Abstract
DNA minor-groove-binding compounds have limited biological applications, in part due to problems with sequence specificity that cause off-target effects. A model to enhance specificity has been developed with the goal of preparing compounds that bind to two AT sites separated by G•C base pairs. Compounds of interest were probed using thermal melting, circular dichroism, mass spectrometry, biosensor-SPR, and molecular modeling methods. A new minor groove binder that can strongly and specifically recognize a single G•C base pair with flanking AT sequences has been prepared. This multi-site DNA recognition mode offers novel design principles to recognize entirely new DNA motifs.
Keywords: Sequence Specificity, Mixed DNA recognition, Rational Design, Heterocyclic Diamidines, Biophysical Analysis
Graphical Abstract
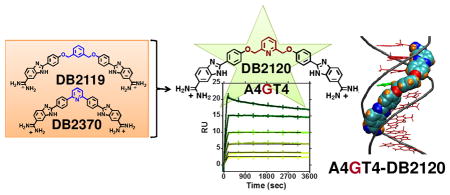
The systematic design and preparation of compounds that can recognize mixed base pair (bp) nucleic acid sequences is a very important goal in DNA small-molecule applications. Such specific recognition would allow new diagnostic applications in vitro while in vivo they could provide new therapeutics and gene-specific probes. This goal has been difficult to reach, however, and to date only polyamides have had significant success in design of compounds for mixed-sequence DNA recognition.1–4 Unfortunately, polyamides can be limited by synthetic costs, aggregation and cell-uptake problems.5,6 In an interesting contrast, relatively simple AT sequence selective minor-groove binders have had good success in therapeutic targeting of DNA in cells ranging from various types of cancers7–13 to bacteria14–16 and parasitic microorganisms.17–22 DNA minor-groove binding heterocyclic diamidines, such as pentamidine and berenil, are examples with a long history of therapeutic use.23–26 DAPI is another heterocyclic diamidine that readily enters cells and is frequently used to stain nuclear DNA in imaging.27 Limited selectivity and off-target effects, however, have restricted the therapeutic applications of these types of compounds, and methods to increase their binding selectivity and affinity are essential for development of improved agents.
The starting points for our development of new sequence-specific agents are synthetic compounds known to recognize AT sequences as well as the extensive literature on A•T specific minor-groove binders. The heterocyclic diamidines described above are all A•T site specific. For recognizing sequences containing G•C bp, new functional groups must be incorporated into the A•T specific agents. Such compounds would be helpful in a number of areas such as targeting the kinetoplast DNA (kDNA) of parasitic microorganisms, where a large number of A•T bp sites are frequently separated by a G•C bp,28–30 as well as transcription factor (TF) inhibition in gene control applications.31 Targeting transcription factors has been recognized as a promising but difficult direction for therapeutic development.32,33 In our initial efforts in this area, compounds with alkyl or alkyl-aromatic linked amidine-benzimidazole-phenyl (ABP) motifs were designed, synthesized and evaluated for binding to A•T sites separated by one or two G•C base pairs.34 The goal of these studies was to determine how linker length and rigidity affect the ability of ABP modules to recognize two A•T sites.
In the progress to our next generation sequence specific minor-groove binders, we have prepared a series of potential mixed-sequence DNA binding molecules from the existing pure AT sequence binders. In the design path to G•C bp recognition by rationally designed small molecules we have incorporated groups that can act as H-bond accepting units in the minor groove of DNA (Fig. 1). Some very encouraging initial success in targeting mixed bp DNA sequences was found with a relatively small, dissymmetric, diamidine compound, DB2277 (Fig. 1), which recognizes a single G in sequences such as AAGTT through an aza-benzimidazole group.35,36 In this report we describe a more general approach by connecting two A•T specific binding modules with H-bond accepting linking units. The results show critical effects of molecular flexibility, position of the linker and heteroatom for mixed sequence-specific DNA recognition.
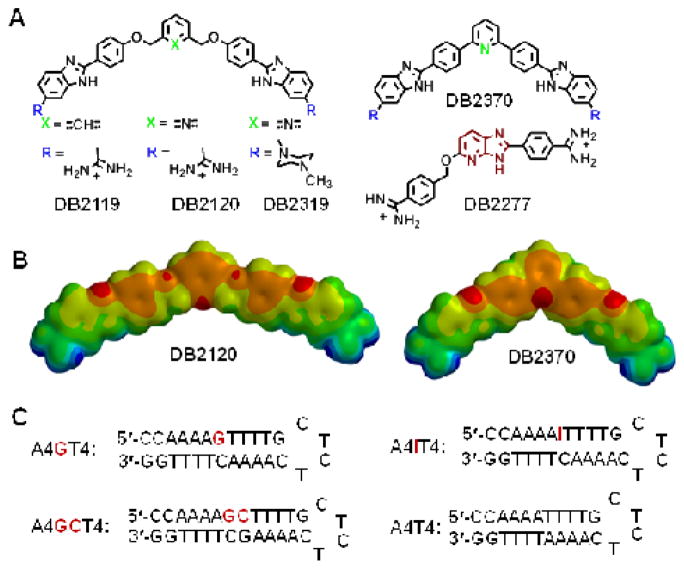
Figure 1.
(A) Chemical structures of mixed sequence targeting compounds. (B) Energy minimized structures of DB2120 and DB2370 at the 6-31G* (p,d) level of theory. In the electrostatic potential maps, red color indicates high electronegativity and blue indicates electron deficient/positively charged regions. (C) DNA hairpin oligomer duplexes used in this study. DNA sequences with 5′-biotin labels were used for SPR studies.
Detailed biophysical studies of the compounds in Figure 1 showed very strong and selective recognition of a G•C bp separating two A•T sites with much weaker recognition of pure A•T sites by a pyridyl-linked compound, DB2120. The rational design and preparation of a non-polyamide compound that can selectively recognize long, mixed bp sequences is a significant step forward in the design of minor-groove binders for potential diagnostic and therapeutic use. While there is much to learn, the new results offer clear directions and leads for the development of a DNA minor groove-binding language.
Compound Design and preparation
The design platform for the compounds in Figure 1 starts with the classical A•T-specific minor-groove binding compound, DB2119.34 The ABP A•T recognition unit was held constant for binding to the flanking A•T sites while other molecular units of DB2119 were varied and the derivatives were evaluated for G•C bp recognition. DB2370 is similar to DB2119 but without the -O-CH2- linker groups and with the central phenyl replaced with a pyridine. It is a direct derivative of a classical triphenyl A•T-specific minor-groove binder. Combinations of these two gives DB2120 with a central pyridine. The DB2120 and DB2370 pair tell us if the pyridine-N can serve as an H-bond acceptor for the minor groove G-NH and whether the interaction is better with or without the flexible -O-CH2-. DB2319 is the same as DB2120 but with the amidine cationic groups changed to methyl piperazinyl which is in Hoechst nuclear DNA staining compounds (Fig. 1). This cationic group change will indicate the contribution of the cations to binding and in cell studies may affect the31 types of cells that can be targeted with this molecular platform. The new bis-benzimidazoles DB2120, DB2319, and DB2370 were synthesized employing standard methodology for coupling the proper substituted phenylenediamine with the appropriate bis- aldehyde; Schemes and experimental details are provided in the supplementary material section.
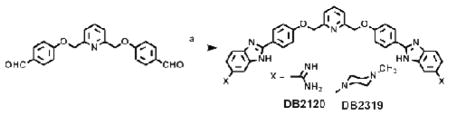
Reagents and conditions
a: 3,4-diaminobenzamidine hydrochloride or 4-(N-methylpiperazin-1-yl)-1,2-phenylenediamine, 1,4-benzoquinone, ethanol, reflux
The compounds in Figure 1 were selected based on Tm screening studies of a large number of synthetic compounds and they illustrate how to incorporate G•C bp recognition into an A•T binding compound. The crucial breakthrough in developing strong and selective G•C bp recognition in this series was achieved with the pyridyl derivative of DB2119, compound DB2120. The phenyl to pyridyl change resulted in an increase in the thermal stability of the single G•C-containing A4GT4 motif (ΔTm = 22 °C; Fig. S1, Table S1, Supplementary material) but a decrease in AT binding (ΔTm = 15 °C). The piperazinyl, DB2319, had some selectivity for G•C bp but weaker binding while DB2370 did not show significant selectivity (ΔTm =19 °C and ΔTm = 24 °C for A4GT4 and A4T4, respectively). These results clearly indicate that the pyridine nitrogen in DB2120 is serving as an H-bond acceptor for the exocyclic amino group of guanine to give a considerable increase in thermal stability of the complex. All compounds showed limited enhancement in thermal stabilities for two G•C-containing sequences such as A4GCT4 (Table S1).
Biosensor-surface plasmon resonance (SPR) is an effective technique to quantitatively monitor biomolecular interactions in real-time to obtain binding affinity, kinetics, and stoichiometry.34–39 Biosensor experiments were conducted with DB2120 based on the Tm screening results and Figure 2 shows sensorgrams of DB2120 with A4GT4, A4IT4 (I = inosine, 2-amino of guanine replaced with H), and A4T4 motifs. It is apparent from these results that the interactions of DB2120 with DNAs with and without a G•C bp are quite distinct. The most striking result is the interaction with the single G•C bp-containing A4GT4, and particularly noteworthy is the very slow dissociation of DB2120 from the DNA complex (Fig. 2A). Even with a quite long experimental dissociation time (~1 h), very little dissociation of DB2120 from the complex is observed under these conditions. Global kinetics fitting yielded a single binding site and an approximate KA = 6.6 × 1010 M−1 (KD = 1.5 × 10−11 M) for DB2120 at 0.1 M Na+. This is an impressive 2000-fold increase compared to the parent compound, DB2119. DB2120 binds to A4T4 in a monomer complex with a 200-fold lower affinity compared to A4GT4, indicating high specificity for the single G•C bp sequence. The sensorgrams of A4T4 show an off-rate that is much faster, and complete dissociation from the complex occurs within the first few minutes of the dissociation phase (Fig. 2C). With the A4GCT4 sequence, no binding is observed for DB2120 under this experimental condition, in agreement with Tm results. When G (A4GT4) is substituted by I (A4IT4) at the minor groove of the mixed-DNA sequence, although the groove width remains almost the same40 the binding affinity of the A4IT4-DB2120 complex decreases by 80-fold (KD = 1.2 × 10−9 M nM). This phenomenon is expected by the interruption of H-bonding between the pyridyl-N and G-NH2 (Fig. 2B) in the A4IT4 complex and confirms the DB2120 to G-NH H-bond.
Figure 2.
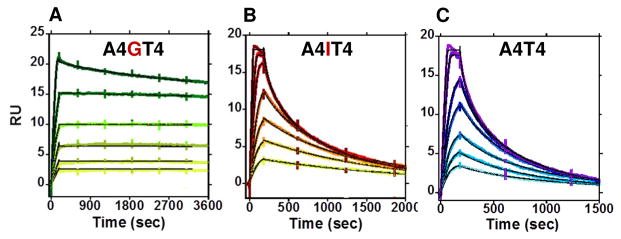
SPR sensorgrams for DB2120 with (A) A4GT4, (B) A4IT4, and (C) A4T4 sequences, respectively. The injected concentrations of DB2120 with A4GT4 are 2, 3, 5, 7, 10 and 15 nM whereas with A4IT4 and A4T4 sequences the injected concentrations are higher; 3, 5, 7, 10, 15, 20, 30 and 5, 7, 10, 15, 20, 30, and 50 nM respectively. The black lines for all three sequences represent global kinetic fitting using a 1:1 interaction model.
In summary, SPR and Tm results converge to indicate very strong and selective monomer binding of DB2120 to AT binding sites separated by a single G•C bp. No other compound from Figure 1 showed significant DNA sequence specific recognition. The SPR results clearly highlight the success and the difficulties (compare DB2120 and DB2370) in the development of G•C specific recognition.
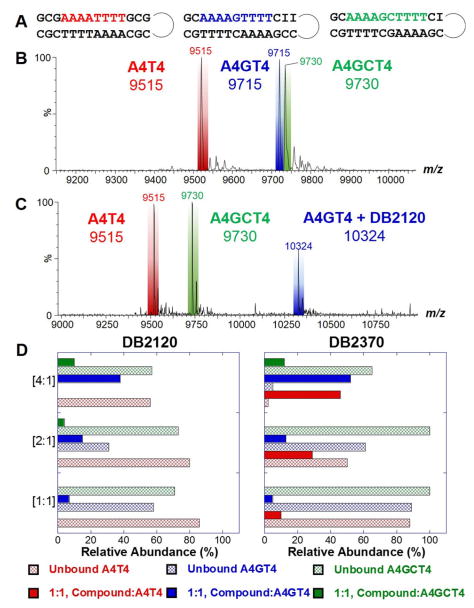
Competition electrospray ionization mass spectrometry (ESI-MS) is a method that we have developed to provide comparative information on the stoichiometry, cooperativity, and relative affinity for DNA complexes.41,42 The use of multiple DNA sequences simultaneously mixed with a small molecule creates a competitive binding environment for comparison of DNA-ligand interactions. Since the free ligand concentration for all of the DNAs is the same, peak intensities of the free, unbound DNA and the DNA-small molecule complexes offer insight into the binding mode and preferred binding site(s). Each titration series contains a pure GC sequence, used as an internal standard, that does not bind ligand. This offers a simple comparison for peak intensities, since the DNA molecular weights are adjusted to be different. To help adjust molecular weights among this set of sequences, substitution of guanine by inosine in the bp(s) nearest the hairpin loop were incorporated (Fig. 3A).
Figure 3.
(A) Sequences used for competition ESI mass spectrometry. Example spectra of (B) free DNA and (C) DNA and DB2120-DNA complexes. (D) Comparison of the peak intensities for DNA and ligand-DNA complexes for a series of titrations of [1:1], [2:1], and [4:1] and are expressed as a mole to mole ratio of ligand to DNAsingle sequence (e.g. [2:1] = 10 μM DB2120 to 5 μM A4T4).
The top spectrum in Figure 3 (Fig. 3B) is an example of free DNA prior to titration. The bottom spectrum (Fig. 3C) illustrates DNA interactions with DB2120 at a [4:1] added ratio. The free A4GT4 sequence is no longer visible and shows that the DNA has formed a 1:1, DB2120-A4GT4 complex, as indicated by the tall peak at m/z 10,324. It is striking that there are no complexes for A4GT4 and A4T4 DNAs at this ratio and is in direct agreement with Tm and SPR experiments. The relative peak intensities for the titrations are compared for DB2120 and DB2370 in Figure 3D and shown as compound ratio versus relative abundance. DB2119 showed relatively weak binding to A4T4 and a small signal for A4GT4 at the [4:1] ratio, in agreement with Tm and SPR results. For DB2370 at a [4:1] ratio, the majority of A4GT4 and A4T4 DNAs are bound with nominal amounts of free DNA, whereas most of the A4GCT4 is left unbound. DB2370 thus has no significant selectivity for A4GT4 over A4T4. All compounds show very poor binding to the two G•C bp sequence (A4GCT4). As with all other methods, DB2120 shows very strong and selective binding for the single G•C-containing sequence, A4GT4.
Circular dichroism (CD) is a powerful technique to detect conformational changes of biomolecules, as well as small molecule binding modes using pattern recognition.43,44 Figure S2 (Supplementary material) shows the titration CD spectra of DB2120 with the A4GT4 sequence. The compound exhibits strong, positive induced CD signals between 300 to 400 nm that have been found to be indicative of minor-groove binding, as expected from the structure. Small and consistent changes in the CD spectral region of DNA (230 to 290 nm) are observed with incremental titration of ligands, indicating only minor conformational changes in DNA upon complex formation
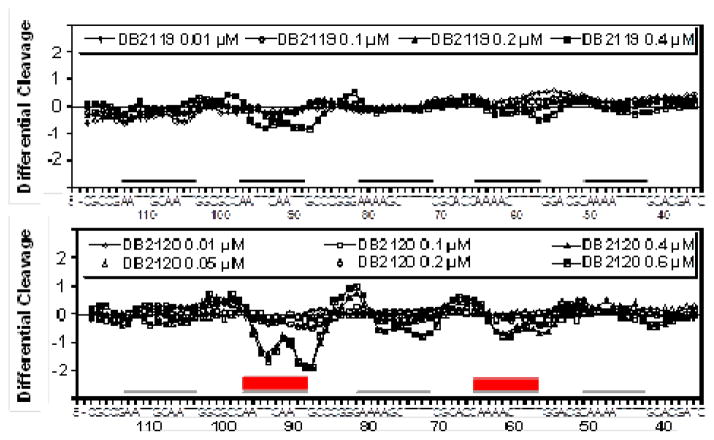
Do these results on sequence specificity by DB2120 hold up with a much longer DNA? To answer this question, binding to a long DNA fragment was also assessed using DNase I footprinting experiments. A DNA fragment containing A4CT4•A4GT4, A4GCT4•A4GCT4, A4T4•A4T4, AATTGCAATT•AATTGCAATT, and AATTCAATT•AATTGAATT sequences (black lines, Fig. 4) was prepared (Experimental Section, Supplementary material) and incubated with increasing concentrations of DB2120 and DB2119 prior to mild DNase I treatment (Fig. 4, experimental gel electrophoresis results in Fig. S3, Supplementary material). The corresponding densitometric analyses for DB2120 and DB2119 (Fig. 4) show very different results for the two compounds that structurally differ in only by replacing a CH atom by N. Strong and selective binding of DB2120 to the AATTCAATT•AATTGAATT site contrasts with the absence of binding to AATTGCAATT•AATTGCAATT. Binding is also seen for DB2120 with A4CT4•A4GT4 but no significant binding to a similar AT sequence without a G•C base pair (A4T4•A4T4). In contrast, DB2119 shows weaker binding to all of the binding sites under the gel conditions, in agreement with the biophysical methods discussed above.
Figure 4.
DNase I footprinting densitometry analysis. Binding of the phenyl (DB2119) and pyridyl (DB2120) compounds to the 3′-end radio-labeled 208bp DNA fragment (experimental details in Supplementary material) containing the five different DNA sequences (A4CT4•A4GT4, A4T4•A4T4, A4GCT4•A4GCT4, AATTCAATT•AATTGAATT, and AATTGCAATT•AATTGCAATT, localized as black lines) was quantified relative to unbound DNA treated with DNase I in similar manner. The primary bound portions of DNA are localized as red boxes. The single G•C bp recognition by the pyridyl compound is visually apparent
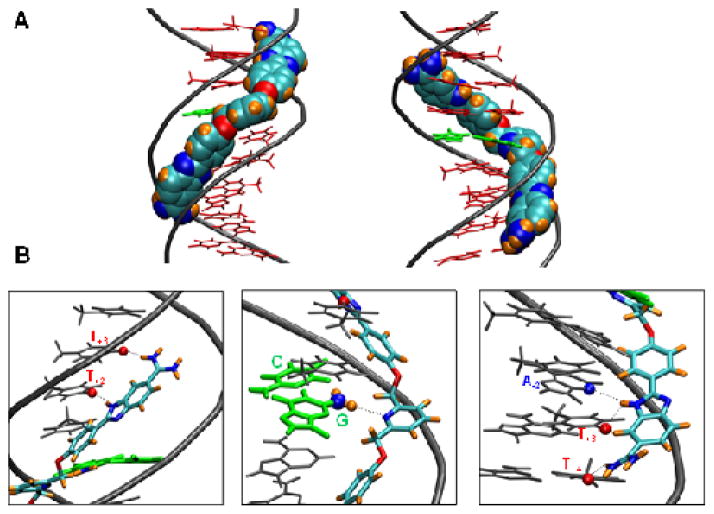
For molecular insights into the structural parameters dictating G•C bp recognition by DB2120, molecular modeling studies were conducted with the A4GT4 sequence using experimentally derived information. Docking analysis reveals that DB2120 complements the DNA minor-groove shape and makes excellent van der Waals interactions with the walls of the minor groove (Fig. 5A). The two terminal ABP modules are oriented parallel to the groove walls, while the central pyridine ring is deeply embedded in the minor groove. As expected, the inner-facing pyridine nitrogen is involved in H-bonding to the central G-NH that extends into the minor-groove (Fig. 5B, middle). Additional H-bonds are formed between the benzimidazole and amidine groups with A•T bp (Fig. 5B, left and right). This orientation provides excellent minor-groove recognition units for both G•C and A•T bp. In addition, the linker flexibility allows molecular twist along the groove that orients the two ABP units in such a way that the individual units on either side of the pyridine ring form strong interactions with both strands of the duplex. The specific interactions observed between DB2120 and the minor groove of A4GT4 dictate the sequence selectivity observed for this compound and provide a rational explanation for the observed mixed-sequence DNA recognition.
Figure 5.
Molecular docking model of DB2120 with the A4GT4 sequence. (A) The model represents the minor (left) and major (right) groove views of DB2120 with A4GT4 sequence. The A•T DNA bases are represented as stick models in red and the central G•C bp is green. The DNA backbone is represented as a tube in grey color. DB2120 is represented in a space-filling model with the carbons light blue, nitrogen dark blue, hydrogens orange and oxygen red. (B) Important interactions between different sections of DB2120 and DNA are illustrated. The central G•C bp is used as the reference for base numbering of the leading and complementary strands. The benzimidazole ring NH and the amidine unit at the top of DB2120 in this orientation forms strong interactions with the carbonyl groups of T+2 and T+3 bases in the minor groove of the leading strand (left). The pyridine nitrogen makes strong interactions with the G-NH2 (ball and stick) in the minor groove (middle). The benzimidazole NH at the bottom end forms bifurcated interactions with A-2 N3 on the 5′-AAACTTT-3′ strand and T+3 carbonyl groups of the 5′-AAAGTTT-3′ strand, respectively. The amidine unit forms strong interactions with the carbonyl group of T+4 of the AAACTTT strand (right). All the interactions contribute to the overall stability of complex formation with the pyridine ring conferring G•C recognition as it was designed to do.
In summary, new ideas for the design of a variety of cell-permeable compounds for sequence specific recognition of DNA are needed to take advantage of the wealth of new and expanding genomic information. The variety is required because of the different uptake characteristic of different cells and the different pharmacokinetics or ADME properties of compounds of different structure.31,45 In this project A•T bp recognition groups were linked by test modules for potential G•C bp recognition with the long-term objective of developing a new paradigm for sequence-specific DNA minor-groove recognition using biologically-compatible heterocyclic cations.
The pyridyl analog of DB2119, DB2120, is a significant advance in the design of compounds that are highly selective for single G•C bp recognition with flanking AT sequences. As noted above, such sequences appear often in the kDNA of parasitic microorganisms as well as TF promoter sites. SPR results clearly show that the slow dissociation rate of DB2120 from the A4GT4 sequence is the primary factor in the strong and selective binding observed. The compound binds rapidly and forms numerous van der Waals and H-bond interactions (Fig. 5) that must be disrupted to dissociate the complex. The absence of the intervening G•C bp in the A4T4 and inosine motifs clearly eliminates the H-bonding interaction between the G-NH2 group and DB2120 and gives faster dissociation and weaker binding. The DNA interactions of DB2120 offer insights that can be integrated into future compound design schemes. The significant loss in binding selectivity by DB2370, due to the absence of the -O-CH2- linker, also indicates that not only the molecular geometry but also the flexibility of DB2120 play very critical roles for the selective recognition of the G-NH2 group. The sequence composition, which primarily governs the local microstructure of the minor groove, also strongly dictates the strength and stoichiometry of binding. While the narrower minor groove of pure A-tract motifs with enhanced negative electrostatic potential offer a more conducive environment for a highly favorable monomer complex formation, the somewhat wider grooves of other A•T or more G•C bp-containing sequences34 may promote strong dimeric complex formation.
The SPR, mass spectrometry, and DNase I footprinting results strongly support mixed bp DNA recognition by DB2120 but not the phenyl derivative, DB2119. This is particularly important since the DNase I cleavage experiments are done with long DNA sequences that are quite different from the shorter segments used in our other biophysical studies. An example densitometry trace of a DNase I footprinting gel that compares DB2119 to results with DB2120 (Fig. 4) shows that AAAACTTTT•AAAAGTTTT and AATTCAATT•AATTGAATT sequences are the strongest binding sites for DB2120 with much weaker binding to two G•C and pure AT sequences.
The molecular model in Figure 5 provides information to explain the strong and specific binding of DB2120 to the A4GT4 site. The pyridyl-N forms a strong and direct H-bond to the G-amino-NH that projects into the minor groove. The two -O-CH2- flexible linking groups of DB2120 allow rotations of the terminal phenyl-benzimidazole-amidine units such that they are able to fit snuggly into the minor groove in the flanking A-tract sequences. Without these groups, DB2370 has poor G•C selectivity. Both benzimidazole-NH and amidine-NH groups form strong, direct H-bonds with C=O groups of thymines in the minor groove (Fig. 5). The amidine cationic charges are well-placed to form ionic interactions with the anionic DNA backbone. The entire DB2120 molecule is involved in optimizing the complex with the DNA minor groove. An electron density calculation (Fig. 1) shows the positive electron density on the amidines and benzimidazole-NH groups and the negative charge density on the pyridyl-N help account for strong binding between DB2120 and the DNA minor groove.
The combined biophysical results reveal that effective recognition of AT-rich sequences with intervening G•C bps can be achieved with heterocyclic cations that have appropriately placed H-bond acceptors flanked by A•T bp recognition modules and appropriate flexibility. This information should assist significantly in the rational design of further potent heterocyclic cations to target larger mixed DNA sequences with more G•C base pairs.
Supplementary Material
Acknowledgments
The work was supported by National Institutes of Health (NIH) grant GM111749 awarded to WDW and DWB and by the Ligue Nationale Contre le Cancer (Comité du Nord) and the IRCL to MHDC. MHDC thanks the IFR114-IMPRT for access to the PMI equipment. SRL thanks the Molecular Basis of Disease Area of Focus (MBDAF) fellowship. The authors thank Carol Wilson for manuscript assistance.
Footnotes
Supplementary material (compound synthetic procedures, materials and methods, UV-Vis thermal spectra, CD spectra, DNase I Footprinting) associated with this article can be found in the online version.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Meier JL, Yu AS, Korf I, Segal DJ, Dervan PB. J Am Chem Soc. 2012;134:17814. doi: 10.1021/ja308888c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dervan PB, Poulin-KerstienATFechter E, Edelson BS. Top Curr Chem. 2005;253:1. [Google Scholar]
- 3.Chavda S, Liu Y, Babu B, Davis R, Sielaff A, Ruprich J, Westrate L, Tronrud C, Ferguson A, Franks A, Tzou S, Adkins C, Rice T, Mackay H, Kluza J, Tahir SA, Lin S, Kiakos K, Bruce CD, Wilson WD, Hartley JA, Lee M. Biochemistry. 2011;19:3127. doi: 10.1021/bi102028a. [DOI] [PubMed] [Google Scholar]
- 4.He G, Vasilieva E, Harris GD, Jr, Koeller KJ, Bashkin JK, Dupureur CM. Biochimie. 2014;102:83. doi: 10.1016/j.biochi.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, Aston K, Koeller KJ, Harris GD, Jr, Rath NP, Bashkin JK, Wilson WD. Org Biomol Chem. 2014;12:7523. doi: 10.1039/c4ob01456a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hargrove AE, Raskatov JA, Meier JL, Montgomery DC, Dervan PB. J Med Chem. 2012;55:5425. doi: 10.1021/jm300380a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Nature. 1997;387:202. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 8.Ueda T, Kakunaga S, Ando M, Yonemori K, Sugiura H, Yamada K, Kawai A. Invest New Drugs. 2014;32:691. doi: 10.1007/s10637-014-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry JA, Le NM, Nguyen B, Howard CM, Bailey SL, Horick SM, Buchmueller KL, Kotecha M, Hochhauser D, Hartley JA, Wilson WD, Lee M. Biochemistry. 2004;43:12249. doi: 10.1021/bi048785z. [DOI] [PubMed] [Google Scholar]
- 10.Munde M, Kumar A, Peixoto P, Depauw S, Ismail MA, Farahat AA, Paul A, Say MV, David-Cordonnier MH, Boykin DW, Wilson WD. Biochemistry. 2014;53:1218. doi: 10.1021/bi401582t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez D, Chou CJ, Latella L, Zeitlin SG, Ku S, Puri PL, Dervan PB, Gottesfeld JM. Cell Cycle. 2006;5:1537. doi: 10.4161/cc.5.14.2913. [DOI] [PubMed] [Google Scholar]
- 12.Patil PC, Satam V, Lee M. Anticancer Agents Med Chem. 2014 Dec 16; Epub ahead of print. [Google Scholar]
- 13.Rizvi MA, Guru S, Naqvi T, Kumar M, Kumbhar N, Akhoon S, Banday S, Singh SK, Bhushan S, Mustafa PG, Shah BA. Bioorg Med Chem Lett. 2014;24:3440. doi: 10.1016/j.bmcl.2014.05.075. [DOI] [PubMed] [Google Scholar]
- 14.Khalaf AI, Waigh RD, Drummond AJ, Pringle B, McGroarty I, Skellern GG, Suckling CJ. J Med Chem. 2004;47:2133. doi: 10.1021/jm031089x. [DOI] [PubMed] [Google Scholar]
- 15.Khalaf AI, Bourdin C, Breen D, Donoghue G, Scott FJ, Suckling CJ, MacMillan D, Clements C, Fox K, Sekibo DAT. Eur J Med Chem. 2012;56:39. doi: 10.1016/j.ejmech.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Zhu W, Wang Y, Li K, Gao J, Huang C-H, Chen CC, Ko TP, Zhang Y, Guo RT, Oldfield E. J Med Chem. 2015;58:1215. doi: 10.1021/jm501449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lombardy RL, Tanious FA, Ramachandran K, Tidwell RR, Wilson WD. J Med Chem. 1996;39:1452. doi: 10.1021/jm9507946. [DOI] [PubMed] [Google Scholar]
- 18.Brendle JJ, Outlaw A, Kumar A, Boykin DW, Patrick DA, Tidwell RR, Werbovetz KA. Antimicrob Agents Chemother. 2002;46:797. doi: 10.1128/AAC.46.3.797-807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson WD, Tanious FA, Mathis A, Tevis D, Hall JE, Boykin DW. Biochimie. 2008;90:999. doi: 10.1016/j.biochi.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thuita JK, Wang MZ, Kagira JM, Denton CL, Paine MF, Mdachi RE, Murilla GA, Ching S, Boykin DW, Tidwell RR, Hall JE, Brun R. PLoS Negl Trop Dis. 2012;6:e1734. doi: 10.1371/journal.pntd.0001734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giordani F, Munde M, Wilson WD, Ismail MA, Kumar A, Boykin DW, Barrett MP. Antimicrob Agents Chemother. 2014;58:1793. doi: 10.1128/AAC.02024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boykin DW, Kumar A, Xiao G, Wilson WD, Bender BC, McCurdy DR, Hall JE, Tidwell RR. J Med Chem. 1998;41:124. doi: 10.1021/jm970570i. [DOI] [PubMed] [Google Scholar]
- 23.Thuita JK, Wang MZ, Kagira JM, Denton CL, Paine MF, Mdachi RE, Murilla GA, Ching S, Boykin DW, Tidwell RR, Hall JE, Brun R. PLoS Negl Trop Dis. 2012;6:e1734. doi: 10.1371/journal.pntd.0001734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paine MF, Wang MZ, Generaux CN, Boykin DW, Wilson WD, De Koning HP, Olson CA, Pohlig G, Burri C, Brun R, Murilla GA, Thuita JK, Barrett MP. Curr Opin Invest Drugs. 2010;11:876. [PubMed] [Google Scholar]
- 25.Cory M, Tidwell RR, Fairley TA. J Med Chem. 1992;35:431. doi: 10.1021/jm00081a003. [DOI] [PubMed] [Google Scholar]
- 26.Kuriakose S, Muleme HM, Onyilagha C, Singh R, Jia P, Uzonna JE. PLoS ONE. 2012;7:e48696. doi: 10.1371/journal.pone.0048696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheeler JR, Gull K, Gluenz E. BMC Biology. 2012;10:1. doi: 10.1186/1741-7007-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marini JC, Levene SD, Crothers DM, Englund PT. Proc Natl Acad Sci USA. 1982;79:7664. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro TA. Proc Natl Acad Sci USA. 1993;90:7809. doi: 10.1073/pnas.90.16.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klingbeil MM, Drew ME, Liu Y, Morris JC, Motyka SA, Saxowsky TT, Wang Z, Englund PT. Protist. 2001;152:255. doi: 10.1078/1434-4610-00066. [DOI] [PubMed] [Google Scholar]
- 31.Munde M, Wang S, Kumar A, Stephens CE, Farahat AA, Boykin DW, Wilson WD, Poon GM. Nucleic Acids Res. 2014;42:1379. doi: 10.1093/nar/gkt955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koehler NA. Curr Opin Chem Biol. 2010;14:331. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darnell JE., Jr Nat Rev Cancer. 2002;2:740. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Chai Y, Kumar A, Tidwell RR, Boykin DW, Wilson WD. J Am Chem Soc. 2012;134:5290. doi: 10.1021/ja211628j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chai Y, Paul A, Rettig M, Wilson WD, Boykin DW. J Org Chem. 2014;79:852. doi: 10.1021/jo402599s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul A, Chai Y, Boykin DW, Wilson WD. Biochemistry. 2015;54:577. doi: 10.1021/bi500989r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen B, Tanious FA, Wilson WD. Methods. 2007;42:150. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Wilson WD. Methods Mol Biol. 2012;613:1. doi: 10.1007/978-1-60327-418-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanjunda R, Munde M, Liu Y, Wilson WD, Wanunu M, Tor Y, editors. Chapter 4 CRC Press-Taylor & Francis Group; Boca Raton, FL: 2011. [Google Scholar]
- 40.Xuan J-C, Weber IT. Nucleic Acids Res. 1992;20:5457. doi: 10.1093/nar/20.20.5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laughlin S, Wang S, Kumar A, Boykin DW, Wilson WD. Anal Bioanal Chem. 2014;406:6441. doi: 10.1007/s00216-014-8044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laughlin S, Wang S, Kumar A, Farahat AA, Boykin DW, Wilson WD. Chem Eur J. 2015;21:5528. doi: 10.1002/chem.201406322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nordén B. Appl Spectrosc Rev. 1978;14:157. [Google Scholar]
- 44.Nordén B, Kubista M, Kurucsev TQ. Rev Biophys. 1992;25:51. doi: 10.1017/s0033583500004728. [DOI] [PubMed] [Google Scholar]
- 45.Wang S, Linde MH, Munde M, Carvalho VD, Wilson WD, Poon GM. J Biol Chem. 2014;289:21605. doi: 10.1074/jbc.M114.575340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.