Abstract
“Antigen surrogates” are synthetic, non-natural molecules that recognize the antigen-binding sites of antibodies. These molecules are of interest as replacements for native antigens as antibody “capture agents” in ELISA-like assays of potential diagnostic utility, for example when the antibody is indicative of a disease state. Antigen surrogates for disease-related antibodies can be mined from one-bead one-compound (OBOC) libraries by first denuding the library of ligands for antibodies present in the serum of control patients or animals, followed by screening the remainder of the library against serum from individuals with a particular disease of interest. Most of the work in this area has been done with peptoids (oligomers of N-alkylated glycine), which provide antibody ligands with only modest affinity and selectivity. Here, we explore the hypothesis that this is due to the “floppiness” of the peptoid backbone by creating libraries of peptoid-like molecules that have conformation-restricting structural elements inserted into their backbones. Indeed, we show here that these libraries can provide high affinity and selectivity antigen surrogates and that this much-improved binding is completely dependent on conformational restriction of the oligomer chain.
Keywords: Type 1 diabetes, NOD mouse, antigen surrogate, peptide tertiary amide, OBOC Library
There is great interest in the discovery of biomarkers for the early diagnosis of disease. A potentially attractive source of such markers is serum antibodies. Many investigators believe that the immune system reacts specifically to a variety of disease states, with the humoral arm of the system producing antibodies that recognize disease-specific molecules as “non-self” antigens.[1] The discovery of these putative disease-specific antibodies is non-trivial. Most approaches involve some type of screen for native antigens or close relatives thereof, for example by comparative screening of serum antibodies from case and control subjects using protein,[2] peptide,[3] lipid,[4] or glycan[5] arrays. More recently, next generation DNA sequencing of antibody-encoding genes or RNAs has been utilized to identify disease-specific antibodies.[6] Our laboratory has explored a different approach in which combinatorial libraries of synthetic, unnatural compounds are screened for “antigen surrogates” that recognize the antigen-binding site of disease-specific antibodies.[7] This process employs a so-called one bead one compound (OBOC) library comprised of hydrophilic beads that each display many copies of a single compound.[8] The beads are first incubated with control sera and ligands for antibodies in these samples are removed from the library (Fig. 1). The remainder of the compounds are then incubated with case sera and antibody ligands identified at this step are further evaluated as potentially interesting antigen surrogates.[7b] If validated, these compounds can serve as capture agents for the antibodies of interest in ELISA-like or similar assays.
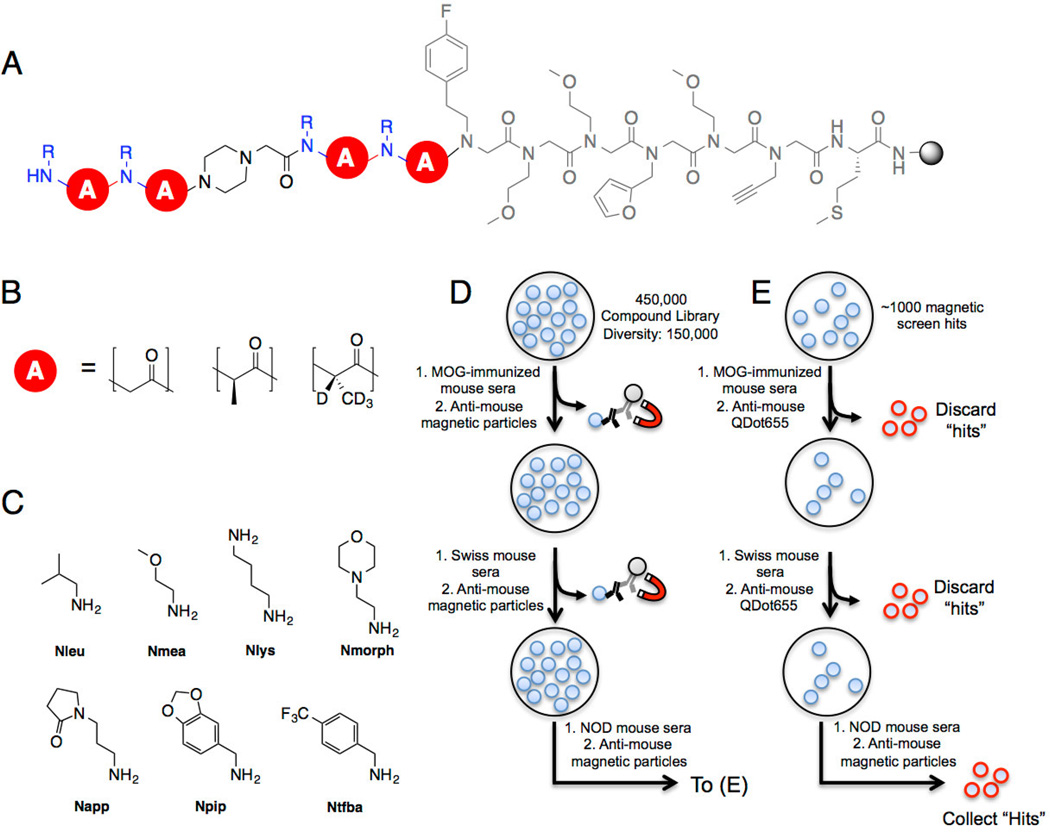
Figure 1. Conformationally constrained PTA-peptoid hybrid library composition and screening.
A) OBOC library design on 90 µm TentaGel macrobeads. The invariant linker is shown in gray. B) The library backbone (red circle) consists of achiral peptoid units or peptide tertiary amide (PTA) units. The R-PTA is deuterated to allow post-screen decoding of stereochemistry by tandem mass spectrometry. C) Seven primary amines were used to install the amide side chains at each position. MMT was used to protect diaminobutane because it is removed under mildly acidic conditions. The total diversity of the library is 150,000 compounds. D) Three copies of the library (450,000 compounds total) were screened against control mouse sera to remove ligands that bound uninteresting IgG or to invariant antibody structural elements. Beads that captured significant antibody were removed via magnetic pull-down after incubation with anti-mouse IgG-coated iron oxide particles. The non-binding beads were incubated with NOD mouse serum in blocking buffer and magnetic anti-mouse IgG and “hit” beads were isolated using a magnetic pull-down. E) The hits isolated from the magnetic screen were stripped of bound protein and incubated with control mouse serum and visualized using red quantum dot-conjugated anti-mouse IgG. Beads that displayed a red halo were removed manually. The remaining beads were incubated with NOD mouse serum and beads displaying a red halo were collected. After cleaving the ligand from each bead, the ligand identity was resolved using tandem MALDI-TOF MS/MS.
Most of the work in this area has employed libraries of peptoids (oligomers of N-alkylated glycine) because diverse libraries of these molecules are easy to construct[9] and peptoids are stable in serum.[10] Peptoid ligands for antibodies enriched in the sera of patients or animals with Alzheimer’s disease (AD) patients, Neuromyelitis Optica (NMO), multiple sclerosis (MS) and Type I diabetes (T1D) have been identified.[7, 11] Unfortunately, these peptoid ligands do not perform well enough to be used as clinical reagents. For human AD and murine T1D, when the peptoid ligands were tested in larger cohorts, significant numbers of false positives were identified, representing off-target binding to non-disease-linked antibodies.[12] Moreover, in most of these cases, the affinity of the peptoid for the antibody was weak. Thus, while peptoid libraries have been a useful vehicle to demonstrate proof of principle of the antigen surrogate approach[7b, 12], it is imperative to identify high affinity ligands for the disease-linked antibodies with little or no off-target binding if this approach is to be of clinical relevance.
Peptoids are quite “floppy” molecules, which likely limits their affinity for antibodies or other proteins and may contribute to off target binding. Thus, our hypothesis was that libraries of more conformationally constrained oligomers might prove to be a superior source of antigen surrogates. Over the last few years, we have developed a number of building blocks that can be employed in place of 2-bromoacetate in the “sub-monomer” synthetic route employed to make peptoids.[13] These new units[14] limit the conformational flexibility of the chain as well as increase its chemical diversity. When used in conjunction with diverse amines, large libraries of peptoid-like, but conformationally constrained, oligomers can be created readily. Here we demonstrate that high affinity ligands with excellent selectivity for antibodies associated with murine T1D[15] can indeed be isolated from libraries of conformationally constrained oligomers.
Results and Discussion
The library employed in this study is shown in Fig. 1A. In addition to 2-bromoacetic acid, both enantiomers of 2-bromopropionic acid were also included in the library.[14e] The presence of a chiral α-carbon together with N-substitution introduces significant conformational restriction due to allylic 1,3 strain effects.[14e] These units are called PTAs (peptide tertiary amides). Since the structures of screening hits must be determined by mass spectrometry, R-2-bromopriopionate and S-2-bromopriopionate-d4[14e] were employed in the synthesis. Because it is difficult to string together more than 2–3 PTA units in high yield due to the difficulty of acylating the hindered amine, a piperidine-containing spacer was inserted after the first two variable units. Another two variable units followed the spacer. Thus, the library contains all possible PTAs, peptoids and combinations thereof. The theoretical diversity is 150,000 compounds.
To remove compounds that recognized the antigen-binding sites of uninteresting antibodies, or the invariant surface of IgGs, the library was first exposed to sera from animals that do not have T1D. Specifically, three copies of the library were incubated with 5 mL of a 100 µg mL−1 (total protein) solution of pooled sera from mice immunized with the 35–55 fragment of myelin oligodendrocyte glycoprotein (MOG(35–55)), a common model for multiple sclerosis (MS)[16]. The beads containing ligands that bound to serum antibodies in these animals were removed from the library by subsequent incubation with an FeO-conjugated secondary antibody and exposure of the beads to a strong magnet.[17] A second control screen was then performed using 100 µg mL−1 of serum from healthy Swiss control mice to more thoroughly clear the library of uninteresting IgG ligands. Once again, compounds capable of binding antibody from these sera were removed magnetically after incubation with magnetic particles coated with anti-mouse IgG antibody.
The beads that did not bind IgG in these first two rounds of screening were then incubated with pooled NOD mouse serum (30 µg mL−1). Again, IgG-binding beads were isolated magnetically. Since the resulting beads numbered in the thousands, presumably due to nonspecific interactions between the magnetic particles and beads during the pull-down (Mendes et al., in preparation), all of the beads were stripped of protein under denaturing conditions and a second round of screening against control and NOD mouse serum was performed, but in this case, IgG-binding beads were visualized by subsequent treatment with secondary antibody conjugated to red QDot655 quantum dots. The beads displaying a bright halo were visualized under a low power fluorescence microscope and picked manually (Fig 1E). All of these compounds were released from the beads and sequenced by tandem MALDI mass spectrometry. Of these 70 final “hits”, only one compound was isolated more than once. The chemical structure of this compound, 1, is shown in Fig 2. Bead screening has a notoriously high false positive rate[18], but we have found that compounds isolated multiple times from a redundant library are almost always bona fide, high-quality ligands for the target protein.[18b] Thus, we focused solely on the single redundant hit, compound 1, in subsequent experiments.
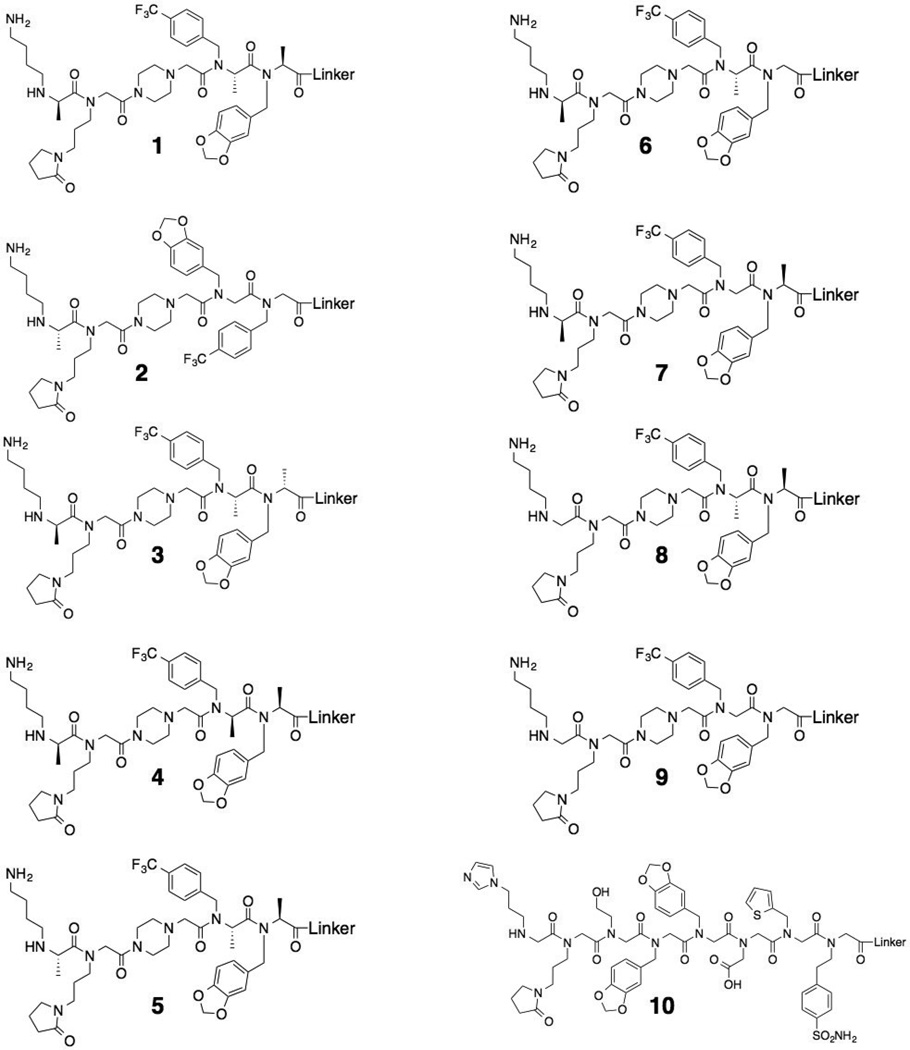
Figure 2. Chemical structures of the compounds used throughout this study.
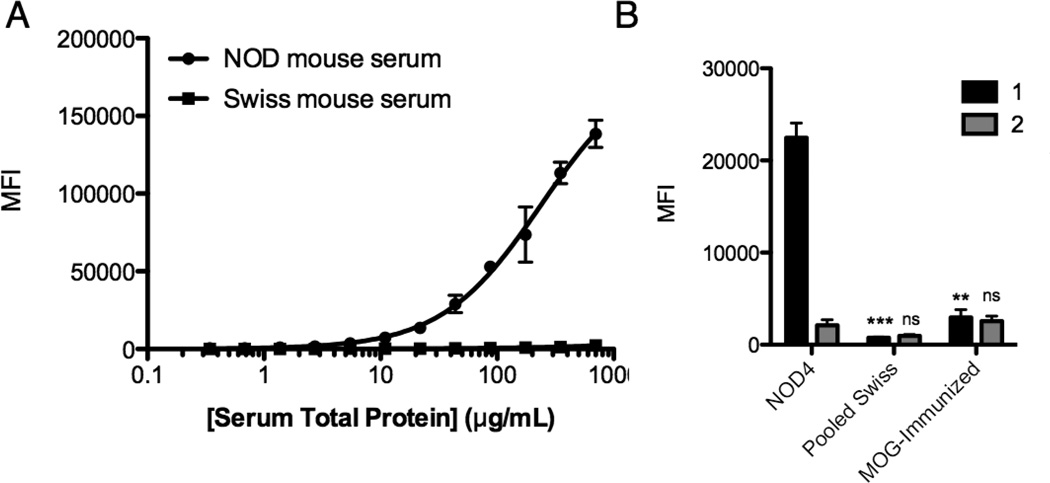
Compound 1 and a scrambled control compound 2 (Fig. 2) were re-synthesized and purified by HPLC. Binding of serum antibodies to these species was analyzed using a multiplexed assay in which the synthetic compound is affixed to the surface of a 10 µm TentaGel bead that is encoded with a particular ratio of blue and orange dyes coupled covalently to the central core of the bead. Multiple beads are incubated with a serum sample. After washing, a secondary antibody labeled with a red dye is added and, after another wash, the beads are analyzed using a flow cytometer, which registers the identity of the bead and quantifies the amount of antibody bound to it.[19] A serum titration revealed that at 500 µg mL−1 serum, 1 retained approximately 60-fold more antibodies from NOD serum than from the Swiss mouse serum tested (Fig. 3A). As shown in Fig. 3B, 2 did not bind significantly more IgG from NOD mouse serum than the Swiss mouse control serum sample. Neither compound interacted significantly with antibodies from the serum containing anti-MOG(35–55) antibodies. These results establish 1 as a genuine ligand for NOD mouse autoantibodies.
Figure 3. Characterization of screening hit 1 and control 2.
A) Serum titration plots generated by incubating a 1:2 serial dilution of NOD or Swiss mouse serum with 10 µm TentaGel beads displaying 1. Binding of serum antibodies was probed using a fluorescent anti-mouse IgG antibody and a flow cytometer to read the fluorescence intensity of each bead. B) Comparison of serum antibody binding to 1 and 2 using sera derived from NOD mice, Swiss mice or MOG(35–55)-immunized C57BL/6 mice at 500 µg mL−1 total protein concentration. In all cases, data are reported as the mean ± s.d. from three experiments. Statistical significance for each ligand was compared to the MFI when incubated with NOD mouse serum and determined using an unpaired t-test: **P < 0.01; ***P < 0.001; ns = not significant.
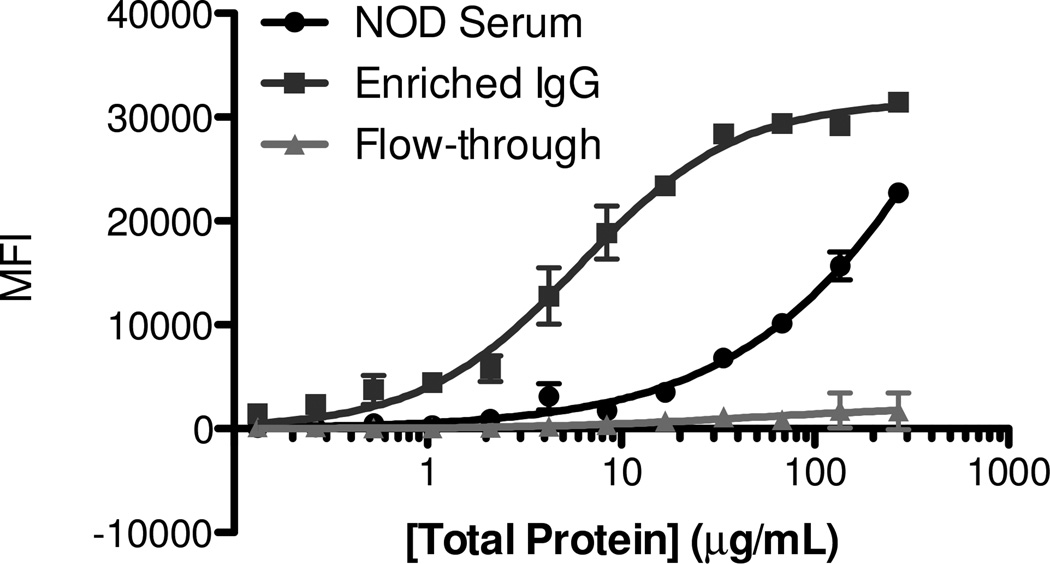
We next determined the “aggregate affinity” of 1 for NOD mouse antibodies that bind to it. Antibodies that recognize compound 1 were purified from serum on a sepharose affinity column displaying compound 1. The bound protein was eluted using a high ionic strength buffer. Since non-IgG serum proteins also eluted from the sepharose column, the IgG antibody concentration in the eluate was determined by Western blotting and comparing to a standard concentration curve obtained using pure IgG (Supplementary Figure S11 & S12). A titration was done with increasing concentrations of this enriched antibody preparation using the flow cytometry assay. The data indicate an “aggregate KD” of 2 ± 1 nM for the complex of compound 1 and the NOD mouse IgG antibodies (Fig. 4). The term aggregate KD is meant to indicate that the number that emerges from this type of a binding experiment reflects an aggregate of the different binding constants of each member of what is undoubtedly a polyclonal antibody population the small molecule. Another caveat is that this measurement is likely to reflect some level of avidity between the surface immobilized small molecule and the bivalent IgG antibodies. In general, we have observed that this effect enhances binding of the small molecule to the antibodies by a factor of 20 on this platform relative to the solution KD.[18b, 19] Therefore, we estimate that the intrinsic aggregate KD of the antibodies•1 complexes is approximately 40 nM.
Figure 4. Binding isotherms generated using affinity-purified antibody.
Antibody from NOD4 serum was purified over a column displaying 1, and binding of the IgG antibody to 1 was measured quantitatively using a flow cytometry-based assay. Beads were incubated with a 2-fold serial dilution of serum, starting from 270 µg mL−1. The eluted antibody fraction exhibited saturating binding to 1 with a Kd of 40 nM.
Our hypothesis going into this study was that the conformational bias provided by the PTA units would “stiffen” the backbone and provide higher affinity antibody binding. To probe this, derivatives of 1 were synthesized that inverted or eliminated the stereocenters, but retained the side chain functionality (see Fig. 2). All eight compounds were linked to the surface of encoded TentaGel beads. The different beads were combined and added to serially diluted NOD mouse serum. After hybridization of a red fluorescent secondary antibody, the beads were analyzed on a flow cytometer.
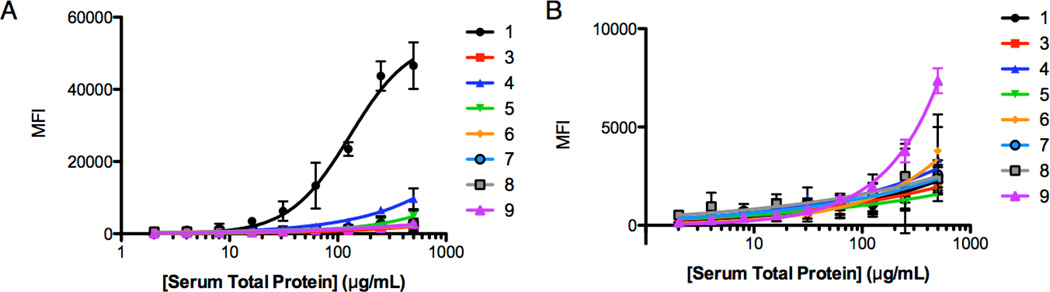
As shown in Fig. 5A, the parent compound 1 retained significantly more NOD mouse antibodies than any of the diastereomeric or des-methyl derivatives, showing clearly that the presence of the three conformationally-restraining chiral centers, and their absolute configuration, play a critical role in high affinity antibody binding.
Figure 5. Effect of stereochemistry on antibody binding.
A) NOD mouse serum titration curve for compounds 1, 3 and 4 generated using a multiplexed flow cytometry assay. B) Compounds 1, 3 and 4 were also tested for binding to Swiss mouse antibodies using the cytometry assay. Note the different axes on the two graphs. Structures of the compounds indicated are shown in Fig. 2.
For example, altering the absolute stereochemistry of any of the three chiral centers (compounds 3,4 and 5; Fig. 2) essentially abolished binding to the antibodies (Fig. 5A). The same was true for all of the des-methyl compounds examined, even when only a single chiral center was eliminated (Figs. 2 and 5A). These data are consistent with the idea that the conformational restriction afforded by the chiral centers in the backbone play a critical role in high affinity and selectivity binding.
It was also notable that when all of these compounds were tested for association with antibodies from healthy mice, which must represent off-target binding, the peptoid 9, lacking all three chiral centers, had the highest level of binding. While the diastereomers of 1 and the single des-methyl derivatives do not bind NOD mouse antibodies, they also have reduced off-target binding to antibodies in the serum of healthy mice relative to the peptoid 9. These data are typical of what we have observed with many different antibody-binding peptoids (unpublished data). These floppy molecules generally have a significantly greater propensity to engage in off-target interactions than the stiffer PTA-containing molecules.
To assess the antibody-binding properties of these molecules more broadly, serum antibody binding to 1 was measured using a large cohort of case and control samples. Serum was collected bi-weekly for seven months from 20 NOD mice, 18 Swiss control mice and 20 non-obese resistant (NOR) mice. NOR mice express disease-related autoantibodies but develop insulitis at a much lower frequency than NOD mice. Each serum sample was diluted to 500 µg mL−1 total protein in a blocking buffer and incubated with TentaGel beads displaying 1. After washing briefly, the beads were probed with fluorescent secondary antibody and the level of antibody binding to the beads was monitored by flow cytometry (Supplementary Figures S13–S15). Table 1 summarizes the mice that, at any blood draw, tested positive for 1-binding IgG antibodies. This criterion was employed because antibody titers rise and fall over time. “Positive” was defined as a level of antibody binding at least five standard deviations above the average level of antibody binding to 1 observed in the control mouse samples. 20% of NOD mice and 40% of NOR mice scored as positive, a modest diagnostic sensitivity. Importantly, none of the control mice provided a positive test (100% diagnostic specificity). The lack of significant binding to antibodies in any of the Swiss mouse serum samples demonstrates the high selectivity of this compound for T1D-related autoantibodies. Conversely, a peptoid ligand, 10, discovered in a previous screen,[11] exhibited off-target binding to antibodies in two Swiss mice, suggesting that conformationally rigid antigen surrogates are more effective diagnostic capture agents by reducing such off-target binding.
Table 1.
Swiss, NOD and NOR mice that express serum antibodies that bind to 1.
| Serum IgG binding to 1 by straina,b |
Serum IgG binding to 10 by straina,c |
|||||
|---|---|---|---|---|---|---|
| Mouse ID |
Swiss | NOD | NOR | Swiss | NOD | NOR |
| 1 | + | |||||
| 2 | + | |||||
| 3 | + | |||||
| 4 | + | + | ||||
| 5 | + | + | + | |||
| 6 | + | |||||
| 7 | ||||||
| 8 | + | + | + | + | ||
| 9 | + | |||||
| 10 | + | + | ||||
| 11 | + | |||||
| 12 | + | + | ||||
| 13 | + | |||||
| 14 | + | |||||
| 15 | + | + | ||||
| 16 | + | |||||
| 17 | + | |||||
| 18 | + | |||||
| 19 | + | + | ||||
| 20 | + | + | ||||
Serum concentration: 500 µg mL−1.
+ indicates that bead MFI was greater than an established five-sigma value.
+ indicates that bead MFI was greater than an established three-sigma value.
Conclusion
Collectively, these studies demonstrate that incorporating rigidifying elements, in this case chiral centers adjacent to N-alkylated amides, into OBOC libraries provides access to high affinity ligands, reducing the need for tedious post-screening affinity optimization efforts. In the context of an unbiased serological screen, a primary screening hit (1) rich in rigidifying PTA units was isolated that bound to T1D-linked antibodies with a Kd of 2 nM when immobilized on TentaGel beads, a value that probably corresponds to approximately 40 nM in solution, though the latter value could not be determined due to the limited amount of purified antibody available. Moreover, this PTA-containing oligomer failed to bind significant antibody from any of the control mice tested, in contrast to a peptoid antigen surrogate isolated as a primary screening hit in previous studies (Table 1). As evidence that the conformation restriction brought about by the chiral center in 1 contributed to its high binding affinity, inversion of a single Cα methyl substituent abolished binding altogether. We expect that screening libraries containing stiffer molecules will provide rapid access to high affinity ligands with enhanced specificity and hope that these “second generation antigen surrogates” will find utility as diagnostic capture agents to detect antibody biomarkers.
Materials and Methods
General Methods
Knorr Amide MBHA resin, Fmoc-protected amino acids, and peptide coupling reagents HBTU and HOBt were purchased from Advanced ChemTech, Oakwood, Anaspec or EMD Millipore. Primary amines used in peptoid synthesis were purchased from Sigma Aldrich, Tetra-Chem Industries, Acros Organics, or Oakwood Products. Solvents and other chemicals were purchased from Sigma-Aldrich, Acros or Fisher Scientific. These chemicals were used without further purification. Deuterated (S)-(—)-2-bromopropionic acid was prepared using the procedure described previously.[14e] All steps involving water used distilled water that was additionally filtered through a Barnstead™ Nanopure™ (Thermo Scientific). Phosphate-buffered saline (10 mM Na2HPO4, 1.8 mM K2HPO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4) was prepared by diluting a 10X stock solution (Corning). Tris-buffered saline (25 mM Tris, 0.15 M NaCl, 0.05% tween, pH 7.4) containing 0.05% tween 20 was prepared by diluting a 20X stock solution (Thermo Scientific). Mice were obtained from Jackson laboratories.
Animal studies
Animal experiments were performed under a protocol approved by the Scripps Florida Institutional Animal Care and Use Committee (IACUC). Blood was collected exclusively from female NOD/LtJ, NOR/LtJ, SWR/J, or C57BL/6 mice as previously described. 22, [20] To obtain serum containing anti-MOG(35–55) antibodies, 7–10 week old C57BL/6 mice were injected with mouse/rat MOG(35–55) peptide (sequence: MEVGWYRSPFSRVVHLYRNGK, Anaspec) conjugated to conjugated to mariculture keyhole limpet hemocyanin (mKLH) along with Titermax® Gold adjuvant (company, CITE us). Blood was collected 8 weeks post-immunization when titers of anti-MOG(35–55) were the highest, as determined using the SensoLyte® Anti-MOG(35–55) mouse/rat IgG Quantitative ELISA Kit (Anaspec).
Synthesis of a PTA-peptoid hybrid library
Library synthesis has been described previously.[18b] After synthesis, side chain protecting groups were removed by washing the library in dichloromethane (DCM) containing 1% trifluoroacetic acid (TFA) and 1% triisopropylsilane (TIS) at room temperature (10 × 5 mL). The deprotected library was washed in DCM (5 × 5 mL) and residual acid was neutralized by washing the in 10% diisopropylethylamine (DIEA) in DMF for 5 min. The beads were washed in DMF (5 × 5 mL) followed by water (10 × 10 mL) and the last water wash proceeded overnight at room temperature. The library was washed in phosphate-buffered saline (PBS) and tris-buffered saline containing 0.05% tween 20 (TBS-T) for 1 h. The library was pre-blocked in PBS StartingBlock™ (Thermo Scientific) for 1 h at 4 °C.
Screening for NOD mouse autoantibody ligands
Pooled sera taken from 8 week old MOG(35–55)-immunized mice was diluted to 100 µg mL−1 in 7 mL of a 1:1 mixture of StartingBlock™ (Thermo Scientific) in PBS, herein referred to as screening buffer. (Thermo Scientific). 7 mL of the diluted sera were added to ≈175 mg of the library and incubated at 4 °C overnight with gentle rotation. The beads were washed with tris-buffered saline containing 0.05% tween 20 (TBS-T, 5 × 10 mL). A magnetic pull-down procedure was used to remove beads containing compounds that bind IgG antibody, and the general pull-down technique was performed as follows. A 1:100 dilution of goat anti-mouse DynaBeads® (Life Technologies) was prepared in 10 mL of screening buffer. This suspension was added to the library and rotated at room temperature. After 1 h, the mixture was transferred to a 50 mL centrifuge tube, and diluted to 40 mL with TBS-T. Eight 62 mm3 N52-grade neodynium magnets were stacked into a cube, wrapped in parafilm and inserted directly into the 50 mL tube. A piece of string was used to suspend the magnet near the top of the tube. The tube was placed on a shaker with modest shaking for ten minutes, and non-magnetized beads were allowed to settle to the bottom. After all non-binders fell to the bottom of the tube, the magnet was removed and placed into a collection tube. The supernatant from the library was quickly transferred to a collection tube to isolate any beads that fell off the magnet when it was removed from the centrifuge tube. The parafilm was removed from the magnet to release beads from the magnetic field, and they were also washed into the collection tube. The remainder of the library was incubated with 7 mL of 100 µg mL−1 of pooled Swiss mouse serum at 4 °C overnight in screening buffer. After washing the library in TBS-T (5 × 10 mL), the library was suspended in a 1:100 dilution of goat anti-mouse antibodies conjugated to DynaBeads(R) for 1 h at room temperature. Compounds that retained significant IgG were removed using the magnetic pull-down method described above. The remaining library beads were then mixed with 5 mL of pooled NOD mouse sera diluted to 30 µg mL−1 in screening buffer. The library was rotated at 4 °C overnight and washed in TBS-T (5 × 10 mL). The magnetic pull-down technique was used to collect beads that bound NOD mouse antibodies. The “hit” beads that were isolated were transferred to a fritted syringe and stripped of bound protein by incubating with 1 mL of trypsin-EDTA (TE) (Life Technologies) for 30 min at 37 °C. The beads were washed with water (3 × 2 mL) and acetonitrile (2 × 2 mL) for 15 min at 37 °C. The beads were washed in a 50% aqueous acetonitrile solution (3 × 2 mL) for 30 min and washed in water (10 × 2 mL) at room temperature. The last water wash was continued overnight. After equilibrating the beads in TBS-T for 1 h and then in screening buffer for 1 h, the beads were incubated with 250 µL of 100 µg mL−1 MOG(35–55)-immunized mouse serum overnight at 4 °C on a rotator. Beads were washed in TBS-T (3 × 1 mL) and incubated with a 1:200 dilution of QDot® 655-conjugated goat F(ab’)2 anti-mouse secondary antibody in screening buffer for 1 h at room temperature on a rotator. The solution was washed with TBS-T (5 × 1 mL) and the beads were visualized on an Olympus IX51 inverted fluorescent microscope using a Prior Lumen200 Lamp. Beads that displayed a red halo were imaged using QImaging software and manually removed with a micropipette. The QDot655-assisted screening procedure was repeated using 250 µL of 100 µg mL−1 pooled Swiss mouse serum. Finally, the QDot655 procedure was repeated using 100 µg mL−1 pooled NOD mouse serum, and hits that were removed were stripped with trypsin and acetonitrile as described, separated individually into the wells of a polypropylene 96-well microtiter plate (Corning) and cleaved from the resin by suspending each bead in 20 µL of a 0.2 N HCl solution containing 50 mg mL−1 cyanogen bromide and reacting at room temperature for 2 h. The HCl solution was removed by evaporation and the residual solid was dissolved in 10 µL of a 40:60 mixture of acetonitrile/water containing 0.1% TFA. Sequence identity was determined using an Applied BioSciences 4800 MALDI-TOF MS/MS using a-cyano-4-hydroxycinnamic acid as matrix.
Synthesis of peptoids and PTA-peptoid hybrids
Compounds 1–5 were synthesized using a modified protocol from Gao and coworkers for PTA-peptoid synthesis.[14e] All compounds were synthesized on 100 mg of Knorr Amide resin (0.44 mmol/g). Fmoc-Cys(Trt)-OH (0.1 g, 0.176 mmol) was loaded onto swelled resin by pre-activating the acid with HBTU (0.066 g, 0.176) and HOBt (0.027 g, 0.176 mmol) and DIEA (0.063 mL, 0.352 mmol) in DMF and then adding it to the resin for 2 h with gentle shaking. The terminal Fmoc group was removed with two washes of 20% piperidine in DMF, the resin was washed in DMF (3 × 3 mL) and successive peptoid units were added using standard peptoid coupling procedures.[9, 18b] For each peptoid unit, 1 mL of 2 M bromoacetic acid (BAA) in DMF was added to the resin followed by 1 mL of 2.5 M diisopropylcarbodiimide (DIC) and the reaction syringe was shaken for 10 min at 37 °C. The resin was washed in DMF (3 × 3 mL) and a 1 M solution of primary amine (or piperazine) was added to the beads and incubated for 1 h at 37 °C. The resin was washed in DMF (3 × 3 mL). For PTA coupling steps, acetylation was performed as follows. (R)-2-bromopropionic acid ((R)-BPA) or (S)-2-bromoprionic acid ((S)-BPA) (20 µL (0.22 mmol) was dissolved in a solution of 20 mg mL−1 bis(trichloromethyl) carbonate (BTC, 0.022 g, 0.075 mmol) in anyhydrous tetrahydrofuran (THF) and pre-cooled to −20 °C in a freezer for 15 min. Meanwhile, the resin was washed consecutively with DMF (5 × 3 mL), dichloromethane (DCM, 5 × 3 mL) and anhydrous THF (5 × 3 mL). DIEA (61 µL, 0.35 mmol) was diluted into 1 mL anhydrous THF and added to the resin. 2,4,6-Trimethylpyridine (60 µL, 0.44 mmol) was added to the cooled BPA/BTC solution and quickly added to the resin slurry, vented, and shaken for 2 h at room temperature until the reaction was complete. The solution remained a pale yellow throughout the incubation period. After acylation was complete, the beads were washed (3 × 3 mL) in THF, DCM and DMF, respectively. A 1 M solution of the appropriate primary amine dissolved in anhydrous DMF was added to the resin and shaken overnight at 60 °C. The resin was washed with DMF (3 × 3 mL). Once the oligomer synthesis was complete, simultaneous deprotection of side chains and cleavage from the resin was accomplished using a 50:49 mixture of TFA/DCM with 1% TIS at 4 °C for 45 min. The solution was separated from the resin and the TFA/DCM was removed by evaporation. The crude compounds were precipitated with cold diethyl ether and pelleted by centrifugation. The pellet was resuspended in 40% aqueous acetonitrile and the compounds were purified on a Vydac reverse-phase C18 column (Grace) fitted onto a Waters 1525 binary high performance liquid chromatography (HPLC) pump equipped with a Waters 2487 Dual Wavelength Absorbance Detector. Ligand identity was confirmed by MALDI-TOF mass spectrometry on a 4800 Plus MALDI TOF/TOF Analyzer (Applied Biosystems) and α-cyano-4-hydroxycinnamic acid as a matrix. Purified compounds were lyophilized and stored at −20°C.
TentaGel bead binding assays
50 mg of 10 µm TentaGel microspheres (0.23 mmol/g) were encoded using Pacific Orange® and Pacific Blue® to create 16 fluorescently distinct populations using a reported protocol.[19] After dye encoding, the beads were left in black 0.5 mL microcentrifuge tubes and ligand immobilization to each population of bead was performed with slight modification from the reported protocol. The beads were washed in DMF (4 × 500 µL) and capped with 20% acetic anhydride for 10 min (no DIEA was added to this solution). The beads were washed (4 × 500 µL DMF), and Fmoc was deprotected in two washes of 20% piperidine for 10 min each. Following four DMF washes, [2-(2-(Fmoc-amino)ethoxy)ethoxy]acetic acid (5 eq) was coupled to the resin using 5 eq each of HBTU and HOBt and activated with DIEA (10 eq). After addition of the activated acid to the beads, the suspension was reacted overnight at room temperature on a rotator. The resin was washed in 20% piperidine in DMF (2 × 500 µL) to remove Fmoc protecting group. The deprotected terminal amine was activated by adding 150 µL 2 M bromoacetic acid and then 150 µL 2.5 M diisopropylcarbodiimide, mixed vigorously for ten minutes and washed with DMF (5 × 500 µL). The ligand was covalently attached via thioalkylation by dissolving the ligand in a 50:50 mixture of PBS:DMF (pH 7.4) to a final concentration of 2.5 mg mL−1 and adding it to the respective fluorescently-encoded bead population. The reaction was mixed overnight at 37 °C with frequent vortexing of the tubes. The beads were washed (3 × 500 µL DMF) and transferred to a MultiScreen™ Solvinert PTFE filter plate (EMD Millipore). The DMF was evacuated and the beads were washed with water (10 × 300 µL). The final water wash continued overnight. The beads were quenched with 150 mM β-mercaptoethanol diluted into PBS for 10 min, washed with PBS (10 × 300 µL), TBS-T (3 × 300 µL) and transferred to a 500 µL centrifuge tube. The suspension of beads was diluted to ~10 mg mL−1, blocked with PBS containing 0.5% BSA and 0.05% tween 20 and stored at 4 °C.
Serum Binding Assays
Binding experiments were performed by diluting serum to the desired concentration in 50 µL of PBS buffer containing 0.5% BSA and 0.05% tween 20 (herein referred to as blocking buffer) in a 0.5 mL microcentrifuge tube. To each serum sample was added 1 µL of a 10 mg mL−1 bead suspension. Binding experiments were performed with constant rotation overnight at 4 °C for titration experiments or for 2 h at room temperature for binding experiments performed for many samples at a single concentration. After incubation, the contents of the microcentrifuge tube were transferred to a 96-well filter plate and washed with TBS-T (2 × 300 µL). To each well of the filter plate was added 150 µL of a 1:200 dilution of Alexa Fluor 647-conjugated goat anti-mouse antibody (Life Technologies) in TBS-T containing 0.5% BSA. After mixing each well by aspiration with a micropipette, the secondary antibody was hybridized for 1 h at room temperature. The beads were washed (2 × 300 µL), taken up in 200 µL of TBS-T and analyzed on a BD™ LSRII flow cytometer (BD Biosciences). The mean fluorescence intensity of each encoded bead population was averaged across 3 independent experiments, and reported as the average MFI ± s.d. of the three runs. Where noted, a three-sigma threshold was established using the MFI from all Swiss mouse serum samples. NOD mouse serum samples that exhibited an MFI greater than the three-sigma threshold were scored as positive and all others as negative.
Affinity-purification of NOD mouse antibodies
Antibodies were enriched from serum by passing serum over an agarose SulfoLink® affinity column (Thermo Scientific) containing covalently immobilized 1. 2 mL of resin slurry was added to a 5 mL fritted syringe and evacuated by centrifugation and washed three times in 2 mL of coupling buffer (50 mM, 5 mM EDTA, pH 8.5). 2 mg of 1 dissolved in PBS was incubated with the column for 45 minutes at room temperature. After washing in PBS containing 1 M NaCl, unreacted iodoacetamide groups were quenched with a solution of 50 mM cysteine in coupling buffer and equilibrated into TBS. Approximately 50 µL of NOD mouse serum was diluted to 500 µL in Gentle Ag/Ab Binding Buffer (Thermo Scientific) and added to the affinity column. The column was rotated for 60 min at room temperature. The column was washed three times with 2 mL TBS and bound IgG was eluted under high salt conditions using Gentle Ag/Ab Elution Buffer pH 6.6 (Thermo Scientific). The IgG was dialyzed overnight in TBS and concentrated to ≈250 µg mL−1 total protein using a 50 kDa molecular weight cut-off spin filter (EMD Millipore). SDS-PAGE gels were stained with Pierce Silver Stain Kit for Mass Spectrometry (Thermo Scientific) and imaged using a ChemiDoc™ XRS+ System (Bio Rad), which also measured band intensities to generate standard curves.
Supplementary Material
Acknowledgements
This research was supported by a grant from the National Institutes of Health (1 DP3 DK094309-01)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson KS, LaBaer J. J. Proteome Res. 2005;4:1123–1133. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a Robinson WH, Steinman L, Utz PJ. Biotechniques. 2002 Dec.(Suppl):66–69. [PubMed] [Google Scholar]; b Robinson WH, Fontoura P, Lee BJ, de Vegvar HE, Tom J, Pedotti R, DiGennaro CD, Mitchell DJ, Fong D, Ho PP, Ruiz PJ, Maverakis E, Stevens DB, Bernard CC, Martin R, Kuchroo VK, van Noort JM, Genain CP, Amor S, Olsson T, Utz PJ, Garren H, Steinman L. Nat Biotechnol. 2003;21:1033–1039. doi: 10.1038/nbt859. [DOI] [PubMed] [Google Scholar]; c Fasolo J, Snyder M. Methods Mol Biol. 2009;548:209–222. doi: 10.1007/978-1-59745-540-4_12. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Gibson DS, Banha J, Penque D, Costa L, Conrads TP, Cahill DJ, O'Brien JK, Rooney ME. J Proteomics. 2010;73:1045–1060. doi: 10.1016/j.jprot.2009.11.013. [DOI] [PubMed] [Google Scholar]; e Lueking A, Possling A, Huber O, Beveridge A, Horn M, Eickoff H, Schuchardt J, Lehrach H, Cahill DJ. Mol. Cell. Proteomics. 2003;2:1342–1349. doi: 10.1074/mcp.T300001-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.a Restrepo L, Stafford P, Magee DM, Johnston SA. Ann Neurol. 2011;70:286–295. doi: 10.1002/ana.22405. [DOI] [PubMed] [Google Scholar]; b Robinson WH, DiGennaro C, Hueber W, Haab B, Kamachi M, Dean E, Fournel S, Fong D, Genovese MC, de Vegvar H, Steiner G, Hirschberg D, Muller S, Pruijn G, van Venrooij W, Smolen J, Brown P, Steinman L, Utz P. Nature Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 4.Kanter JL, Narayana S, Ho PP, Catz I, Warren KG, Sobel RA, Steinman L, Robinson WH. Nat Med. 2006;12:138–143. doi: 10.1038/nm1344. [DOI] [PubMed] [Google Scholar]
- 5.Wang D, Bhat R, Sobel RA, Huang W, Wang LX, Olsson T, Steinman L. Drug Dev Res. 2014;75:172–188. doi: 10.1002/ddr.21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a Tan YC, Kongpachith S, Blum LK, Ju CH, Lahey LJ, Lu DR, Cai X, Wagner CA, Lindstrom TM, Sokolove J, Robinson WH. Arthritis Rheumatol. 2014;66:2706–2715. doi: 10.1002/art.38754. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tan YC, Blum LK, Kongpachith S, Ju CH, Cai X, Lindstrom TM, Sokolove J, Robinson WH. Clin Immunol. 2014;151:55–65. doi: 10.1016/j.clim.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Lavinder JJ, Wine Y, Giesecke C, Ippolito GC, Horton AP, Lungu OI, Hoi KH, DeKosky BJ, Murrin EM, Wirth MM, Ellington AD, Dorner T, Marcotte EM, Boutz DR, Georgiou G. Proc Natl Acad Sci U S A. 2014;111:2259–2264. doi: 10.1073/pnas.1317793111. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O'Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR, Becker J, Benjamin B, Blakesley R, Bouffard G, Brooks S, Coleman H, Dekhtyar M, Gregory M, Guan X, Gupta J, Han J, Hargrove A, Ho SL, Johnson T, Legaspi R, Lovett S, Maduro Q, Masiello C, Maskeri B, McDowell J, Montemayor C, Mullikin J, Park M, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Thomas J, Thomas P, Vemulapalli M, Young A. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. Nature biotechnology. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Reddy MM, Wilson R, Wilson J, Connell S, Gocke A, Hynan L, German D, Kodadek T. Cell. 2011;144:132–142. doi: 10.1016/j.cell.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Raveendra B, Hao W, Baccala R, Reddy MM, Schilke J, Bennett JL, Theofiliopolous AN, Kodadek T. Chem. & biol. 2013;20:350–359. doi: 10.1016/j.chembiol.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 9.a Figliozzi GM, Goldsmith R, Ng SC, Banville SC, Zuckermann RN. Methods Enzymol. 1996;267:437–447. doi: 10.1016/s0076-6879(96)67027-x. [DOI] [PubMed] [Google Scholar]; b Alluri PG, Reddy MM, Bachhawat-Sikder K, Olivos HJ, Kodadek T. J. Am. Chem. Soc. 2003;125:13995–14004. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 10.Miller SM, Simon RJ, Ng S, Zuckermann RN, Kerr JM, Moos WH. Bioorg. Med. Chem. Lett. 1994;4:2657–2662. [Google Scholar]
- 11.Doran TM, Simanski S, Kodadek T. ACS Chem. Biol. 2015;10:401–412. doi: 10.1021/cb5007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kodadek T. Chem. & biol. 2014;21:1066–1074. doi: 10.1016/j.chembiol.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. J. Amer. Chem. Soc. 1992;114:10646–10647. [Google Scholar]
- 14.a Aditya A, Kodadek T. ACS Comb Sci. 2012;14:164–169. doi: 10.1021/co200195t. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Aquino C, Sarkar M, Chalmers MJ, Mendes K, Kodadek T, Micalizio G. Nature Chem. 2011;4:99–104. doi: 10.1038/nchem.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Suwal S, Kodadek T. Org. Biomol. Chem. 2013;11:2088–2092. doi: 10.1039/c3ob27476d. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Suwal S, Kodadek T. Organic & biomolecular chemistry. 2014;12:5831–5834. doi: 10.1039/c4ob00829d. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Gao Y, Kodadek T. Chem. & biol. 2013;20:360–369. doi: 10.1016/j.chembiol.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson MS, Bluestone JA. Annu Rev Immunol. 2005;23:447–485. doi: 10.1146/annurev.immunol.23.021704.115643. [DOI] [PubMed] [Google Scholar]
- 16.Racke MK. Curr Protoc Neurosci. 2001 doi: 10.1002/0471142301.ns0907s14. Chapter 9, Unit9 7. [DOI] [PubMed] [Google Scholar]
- 17.Astle JM, Simpson LS, Huang Y, Reddy MM, Wilson R, Connell S, Wilson J, Kodadek T. Chem. & biol. 2010;17:38–45. doi: 10.1016/j.chembiol.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a Lian W, Upadhyaya P, Rhodes CA, Liu Y, Pei D. J Am Chem Soc. 2013;135:11990–11995. doi: 10.1021/ja405106u. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Doran TM, Gao Y, Mendes K, Dean S, Simanski S, Kodadek T. ACS Comb. Sci. 2014;16:259–270. doi: 10.1021/co500030f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doran TM, Kodadek T. ACS Chem. Biol. 2014;9:339–346. doi: 10.1021/cb400806r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golde WT, Gollobin P, Rodriguez LL. Lab Anim. (NY) 2005;34:39–43. doi: 10.1038/laban1005-39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.