of material and minor components in a mixture of glycans. For example, de Boer et al conjugated AA and AB tags to free glycans and printed the glycan conjugates [67]. They also developed a surface plasmon resonance (SPR) based natural glycan microarray based on the same chemistry [72]. Similarly, glycan-diaminopyridine (DAP) conjugates were prepared and also printed [66]. These conjugates possess only primary or secondary arylamines as functional groups, which are less reactive and can only be printed onto epoxy coated slides. The glycan-AEAB conjugates, however, can be more efficiently printed on to both NHS and epoxy slides through the alkylamine [68,73].
As described above, the two major technical challenges in Functional Glycomics are the high throughput sequencing and synthesis of glycans. With the help of bifunctional fluorescent tags and multidimensional HPLC, a large number of relatively pure natural glycan structures can be obtained. Various HPLC columns with different separation mechanisms could be combined sequentially for ultimate resolution of individual glycans. These include size exclusive chromatography (SEC), amino normal phase column, hydrophilic interaction liquid chromatography (HILIC) column, strong anion exchange (SAX) and weak anion exchange (WAX) columns, C18-reverse phase and porous graphitized carbon (PGC)-reverse phase columns. Usually two or three columns can be combined to resolve a labeled glycan mixture to mostly individual structures. For a very complex mixture, SEC column is often applied at the beginning to give several fractions with different sizes of glycans. Preparative or semi-preparative normal phase or SAX columns can be used to provide the major resolution and separate a mixture to many fractions. These fractions can be finely separated by analytical PGC and/or C18 columns. This sequence results in salt-free individual glycans in nmol to μmol range, which can be characterized and immobilized on the glycan microarray.
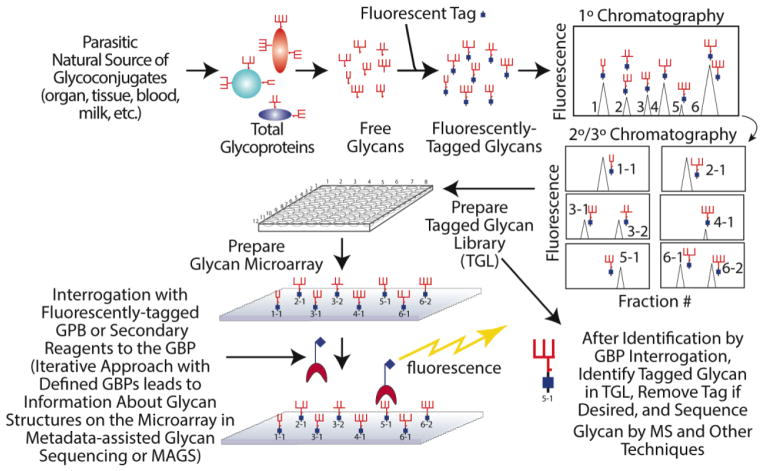
While detailed structural analysis of all of these glycans would be formidable, it is only necessary for those glycans that show positive and consistent binding towards GBPs. This strategy, termed “Shotgun Glycomics” [27], aims to directly study the structures and functions of relevant glycans while bypassing the technical bottlenecks (Fig. 4). This strategy has been applied on a number of systems. The shotgun glycan microarray of gangliosides revealed a ganglioside lactone as a potential epitope when sera from patients with Lyme disease were screened [27]. Human milk glycan microarrays were prepared and analyzed with many GBPs and viruses [74–76]. Through a combination of microarray-based Metadata-Assisted Glycan Sequencing (MAGS) [3] and sequential MSn analysis, a number of distinct human milk glycans were found to be ligands for different strains of rotavirus, and many of the biologically relevant glycans had never been previously identified in human milk. From pig lung tissue, a shotgun glycan microarray was prepared to study the binding of influenza virus to natural glycan ligands [77]. van Diepen et al [78] prepared schistosomes from different life stages to study anti-glycan antibody responses in infected people. Differential anti-glycan immune responses were observed for different groups of people.
Figure 4.
The general strategy for preparing fluorescently-tagged glycans from natural sources for generation of shotgun glycan microarrays.
Enzymatic modification of fluorescently tagged natural glycans has also proven to be an efficient method to build specialized glycan microarrays. The fluorescent/functional tag greatly simplified the assay of enzymatic reactions and preparation and immobilization of glycans. A mannose-6-phosphate (Man-6-P) array was prepared through this strategy and utilized to study the specificities of the Mannose-6-P receptors [79–81]. Similarly, a modified sialyl glycan array was constructed for the analysis of sialic acid-binding lectins and viruses [82,83].
Summary
The ability to fluorescently tagged free glycans has revolutionized analyses of their structures and quantification. Such tagging approaches are relatively unique to glycans, since other classes of macromolecules, e.g. peptides and nucleic acids, normally lack reactive aldehydes. The use of fluorescent tags has permitted separation and quantitative analyses of glycans from natural sources, such as serum glycoproteins, and has now even been automated [84–86]. Combining the ability of fluorescent-tagging of glycans with glycan separation and purification, as for shotgun glycomics, allows the production of libraries of natural glycans for immobilization onto glycan microarrays and conjugation to other types of surfaces and biomolecules. Natural glycans are structurally diverse and are more biologically relevant for functional glycomics study. Nevertheless, more novel methods are still in great need, especially for O-glycans, GAGs, and GPI-anchors. Such microarray technologies have proven to be a practical strategy to study protein-glycan interactions systematically and are expected to translate to more applications in the future.
Acknowledgments
The work of the authors in this area is supported by NIH Grant P41GM103694 to RDC.
References
- 1.Cummings RD, Pierce JM. The Challenge and Promise of Glycomics. Chem Biol (Oxford, U K) 2014;21 (1):1–15. doi: 10.1016/j.chembiol.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat Chem Biol. 2006;2(5):238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 3.Smith DF, Cummings RD. Application of Microarrays for Deciphering the Structure and Function of the Human Glycome. Mol Cell Proteomics. 2013;12(4):902–912. doi: 10.1074/mcp.R112.027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taniguchi N, Hancock W, Lubman DM, Rudd PM. The Second Golden Age of Glycomics: From Functional Glycomics to Clinical Applications. J Proteome Res. 2009;8(2):425–426. doi: 10.1021/pr801057j. [DOI] [PubMed] [Google Scholar]
- 5.Cummings RD. The repertoire of glycan determinants in the human glycome. Mol Biosyst. 2009;5(10):1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 6.Gahoi N, Ray S, Srivastava S. Array-based proteomic approaches to study signal transduction pathways: Prospects, merits and challenges. Proteomics. 2015;15(2–3):218–231. doi: 10.1002/pmic.201400261. [DOI] [PubMed] [Google Scholar]
- 7.Marino K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: A path through the maze. Nat Chem Biol. 2010;6(10):713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- 8.Zaia J. Mass spectrometry and glycomics. OMICS. 2010;14(4):401–418. doi: 10.1089/omi.2009.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen PH, Karlsson NG, Kolarich D, Packer NH. Structural analysis of N- and O-glycans released from glycoproteins. Nature protocols. 2012;7(7):1299–1310. doi: 10.1038/nprot.2012.063. [DOI] [PubMed] [Google Scholar]
- 10.Lundborg M, Widmalm G. Structural analysis of glycans by NMR chemical shift prediction. Analytical chemistry. 2011;83(5):1514–1517. doi: 10.1021/ac1032534. [DOI] [PubMed] [Google Scholar]
- 11.North SJ, Hitchen PG, Haslam SM, Dell A. Mass spectrometry in the analysis of N-linked and O-linked glycans. Curr Opin Struct Biol. 2009;19(5):498–506. doi: 10.1016/j.sbi.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toukach FV, Ananikov VP. Recent advances in computational predictions of NMR parameters for the structure elucidation of carbohydrates: methods and limitations. Chem Soc Rev. 2013;42(21):8376–8415. doi: 10.1039/c3cs60073d. [DOI] [PubMed] [Google Scholar]
- 13.Wang LX, Davis BG. Realizing the Promise of Chemical Glycobiology. Chem Sci. 2013;4(9):3381–3394. doi: 10.1039/C3SC50877C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castagner B, Seeberger PH. Automated solid phase oligosaccharide synthesis. Top Curr Chem. 2007;278:289–309. [Google Scholar]
- 15.Hsu CH, Hung SC, Wu CY, Wong CH. Toward Automated Oligosaccharide Synthesis. Angew Chem, Int Ed. 2011;50(50):11872–11923. doi: 10.1002/anie.201100125. [DOI] [PubMed] [Google Scholar]
- 16.Marino PA, Muthana SM, Gildersleeve JC. Glycan microarrays: Powerful tools for biomarker discovery. Cancer Biomarkers. 2014;14(1):29–41. doi: 10.3233/CBM-130383. [DOI] [PubMed] [Google Scholar]
- 17.Smith DF, Song X, Cummings RD. Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods in enzymology. 2010;480:417–444. doi: 10.1016/S0076-6879(10)80033-3. [DOI] [PubMed] [Google Scholar]
- 18.Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- 19.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(49):17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat Chem Biol. 2006;2(5):238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 21.Park S, Shin I. Fabrication of carbohydrate chips for studying protein-carbohydrate interactions. Angew Chem Int Ed Engl. 2002;41(17):3180–3182. doi: 10.1002/1521-3773(20020902)41:17<3180::AID-ANIE3180>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 22.Song X, Heimburg-Molinaro J, Cummings RD, Smith DF. Chemistry of natural glycan microarrays. Curr Opin Chem Biol. 2014;18:70–77. doi: 10.1016/j.cbpa.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvarez RA, Blixt O. Identification of ligand specificities for glycan-binding proteins using glycan arrays. Methods in enzymology. 2006;415:292–310. doi: 10.1016/S0076-6879(06)15018-1. [DOI] [PubMed] [Google Scholar]
- 24.Rillahan CD, Paulson JC. Glycan microarrays for decoding the glycome. Annu Rev Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han X, Zheng Y, Munro CJ, Ji Y, Braunschweig AB. Carbohydrate nanotechnology: hierarchical assembly using nature’s other information carrying biopolymers. Curr Opin Biotechnol. 2014;34C:41–47. doi: 10.1016/j.copbio.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Song X, Lasanajak Y, Rivera-Marrero C, Luyai A, Willard M, Smith DF, Cummings RD. Generation of a natural glycan microarray using 9-fluorenylmethyl chloroformate (FmocCl) as a cleavable fluorescent tag. Analytical biochemistry. 2009;395(2):151–160. doi: 10.1016/j.ab.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song X, Lasanajak Y, Xia B, Heimburg-Molinaro J, Rhea JM, Ju H, Zhao C, Molinaro RJ, Cummings RD, Smith DF. Shotgun glycomics: a microarray strategy for functional glycomics. Nature methods. 2011;8(1):85–90. doi: 10.1038/nmeth.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palma AS, Feizi T, Childs RA, Chai W, Liu Y. The neoglycolipid (NGL)-based oligosaccharide microarray system poised to decipher the meta-glycome. Curr Opin Chem Biol. 2014;18:87–94. doi: 10.1016/j.cbpa.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muthana SM, Gildersleeve JC. Glycan microarrays: powerful tools for biomarker discovery. Cancer Biomark. 2014;14(1):29–41. doi: 10.3233/CBM-130383. [DOI] [PubMed] [Google Scholar]
- 30.Breitling J, Aebi M. N-linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2013;5(8):a013359. doi: 10.1101/cshperspect.a013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanley P, Schachter H, Taniguchi N. Essentials of Glycobiology. Cold Spring Harbor Laboratories Press; 2009. N-Glycans (Chapter 8) [Google Scholar]
- 32.Tarentino AL, Plummer TH, Jr, Maley F. The release of intact oligosaccharides from specific glycoproteins by endo-beta-N-acetylglucosaminidase H. The Journal of biological chemistry. 1974;249(3):818–824. [PubMed] [Google Scholar]
- 33.Elder JH, Alexander S. endo-beta-N-acetylglucosaminidase F: endoglycosidase from Flavobacterium meningosepticum that cleaves both high-mannose and complex glycoproteins. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(15):4540–4544. doi: 10.1073/pnas.79.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plummer TH, Jr, Tarentino AL. Purification of the oligosaccharide-cleaving enzymes of Flavobacterium meningosepticum. Glycobiology. 1991;1(3):257–263. doi: 10.1093/glycob/1.3.257. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi N, Nishibe H. Some characteristics of a new glycopeptidase acting on aspartylglycosylamine linkages. Journal of biochemistry. 1978;84(6):1467–1473. doi: 10.1093/oxfordjournals.jbchem.a132270. [DOI] [PubMed] [Google Scholar]
- 36.Taga EM, Waheed A, Van Etten RL. Structural and chemical characterization of a homogeneous peptide N-glycosidase from almond. Biochemistry. 1984;23(5):815–822. doi: 10.1021/bi00300a006. [DOI] [PubMed] [Google Scholar]
- 37.Tretter V, Altmann F, Marz L. Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached alpha 1----3 to the asparagine-linked N-acetylglucosamine residue. European journal of biochemistry / FEBS. 1991;199(3):647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x. [DOI] [PubMed] [Google Scholar]
- 38.Yosizawa Z, Sato T, Schmid K. Hydrazinolysis of alpha-1-acid glycoprotein. Biochim Biophys Acta. 1966;121(2):417–420. doi: 10.1016/0304-4165(66)90134-6. [DOI] [PubMed] [Google Scholar]
- 39.Nakakita S, Sumiyoshi W, Miyanishi N, Hirabayashi J. A practical approach to N-glycan production by hydrazinolysis using hydrazine monohydrate. Biochem Biophys Res Commun. 2007;362(3):639–645. doi: 10.1016/j.bbrc.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y, Mechref Y, Novotny MV. Microscale nonreductive release of O-linked glycans for subsequent analysis through MALDI mass spectrometry and capillary electrophoresis. Analytical chemistry. 2001;73(24):6063–6069. doi: 10.1021/ac015534c. [DOI] [PubMed] [Google Scholar]
- 41.Yu G, Zhang Y, Zhang Z, Song L, Wang P, Chai W. Effect and limitation of excess ammonium on the release of O-glycans in reducing forms from glycoproteins under mild alkaline conditions for glycomic and functional analysis. Analytical chemistry. 2010;82(22):9534–9542. doi: 10.1021/ac102300r. [DOI] [PubMed] [Google Scholar]
- 42.Song X, Ju H, Zhao C, Lasanajak Y. Novel strategy to release and tag N-glycans for functional glycomics. Bioconjugate chemistry. 2014;25(10):1881–1887. doi: 10.1021/bc500366v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endo Y, Kobata A. Partial purification and characterization of an endo-alpha-N-acetylgalactosaminidase from the culture of medium of Diplococcus pneumoniae. Journal of biochemistry. 1976;80(1):1–8. doi: 10.1093/oxfordjournals.jbchem.a131240. [DOI] [PubMed] [Google Scholar]
- 44.Carlson DM. Oligosaccharides isolated from pig submaxillary mucin. The Journal of biological chemistry. 1966;241(12):2984–2986. [PubMed] [Google Scholar]
- 45.Carlson DM. Structures and immunochemical properties of oligosaccharides isolated from pig submaxillary mucins. The Journal of biological chemistry. 1968;243(3):616–626. [PubMed] [Google Scholar]
- 46.Wang C, Fan W, Zhang P, Wang Z, Huang L. One-pot nonreductive O-glycan release and labeling with 1-phenyl-3-methyl-5-pyrazolone followed by ESI-MS analysis. Proteomics. 2011;11(21):4229–4242. doi: 10.1002/pmic.201000677. [DOI] [PubMed] [Google Scholar]
- 47.Zauner G, Koeleman CA, Deelder AM, Wuhrer M. Mass spectrometric O-glycan analysis after combined O-glycan release by beta-elimination and 1-phenyl-3-methyl-5-pyrazolone labeling. Biochim Biophys Acta. 2012;1820(9):1420–1428. doi: 10.1016/j.bbagen.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 48.Merry AH, Neville DCA, Royle L, Matthews B, Harvey DJ, Dwek RA, Rudd PM. Recovery of intact 2-aminobenzamide-labeled o-glycans released from glycoproteins by hydrazinolysis. Anal Biochem. 2002;304(1):91–99. doi: 10.1006/abio.2002.5620. [DOI] [PubMed] [Google Scholar]
- 49.Patel T, Bruce J, Merry A, Bigge C, Wormald M, Parekh R, Jaques A. Use of hydrazine to release in intact and unreduced form both N- and O-linked oligosaccharides from glycoproteins. Biochemistry. 1993;32(2):679–693. doi: 10.1021/bi00053a037. [DOI] [PubMed] [Google Scholar]
- 50.Chai W, Feizi T, Yuen CT, Lawson AM. Nonreductive release of O-linked oligosaccharides from mucin glycoproteins for structure/function assignments as neoglycolipids: application in the detection of novel ligands for E-selectin. Glycobiology. 1997;7(6):861–872. doi: 10.1093/glycob/7.6.861. [DOI] [PubMed] [Google Scholar]
- 51.Miura Y, Kato K, Takegawa Y, Kurogochi M, Furukawa J, Shinohara Y, Nagahori N, Amano M, Hinou H, Nishimura S. Glycoblotting-assisted O-glycomics: ammonium carbamate allows for highly efficient o-glycan release from glycoproteins. Analytical chemistry. 2010;82(24):10021–10029. doi: 10.1021/ac101599p. [DOI] [PubMed] [Google Scholar]
- 52.Prasanphanich NS, Song X, Heimburg-Molinaro J, Luyai A, Lasanajak Y, Cutler C, Smith DF, Cummings RD. An Intact Reducing Glycan Promotes the Specific Immune Response to Lacto-N-neotetraose-BSA Neoglycoconjugates. Bioconjugate chemistry. 2015 doi: 10.1021/acs.bioconjchem.5b00036. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito M, Yamagata T. A novel glycosphingolipid-degrading enzyme cleaves the linkage between the oligosaccharide and ceramide of neutral and acidic glycosphingolipids. The Journal of biological chemistry. 1986;261(30):14278–14282. [PubMed] [Google Scholar]
- 54.Ito M, Yamagata T. Purification and characterization of glycosphingolipid-specific endoglycosidases (endoglycoceramidases) from a mutant strain of Rhodococcus sp Evidence for three molecular species of endoglycoceramidase with different specificities. The Journal of biological chemistry. 1989;264(16):9510–9519. [PubMed] [Google Scholar]
- 55.Ashida H, Yamamoto K, Kumagai H, Tochikura T. Purification and characterization of membrane-bound endoglycoceramidase from Corynebacterium sp. European journal of biochemistry / FEBS. 1992;205(2):729–735. doi: 10.1111/j.1432-1033.1992.tb16836.x. [DOI] [PubMed] [Google Scholar]
- 56.Ishibashi Y, Nakasone T, Kiyohara M, Horibata Y, Sakaguchi K, Hijikata A, Ichinose S, Omori A, Yasui Y, Imamura A, Ishida H, Kiso M, Okino N, Ito M. A novel endoglycoceramidase hydrolyzes oligogalactosylceramides to produce galactooligosaccharides and ceramides. The Journal of biological chemistry. 2007;282(15):11386–11396. doi: 10.1074/jbc.M608445200. [DOI] [PubMed] [Google Scholar]
- 57.Wiegandt H, Baschang G. the Isolation of the Sugar Portion of Glycosphingolipids by Ozonolysis and Fragmentation. Z Naturforsch B. 1965;20:164–166. [PubMed] [Google Scholar]
- 58.Hakomori SI. Release of carbohydrates from sphingoglycolipid by osmium-catalyzed periodate oxidation followed by treatment with mild alkali. J Lipid Res. 1966;7(6):789–792. [PubMed] [Google Scholar]
- 59.Song X, Smith DF, Cummings RD. Nonenzymatic release of free reducing glycans from glycosphingolipids. Anal Biochem. 2012;429:82–87. doi: 10.1016/j.ab.2012.06.029. (Copyright (C) 2013 American Chemical Society (ACS). All Rights Reserved.) [DOI] [PubMed] [Google Scholar]
- 60.Noti C, de Paz JL, Polito L, Seeberger PH. Preparation and use of microarrays containing synthetic heparin oligosaccharides for the rapid analysis of heparin-protein interactions. Chemistry. 2006;12 (34):8664–8686. doi: 10.1002/chem.200601103. [DOI] [PubMed] [Google Scholar]
- 61.Park TJ, Lee MY, Dordick JS, Linhardt RJ. Signal amplification of target protein on heparin glycan microarray. Analytical biochemistry. 2008;383(1):116–121. doi: 10.1016/j.ab.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hase S, Ikenaka T, Matsushima Y. Structure analyses of oligosaccharides by tagging of the reducing end sugars with a fluorescent compound. Biochem Biophys Res Commun. 1978;85(1):257–263. doi: 10.1016/s0006-291x(78)80037-0. [DOI] [PubMed] [Google Scholar]
- 63.Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Analytical biochemistry. 1995;230(2):229–238. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- 64.Anumula KR. Single tag for total carbohydrate analysis. Analytical biochemistry. 2014;457:31–37. doi: 10.1016/j.ab.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 65.Xia B, Kawar ZS, Ju T, Alvarez RA, Sachdev GP, Cummings RD. Versatile fluorescent derivatization of glycans for glycomic analysis. Nature methods. 2005;2(11):845–850. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 66.Song X, Xia B, Lasanajak Y, Smith DF, Cummings RD. Quantifiable fluorescent glycan microarrays. Glycoconjugate journal. 2008;25(1):15–25. doi: 10.1007/s10719-007-9066-8. [DOI] [PubMed] [Google Scholar]
- 67.de Boer AR, Hokke CH, Deelder AM, Wuhrer M. General microarray technique for immobilization and screening of natural glycans. Analytical chemistry. 2007;79(21):8107–8113. doi: 10.1021/ac071187g. [DOI] [PubMed] [Google Scholar]
- 68.Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 2009;16(1):36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luyai A, Lasanajak Y, Smith DF, Cummings RD, Song X. Facile Preparation of Fluorescent Neoglycoproteins Using p-Nitrophenyl Anthranilate as a Heterobifunctional Linker. Bioconjugate chemistry. 2009;20(8):1618–24. doi: 10.1021/bc900189h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song X, Johns BA, Ju H, Lasanajak Y, Zhao C, Smith DF, Cummings RD. Novel cleavage of reductively aminated glycan-tags by N-bromosuccinimide to regenerate free, reducing glycans. ACS chemical biology. 2013;8(11):2478–2483. doi: 10.1021/cb400513k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamada K, Hirabayashi J, Kakehi K. Analysis of O-Glycans as 9-Fluorenylmethyl Derivatives and Its Application to the Studies on Glycan Array. Anal Chem (Washington, DC, U S) 2013;85:3325–3333. doi: 10.1021/ac303771q. [DOI] [PubMed] [Google Scholar]
- 72.de Boer AR, Hokke CH, Deelder AM, Wuhrer M. Serum antibody screening by surface plasmon resonance using a natural glycan microarray. Glycoconjugate J. 2008;25(1):75–84. doi: 10.1007/s10719-007-9100-x. [DOI] [PubMed] [Google Scholar]
- 73.Song X, Lasanajak Y, Xia B, Smith DF, Cummings RD. Fluorescent glycosylamides produced by microscale derivatization of free glycans for natural glycan microarrays. ACS chemical biology. 2009;4(9):741–750. doi: 10.1021/cb900067h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu Y, Lasanajak Y, Song X, Hu L, Ramani S, Mickum ML, Ashline DJ, Prasad BV, Estes MK, Reinhold VN, Cummings RD, Smith DF. Human milk contains novel glycans that are potential decoy receptors for neonatal rotaviruses. Molecular & cellular proteomics : MCP. 2014;13(11):2944–2960. doi: 10.1074/mcp.M114.039875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ashline DJ, Yu Y, Lasanajak Y, Song X, Hu L, Ramani S, Prasad V, Estes MK, Cummings RD, Smith DF, Reinhold VN. Structural characterization by multistage mass spectrometry (MSn) of human milk glycans recognized by human rotaviruses. Molecular & cellular proteomics : MCP. 2014;13(11):2961–2974. doi: 10.1074/mcp.M114.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu Y, Mishra S, Song X, Lasanajak Y, Bradley KC, Tappert MM, Air GM, Steinhauer DA, Halder S, Cotmore S, Tattersall P, Agbandje-McKenna M, Cummings RD, Smith DF. Functional glycomic analysis of human milk glycans reveals the presence of virus receptors and embryonic stem cell biomarkers. The Journal of biological chemistry. 2012;287(53):44784–44799. doi: 10.1074/jbc.M112.425819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Byrd-Leotis L, Liu R, Bradley KC, Lasanajak Y, Cummings SF, Song X, Heimburg-Molinaro J, Galloway SE, Culhane MR, Smith DF, Steinhauer DA, Cummings RD. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(22):E2241–2250. doi: 10.1073/pnas.1323162111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Diepen A, Smit CH, van Egmond L, Kabatereine NB, de Moira AP, Dunne DW, Hokke CH. Differential anti-glycan antibody responses in Schistosoma mansoni-infected children and adults studied by shotgun glycan microarray. PLoS Neglected Trop Dis. 2012;6(11):e1922. doi: 10.1371/journal.pntd.0001922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song X, Lasanajak Y, Olson LJ, Boonen M, Dahms NM, Kornfeld S, Cummings RD, Smith DF. Glycan microarray analysis of P-type lectins reveals distinct phosphomannose glycan recognition. The Journal of biological chemistry. 2009;284(50):35201–35214. doi: 10.1074/jbc.M109.056119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bohnsack RN, Song X, Olson LJ, Kudo M, Gotschall RR, Canfield WM, Cummings RD, Smith DF, Dahms NM. Cation-independent mannose 6-phosphate receptor: a composite of distinct phosphomannosyl binding sites. The Journal of biological chemistry. 2009;284(50):35215–35226. doi: 10.1074/jbc.M109.056184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olson LJ, Castonguay AC, Lasanajak Y, Peterson FC, Cummings RD, Smith DF, Dahms NM. Identification of a fourth mannose 6-phosphate binding site in the cation-independent mannose 6-phosphate receptor. Glycobiology. 2015 Jan 8; doi: 10.1093/glycob/cwv001. pii: cwv001 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Padler-Karavani V, Song X, Yu H, Hurtado-Ziola N, Huang S, Muthana S, Chokhawala HA, Cheng J, Verhagen A, Langereis MA, Kleene R, Schachner M, de Groot RJ, Lasanajak Y, Matsuda H, Schwab R, Chen X, Smith DF, Cummings RD, Varki A. Cross-comparison of protein recognition of sialic acid diversity on two novel sialoglycan microarrays. The Journal of biological chemistry. 2012;287(27):22593–22608. doi: 10.1074/jbc.M112.359323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song X, Yu H, Chen X, Lasanajak Y, Tappert MM, Air GM, Tiwari VK, Cao H, Chokhawala HA, Zheng H, Cummings RD, Smith DF. A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. The Journal of biological chemistry. 2011;286(36):31610–31622. doi: 10.1074/jbc.M111.274217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campbell MP, Ranzinger R, Lutteke T, Mariethoz J, Hayes CA, Zhang J, Akune Y, Aoki-Kinoshita KF, Damerell D, Carta G, York WS, Haslam SM, Narimatsu H, Rudd PM, Karlsson NG, Packer NH, Lisacek F. Toolboxes for a standardised and systematic study of glycans. BMC Bioinformatics. 2014;15 (Suppl 1):S9. doi: 10.1186/1471-2105-15-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tharmalingam T, Adamczyk B, Doherty MA, Royle L, Rudd PM. Strategies for the profiling, characterisation and detailed structural analysis of N-linked oligosaccharides. Glycoconjugate journal. 2013;30(2):137–146. doi: 10.1007/s10719-012-9443-9. [DOI] [PubMed] [Google Scholar]
- 86.Marino K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nat Chem Biol. 2010;6(10):713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]