Abstract
Regulation of protein abundance is a critical aspect of cellular function, organism development, and aging. Alternative splicing may give rise to multiple possible proteoforms of gene products where the abundance of each proteoform is independently regulated. Understanding how the abundances of these distinct gene products change is essential to understanding the underlying mechanisms of many biological processes. Bottom-up proteomics mass spectrometry techniques may be used to estimate protein abundance indirectly by sequencing and quantifying peptides that are later mapped to proteins based on sequence. However, quantifying the abundance of distinct gene products is routinely confounded by peptides that map to multiple possible proteoforms. In this work, we describe a technique that may be used to help mitigate the effects of confounding ambiguous peptides and multiple proteoforms when quantifying proteins. We have applied this technique to visualize the distribution of distinct gene products for the whole proteome across 11 developmental stages of the model organism Caenorhabditis elegans. The result is a large multidimensional dataset for which web-based tools were developed for visualizing how translated gene products change during development and identifying possible proteoforms. The underlying instrument raw files and tandem mass spectra may also be downloaded. The data resource is freely available on the web at http://www.yeastrc.org/wormpes/.
Keywords: Proteoform, Proteomics, Visualization, Database, Caenorhabditis elegans, Development, Protein separation, SDS-PAGE
Graphical abstract
Introduction
Bottom-up shotgun proteomics is a widely used technique for identifying peptides and, indirectly by inference, proteins present in biological samples. Broad adoption of this technique was facilitated by the advent of SEQUEST [1] (and the availability of new genome sequences), which greatly streamlined the interpretation of tandem mass spectra. By searching spectra against a list of candidate peptides taken from a database of possible protein sequences, SEQUEST provided an unprecedented ability to quickly and easily identify proteins present in a protein mixture.
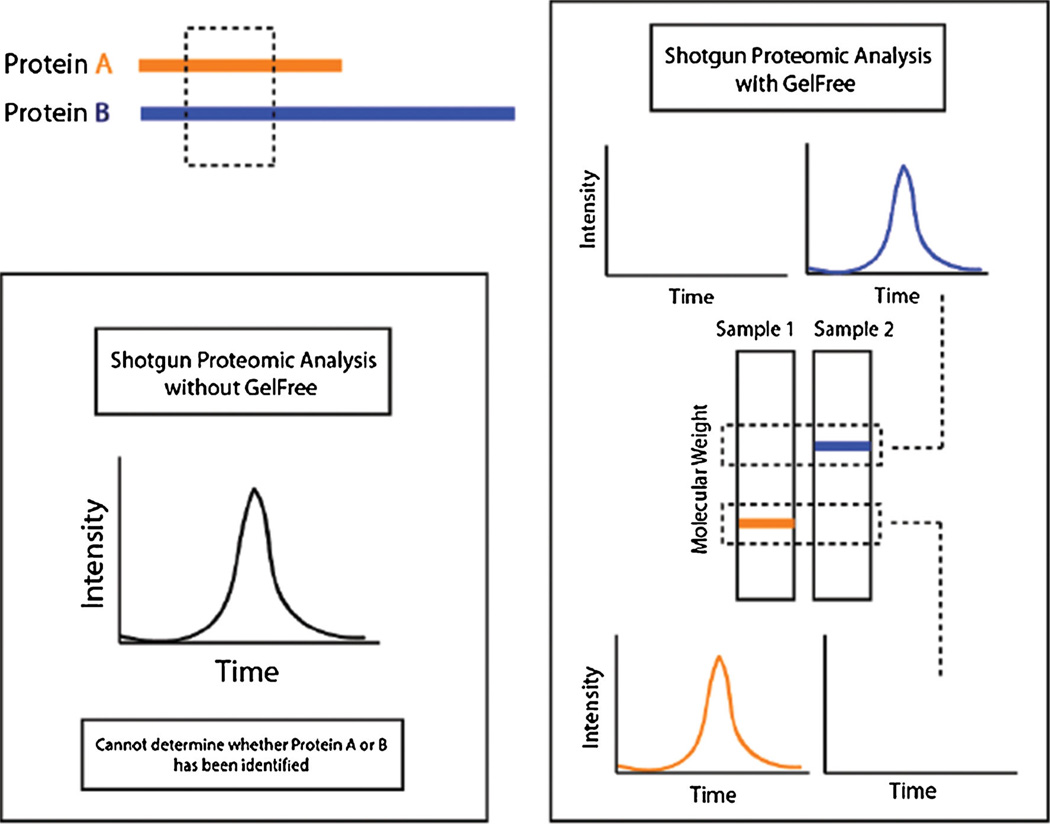
However, matching spectra to sequences present in a database, by its very nature, has practical considerations that may complicate the interpretation of the data in a biological context. In samples from complex proteomes, identified peptides commonly match multiple gene products or proteoforms that may be present in the sequence database, and choosing which gene products or proteoforms are represented by an ambiguous peptide may not be possible (Figure 1, left panel). This is a particular issue when attempting to identify distinct proteoforms such as those resulting from alternative splicing or a post-translational modification because any peptide mapping to one variant is very likely to match others.
Figure 1.
A depiction of how the Gelfree mass fractionation helps mitigate the confounding effects of ambiguous peptides. The left panel illustrates how discerning between proteins is not possible given an ambiguous peptide in a sample containing both proteins. The right panel illustrates how separating the proteins by mass using Gelfree prior to analysis helps eliminate the ambiguity by ensuring that the sampled peptide can only be from a protein from the respective mass range
Increasingly, proteomics studies are focusing not only on the identification of proteins but also on the differences in the proteome between biological samples. Multiple techniques have been developed to quantify proteins in bottom-up shotgun proteomics experiments—largely encompassed by methods that require introduction of internal reference standards (such as SILAC [2], ITRAQ [3], and ICAT [4]), and so-called “label-free” methods that do not (such as spectral counting). Spectral counting, which uses a metric based simply on the number of observations for all peptides mapping to a given protein in an experiment, is a widely-used and computationally inexpensive technique for comparing differences between samples [5–9]. However, the problem of ambiguous peptides is compounded when attempting to quantify distinct gene products or proteoforms using spectral counting. Given peptides, not proteins, are being measured, and given no clear way to determine which proteoforms containing that peptide are contributing spectrum counts for that peptide, how can one reliably estimate the presence of each of those proteoforms using this method?
A technique that assigns these ambiguous peptides to distinct gene products or proteoforms using bottom-up proteomics was developed and applied as part of the modENCODE project [10], which aimed to fill in the gaps in the genome annotation for Caenorhabditis elegans. This technique uses Gelfree fractionation [11] to separate the endogenous proteins in a sample by mass before analysis by mass spectrometry so that identified peptides that map to multiple gene products with distinct masses may be attributed specifically to the gene product with the correct mass for the fraction (Figure 1, right panel). For the modENCODE study, this technique was applied separately to whole proteomes of 11 distinct developmental stages of Caenorhabditis elegans, resulting in a rich, multidimensional dataset that could conceivably be used to not only confirm the presence of distinct gene products or proteoforms but also to estimate and compare quantities of those gene products or proteoforms between developmental stages using spectral counting.
Given the complexity of the data, tools designed to help interpret the SEQUEST results in a biologically meaningful context are essential for efficient discovery and proteogenomic analysis. To this end, we constructed a database and web application that allow searching, visualizing, and downloading the data. Spectral counting-based analysis was performed, and the web application provides tools for identifying distinct proteoforms and interrogating how the quantities of those proteoforms may change with respect to developmental stage. The web site and all raw data are freely available at http://www.yeastrc.org/wormpes/.
Methods
Sample Preparation and Mass Spectrometry Analysis
Eleven developmental stages of C. elegans were analyzed—N2 embryo, N2 L1, N2 L2, N2 L3, N2 L4, N2 YA, N2 dauer, spe-9L4, spe-9 YA, spe-9 adult, and him-8. Each developmental stage was grown on agar plates at 20°C seeded with the NA22 strain of E. coli. [12], sucrose floated, lysed in the presence of protease inhibitors (Roche Diagnostics, Indianapolis, IN, USA) and centrifuged to separate insoluble and soluble fractions. A 200 µg soluble lysate of each developmental stage was reduced with 5 mM DTT (Sigma, St. Louis, MO) in 30 uL Gelfree sample buffer (125 mM Tris, 4% SDS, 0.025% bromophenol blue, pH 7) and vortexed and heated to 50°C for 10min. The samples were then cooled to room temperature, alkylated with 15 mM IAA (Sigma) and incubated at room temperature in the dark for 10min. The samples were separated into 15 molecular weight fractions ranging from3.5 to 500 kDa using the Gelfree 8100 fractionation system (Protein Discovery/Expedeon). Twelve fractions were collected from the mid-range Gelfree cartridge (3.5–100 kDa) and three fractions were collected from the high-range Gelfree cartridge (3.5–500 kDa).
Approximate molecular weight range based on visualization of SDS-PAGE of fractions with molecular weight marker:
fraction 1 (3.5–15 kD)
fraction 2 (13–17 kD)
fraction 3 (15–20 kD)
fraction 4 (15–25 kD)
fraction 5 (17–30 kD)
fraction 6 (23–35 kD)
fraction 7 (30–42 kD)
fraction 8 (35–50 kD)
fraction 9 (40–57 kD)
fraction 10 (50–57 kD)
fraction 11 (55–77 kD)
fraction 12 (70–100 kD)
fraction 15 (120–200 kD)
fraction 16 (190–250 kD)
Each fraction was trypsin (Promega, Madison, WI) digested. SDS was removed with SDS removal columns (Pierce, Rockville, Il, USA) and salts were removed with MCX columns (Waters, Milford, MA, USA). The peptides from each fraction were analyzed using a 35 cm fused silica 75 µm column and a 4 cm fused silica Kasil1 (PQ Corporation, Malvern, PA, USA) frit trap loaded with Jupiter C12 reverse phase resin (Phenomenex, Torrance, CA, USA)with a 120-min LC-MS/MS run on a Thermo LTQ-Orbitrap Velos mass spectrometer coupled with an Eksigent nanoLC 2D. A biological and analytical replicate was performed for each sample.
Accurate masses were assigned using Bullseye [13] and peptides were identified using SEQUEST searched against a FASTA protein sequence database comprising Wormbase wormpep (WS229) [14], RNA-seq-based predictions [10, 15], and gene predictions and translated C. briggsae intergenic ORFs as described in Merrihew et al. [16]. P-values and q-values were assigned to PSMs and peptides on a per-fraction basis using Percolator [17].
To guard against the effective increase in false discovery rate (FDR) associated with combining multiple datasets that are each filtered on q-value, we calculated a single q-value for each distinct peptide in the dataset that is meant to be the minimum false discovery rate at which we may confidently consider the peptide to be present in the whole dataset. We ranked all the target and decoy PSMs by P-value from every run together as calculated by Percolator in their respective MS/MS runs, eliminated all but the top-scoring PSM for each distinct peptide, and used the decoys as an empirical null for the targets. Specifically, we computed a decoy-based P-value for each target peptide (i.e., the ratio of decoys that score better than the target score), and then converted the resulting P-values to q-values using qvality [18]. Only peptides with a q-value ≤0.01 using this method were considered for spectral counting.
Normalized Spectrum Count (NSC)
Calculating NSC
We used a normalized spectrum count (NSC) as a measure of the protein signal. To calculate the NSC, we first calculated the ratio of all PSMs attributable to a protein (NSCratio) by dividing the number of PSMs for that protein (Sp) by the total number of PSMs for all proteins in that condition (St). That is:
NSCratio will typically be a very small decimal. For example, in a condition with 20,000 PSMs with 10 attributable to a protein of interest, NSCratio would be 5E-4. Comparing changes between very small decimals may not be intuitive to end users. To aid in interpreting the data, we converted the NSCratio into an integer that preserves the fold change between different NSCratio values between comparable conditions. This was done by dividing the NSCratio calculated for all proteins in each separate comparable condition by the minimum NSCratio found for all proteins across all comparable conditions (NSCmin ratio) and rounding to the nearest integer:
So, given an NSCratio for a protein in three conditions of 5E-9, 4E-6, and 2E-7 and a NSCmin ratio of 1E-9, the NSC would be calculated as 5, 4000, and 200, respectively.
NSC was calculated for all proteins separately for each developmental stage, such that the abundances may be compared between developmental stages. To calculate the NSCratio for a protein for a developmental stage, Sp is the sum total of PSMs for that protein across all fractions (including all replicates) and St is the sum total of all PSMs for all proteins across all fractions (including all replicates). Then, to calculate NSC, all NSCratio values are divided by NSCmin ratio, which is the minimum NSCratio calculated for all proteins across all developmental stages. (Only peptides with a whole-dataset q-value≤ 0.01 and PSMs with a q-value≤0.01 as calculated by the Percolator algorithm were considered).
The same method was used to compute NSC values for proteins for individual mass fractions. NSCratio was calculated where Sp is the sum total of PSMs for that protein in that mass fraction across all developmental stages, and St is the sum total of PSMs for all proteins in that fraction across all developmental stages. NSC was then calculated using an NSCmin ratio that was the minimum NSCratio calculated for all proteins across all fractions.
To compare spectrum counts between combinations of developmental stage and mass fraction, NSCratio was calculated where Sp was the sum total of PSMs for a protein using all replicate runs of that specific developmental stage and mass fraction, and St was the sum total of PSMs for all proteins in those runs. NSC was then calculated using an NSCmin ratio that was the minimum NSCratio calculated for all proteins across all possible combinations of developmental stage and mass fraction.
Considerations for NSC
It is important to note that we are not performing any quantitative comparisons. We are only using NSC values to make qualitative comparisons of the same protein between samples. Properties of proteins, such as protein length or performance of tryptic peptides specific to a protein in the mass spectrometer, may have significant effects on spectrum counts for a given protein that are independent of the amount of protein. The NSAF score [5] was developed to account for protein length by dividing the spectrum count for each protein by the protein’s length to calculate a spectrum abundance factor (SAF), then dividing this SAF by the sum of the SAF calculated for all other proteins in the run to arrive at a normalized SAF (NSAF). However, NSAF ignores the variable peptide performance resulting from different possible tryptic peptides between separate proteins. Additionally, we were not wholly confident in the true sequence lengths of the detected proteins as we may be unknowingly detecting alternate splice variants and proteoforms that are posttranslationally modified. Given these two factors, we chose to exclude protein length from the calculation of NSC to avoid the implication that NSC values may be legitimately compared between separate proteins.
An inherent limitation in most (if not all) methods that use spectral counting is that deviation in conditions (or experimental design) between compared samples may introduce inherent biases for classes of proteins that are not a function of the biology as much as they are a function of the methods themselves (e.g., biases that enrich for size or hydrophobicity). These biases may invalidate comparison between samples by sufficiently altering the likelihood of sampling a particular protein (and thus its spectral counts) based solely on non-meaningful attributes of that protein. In this dataset, we use NSC to compare gene products across developmental stages and across separate mass fractions. While comparing spectrum counts across developmental stages should not be subject to these artificial biases, comparing spectrum counts across separate mass fractions from the Gelfree separation may have biases in terms of the complement of expected proteins in the fraction, and so may impact the likelihood of sampling a given protein. When comparing directly between mass fractions, users should not consider the NSC a direct comparison of abundance between those fractions but rather a crude proxy of how enriched the individual fractions are for the protein of interest.
Web Site and Database Implementation
A relational database was designed (schema available upon request) and implemented using the MySQL (http://www.mysql.com/) relational database management system (RDBMS). Code was written using Java (http://www.java.com/) to process the data files resulting from the mass spectrometry data analysis and populate the database. A web application was developed using Java, HTML, CSS, and Javascript on the Apache Tomcat (http://tomcat.apache.org/) Java servlet container and the Struts application framework (http://struts.apache.org/). The database and web application are run on Intel-based servers running Red Hat Enterprise Linux (RHEL) 6.4 (http://www.redhat.com/).
Blast [19] (blastp: 2.2.25+) was installed on multiple RHEL servers to support user-driven searching of the dataset by sequence. The FASTA file used to search the MS/MS data was used to build the Blast sequence database. A Jobcenter [20] client module for executing Blast was developed and installed on the Blast servers and linked to an in-house installation of Jobcenter to support distributed execution of user-driven Blast requests from the web application.
Results and Discussion
The dataset comprises 698 MS/MS runs from which 4,732,473 PSMs were identified (individual q-value≤0.01) for 39,563 distinct peptides (whole-dataset q-value≤0.01) mapping to 28,740 protein sequences from the FASTA file used to search the data. Of the 39,563 peptides, 8725 map uniquely to a single protein sequence, and of the 39,563 peptides, 2748 do not map to any protein found in Wormbase, but map to 1273 protein sequences that are the result of RNA-seq or computational prediction (see the “Methods” section). Given the large, multidimensional nature of the data (each run being a biological or technical replicate of a combination of developmental stage and mass fraction), a database and web-based interface were constructed to collate the data, help find proteins of interest, visualize how abundances of those proteins (and their possible proteoforms) may change as a function of developmental stage, and view the underlying, supporting mass spectrometry data.
Searching for Proteins
Users may search for proteins by using query strings (such as common name, accession string, or keyword) or by protein sequence using Blastp. Searching using query string effectively limits the possible results to those proteins found in Wormbase because those are the only annotated proteins in the dataset. However, many proteins in the dataset are the result of RNA-seq or computational prediction and have no commonly known names or annotations. To solve this, a system for searching by sequence with Blastp was set up (see the “Methods” section) and a novel interface for visualizing Blast results was constructed that colors hits based on confidence and clusters the search results based on where they physically map to the query sequence. This approach will tend to cluster matching proteoforms together as easily distinguishable groups and aid users in interpreting the results and selecting possible proteins of interest. From either search method, users may click on the names of proteins to visualize comparative protein abundance and proteomics data associated with that protein.
Visualizing NSC Abundance
Three tools were developed to visualize the distribution of proteoforms across fraction and condition—NSC bar chart, which provides a one-dimensional view for comparing NSC protein values as a function of developmental stage or mass fraction (Figure 2); Protein Heat Map, which provides a two-dimensional view for comparing NSC protein values as a function of developmental stage and mass fraction (Figure 3); and Peptide Coverage Heat Map, which visualizes how the detection of particular peptides in a protein changes as a function of developmental stage or mass fraction (Figure 4).
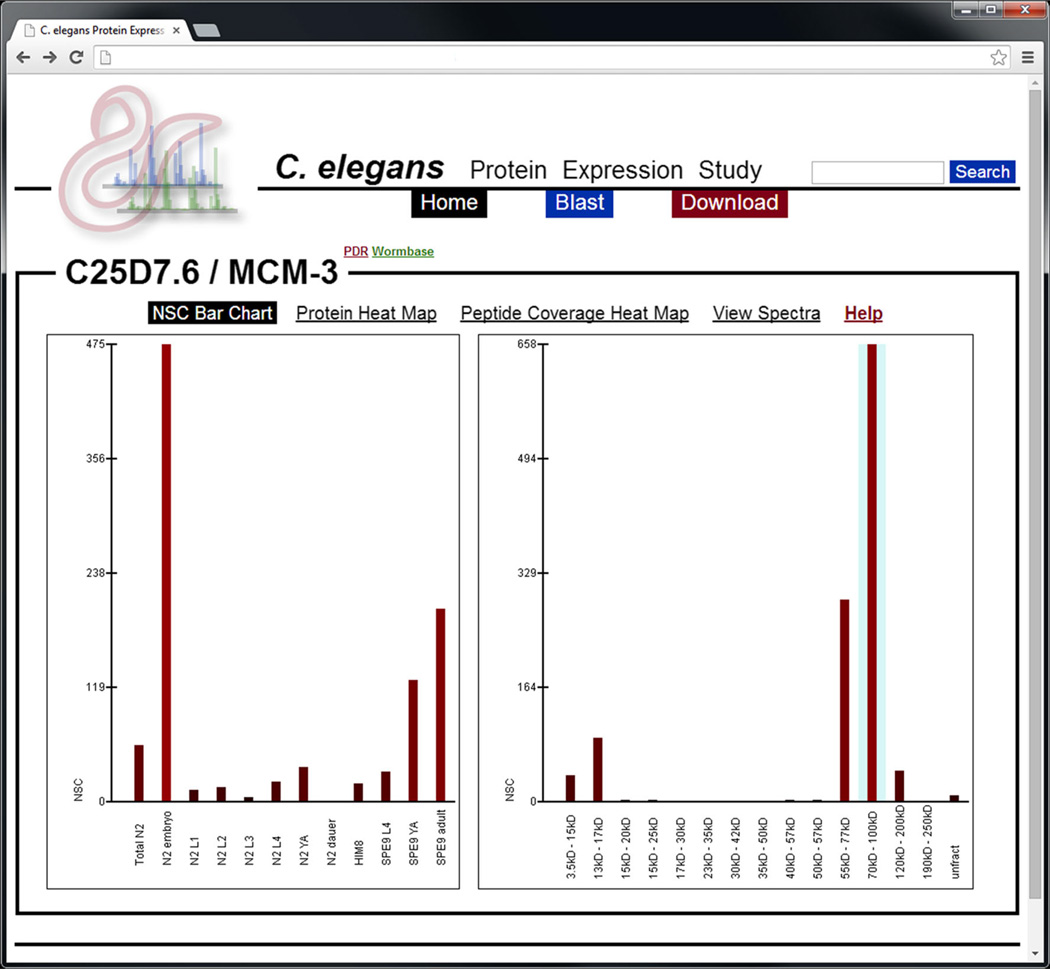
Figure 2.
A screenshot depicting the NSC bar chart for the MCM-3 protein—a protein known to affect embryonic viability [22]. The y-axis is NSC and the x-axis is either developmental stage (left panel) or mass fraction (right panel). The blue shaded area in the right-hand graph indicates the expected mass fraction for this protein. Bars may be clicked on to view peptides, PSMs, and spectra associated with those spectral counts. In this example, NSC is highest in the embryonic developmental stage and highest in the expected mass fraction
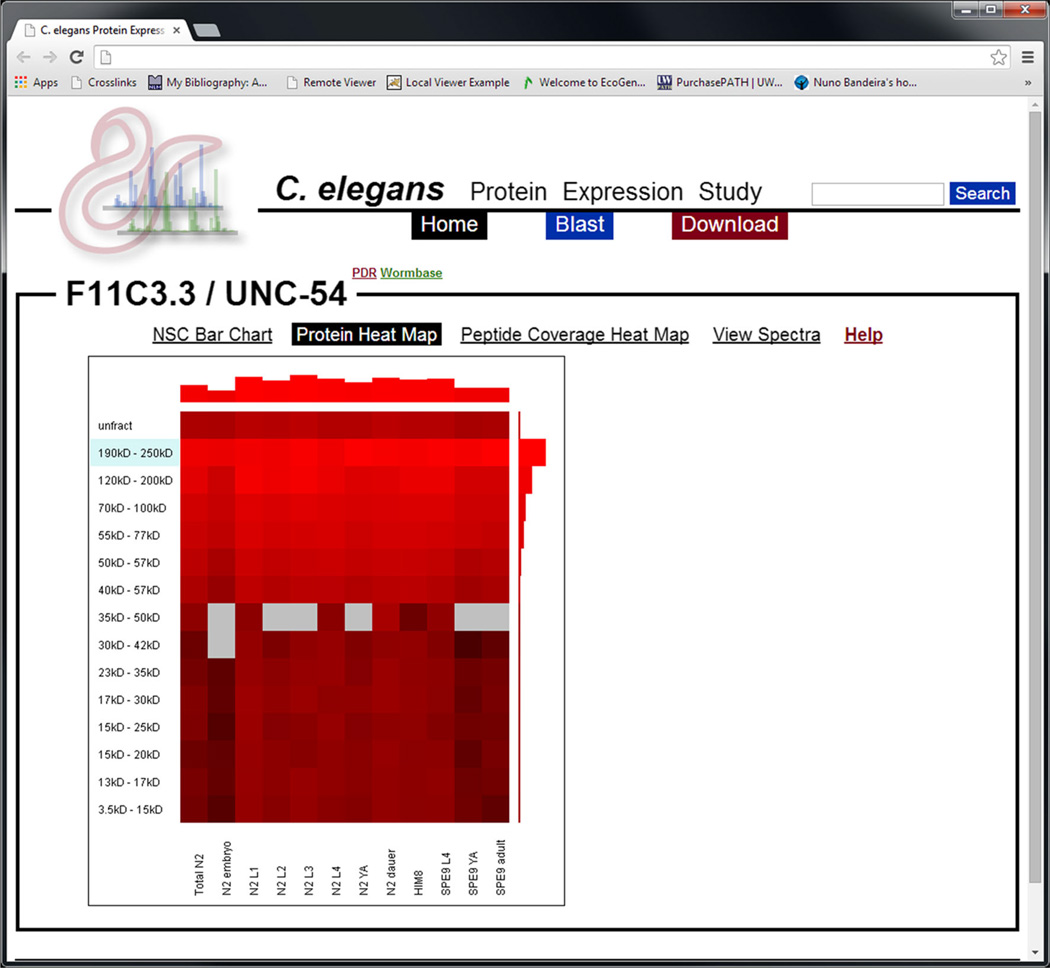
Figure 3.
A screenshot depicting the protein heat map forUNC-54, the primary myosin heavy chain found in C. elegans. The y-axis is mass fraction and the x-axis is developmental stage. Brighter red indicates higher relative protein abundance as measured in NSC. Grey regions indicate that no PSMs were observed in that mass fraction/developmental stage combination. The blue-shaded mass fraction indicates the expected mass fraction for the protein. Each box may be clicked on to view peptides, PSMs, and spectra associated with those spectral counts. The bar graph at the top and right-hand side indicates the total abundance of the respective developmental stage or mass fraction
Figure 4.
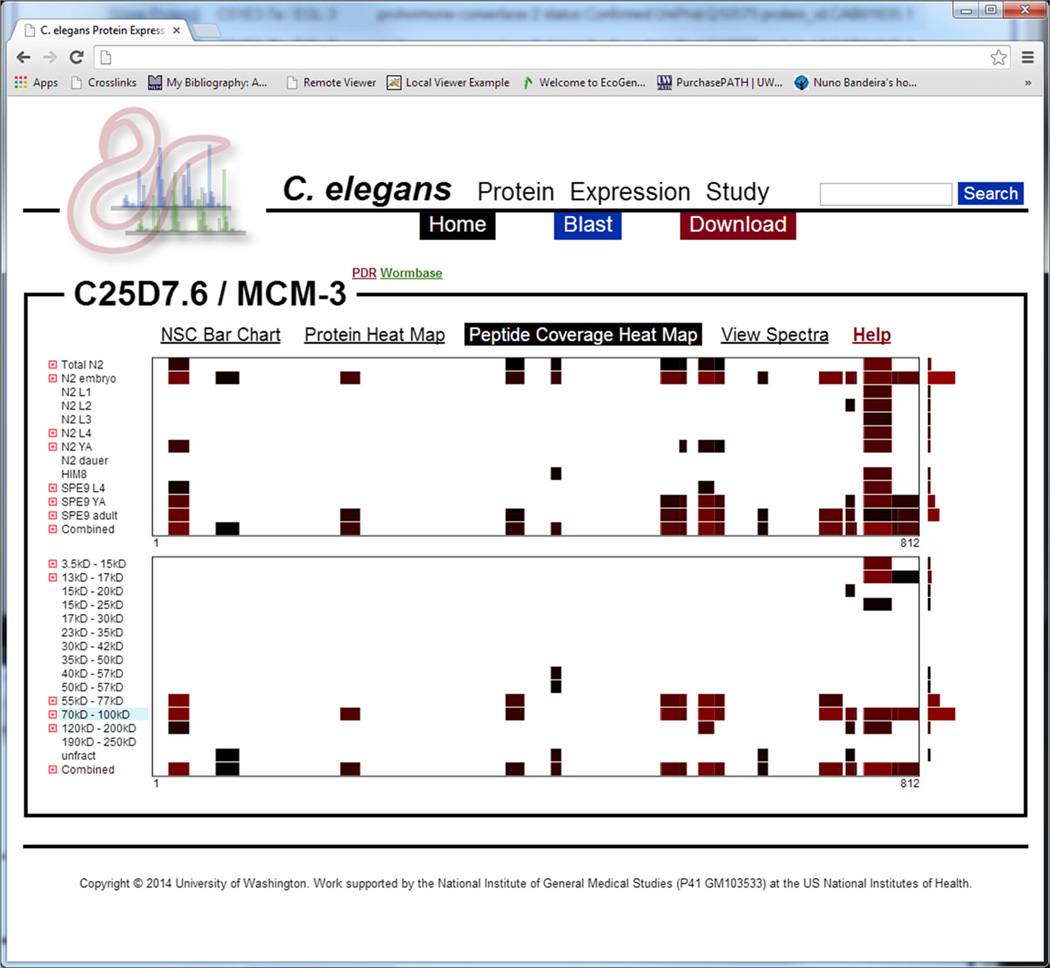
A screenshot depicting the peptide coverage heat map for the MCM-3 protein—a protein known to affect embryonic viability. The x-axis represents the protein’s sequence laid out left-to-right from N- to C-terminus. The y-axis in the top graphic represents developmental stage and the y-axis in the bottom graphic represents mass fraction. The blue-shaded fraction represents the expected mass fraction for this protein. Each colored segment represents an area of sequence coverage specific to the respective developmental stage or mass fraction, and the color indicates the abundance of that observed peptide in NSC (brighter red indicates higher abundance). The bar-graph to the right of each section indicates the total abundance of protein for its respective row
NSC Bar Chart
The NSC bar chart makes use of a simple bar graph to compare NSC signal by showing how the total NSC of all peptides that map to a given protein change with respect to developmental stage. However, some peptides may map (by sequence) to multiple proteoforms and if other proteoforms are present, it is not simple to determine which (if any) of the peptides that map to the current protein were detected as a result of the presence of one or more of the other proteoforms. To help determine if (and to what degree) confounding proteins may be present, a bar graph comparing NSC between mass fractions is also presented that shows whether or not PSMs for peptides mapping to the current protein were detected in mass fractions other than the expected mass fraction for this protein’s calculated mass (expected fraction is shaded blue). Detection of peptides in other fractions may indicate the presence of proteoforms (previously known or unknown), protein degradation products, or that the accepted protein sequence is incorrect. In the case of signal present only in the expected mass fraction, caution should still be used as multiple proteoforms of a protein may have similar masses that cannot be distinguished by mass fraction.
Hovering the mouse pointer over any of the bars will show the raw and normalized spectrum counts being represented. The bars may be clicked on to view the peptides, PSMs, and spectra associated with those spectral counts. Each PSM is annotated with both the developmental stage and mass fraction in which it was observed in order to further interrogate the presence and effects of possible proteoforms.
Protein Heat Map
The protein heat map visualizes protein NSC with respect to both developmental stage and mass fraction simultaneously and is designed to further interrogate the presence and character of possible proteoforms—and help mitigate the effects of those proteoforms when interpreting NSC. With the heat map it is not only possible to see in which mass fractions peptides mapping to a given protein were detected but also how the NSC in each of those mass fractions is different with respect to developmental stage. In the heat map, brighter red represents a higher NSC and grey represents the lack of detected PSMs for that developmental stage/mass fraction combination. Red boxes outside the expected mass fraction may indicate the presence of peptides also matching to proteoforms. Differences between mass fractions in the pattern of NSC with respect to developmental stage may additionally suggest the presence of proteoforms whose abundances are differentially regulated with respect to developmental stage. Additionally, the confounding effects of multiple proteoforms may be mitigated somewhat by examining only the pattern of NSC in the expected mass fraction for the protein of interest.
Red squares in the heat map may be hovered over with the mouse pointer to view the raw and normalized spectral counts, and red squares may be clicked on to view peptides, PSMs, and spectra found for the specific developmental stage/mass fraction combination. A bar graph is present at the top and right side of the heat map that represents the total NSC for each developmental stage and mass fraction, respectively. Each bar may also be hovered over to view spectral counts and clicked to view peptides, PSMs, and spectra.
Peptide Coverage Heat Map
The peptide coverage heat map attempts to provide still further insight into proteoforms by providing a visual comparison of individual peptides that map to a given protein as a function of developmental stage or biochemical fraction. This view uses the Mason viewer [21] to lay out the protein sequence coverage as a row by drawing rectangles along the horizontal axis (where the left and right edges are the N- and C-termini) that represent which segments of the protein are covered by identified peptides. The colors of the rectangles are shades of red, such that brighter red indicates a higher NSC. The software then stacks the rows vertically using the same scale so that patterns of sequence coverage may be easily compared between different stages or fractions. Where multiple peptides overlap and map to the same position in the protein, the cumulative NSC for peptides mapping to a given protein position are used to determine shading. In this case, distinct peptides may also be viewed by expanding a developmental stage or mass fraction by clicking the icon to the left of the row label.
Using this view, it is simple to see how patterns of protein coverage change between stages or fractions. Differences in this pattern may be the result of detecting proteoforms with overlapping peptides and provide some insight into the sequence composition of those proteoforms. It is also possible to review which peptides are contributing most significantly to the spectral count for a given protein, and in which mass fractions those specific peptides are most significantly represented.
All segments of protein coverage may be hovered over with the mouse pointer to view position in the protein, raw spectrum count, and NSC. Where peptides overlap, a row for a given stage or fraction may be expanded to view individual peptides. Individual peptides may be clicked on to view sequence, PSMs, and spectra associated with that peptide.
Application to a Biological Example
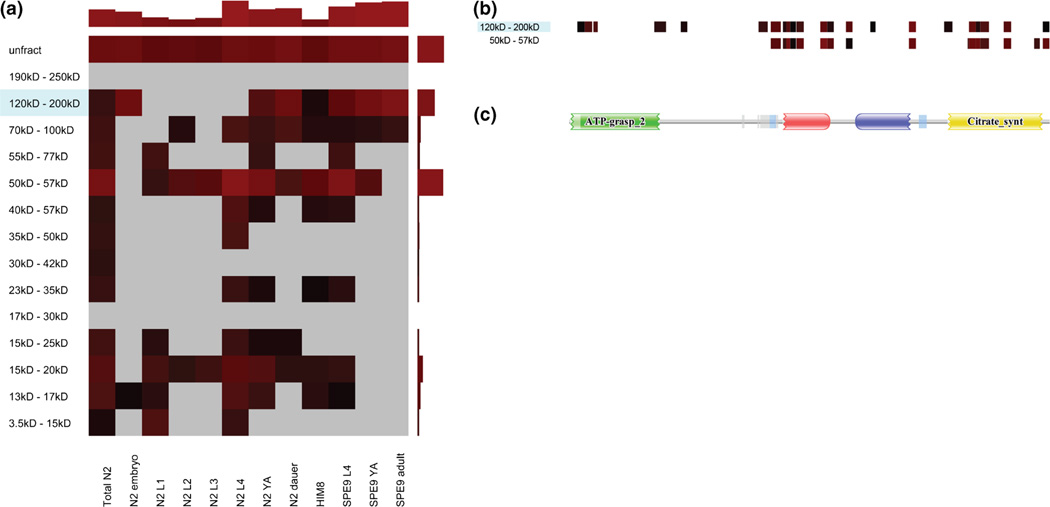
As an illustration of how these views may be applied to proteogenomic analysis, we provide an example in Figure 5 that suggests a possible, unknown proteoform of a specific ATP-citrate synthase (D1005.1) that may be differently expressed in different developmental stages. The protein heat map shows that peptides mapping to this protein are found in distinct mass fractions, and peptides mapping to those respective fractions are represented in different developmental stages (Figure 5a). Additionally, the peptide coverage heat map suggests that the proteoform in the lighter mass fraction may be missing the N-terminus of the protein (Figure 5b), which corresponds to a known domain in the protein (Figure 5c). Although not definitive, these data suggest that further biological characterization of the gene products from D1005.1 may be warranted.
Figure 5.
An example protein heat map and peptide coverage heat map for D1005.1 (a probable ATP-citrate synthase), which may have two possible proteoforms. (a) The protein heat map for the protein D1005.1. D1005.1 has an estimated molecular weight of 121.6 kD, and the gene coding for D1005.1 has no known splice variants (according to WormBase). The NSC for this protein is relatively high in its expected mass fraction (indicated by blue shading); however, it is higher in the 50–57 kD mass fraction. This may indicate some highly sampled peptides that map to D1005.1 also map to another proteoform with a lower mass. Also of note is that no PSMs were found in the lower mass fraction for the embryo developmental stage, whereas a NSC of 108 was calculated for D1005.1 in the higher mass fraction for the same developmental stage. Alternatively, no PSMs were found for D1005.1 in the N2L4 developmental stage in the higher mass fraction, whereas a NSC of 277 was calculated for this stage in the lower mass fraction. This indicates the possibility that each of the two proteoforms is regulated differently with regard to developmental stage. (b) The peptide coverage heat map for D1005.1 for the 50–57 kD and 120–200 kD mass fractions, which shows where the peptides found in the respective mass fractions map to the protein sequence. The lower mass fraction is missing N-terminal peptides found in the higher mass fraction. (c) A domain image generated by PFAM [23] for D1005.1. The missing N-terminal peptides largely correspond to a predicted ATP grasp domain
Viewing Underlying MS/MS Data
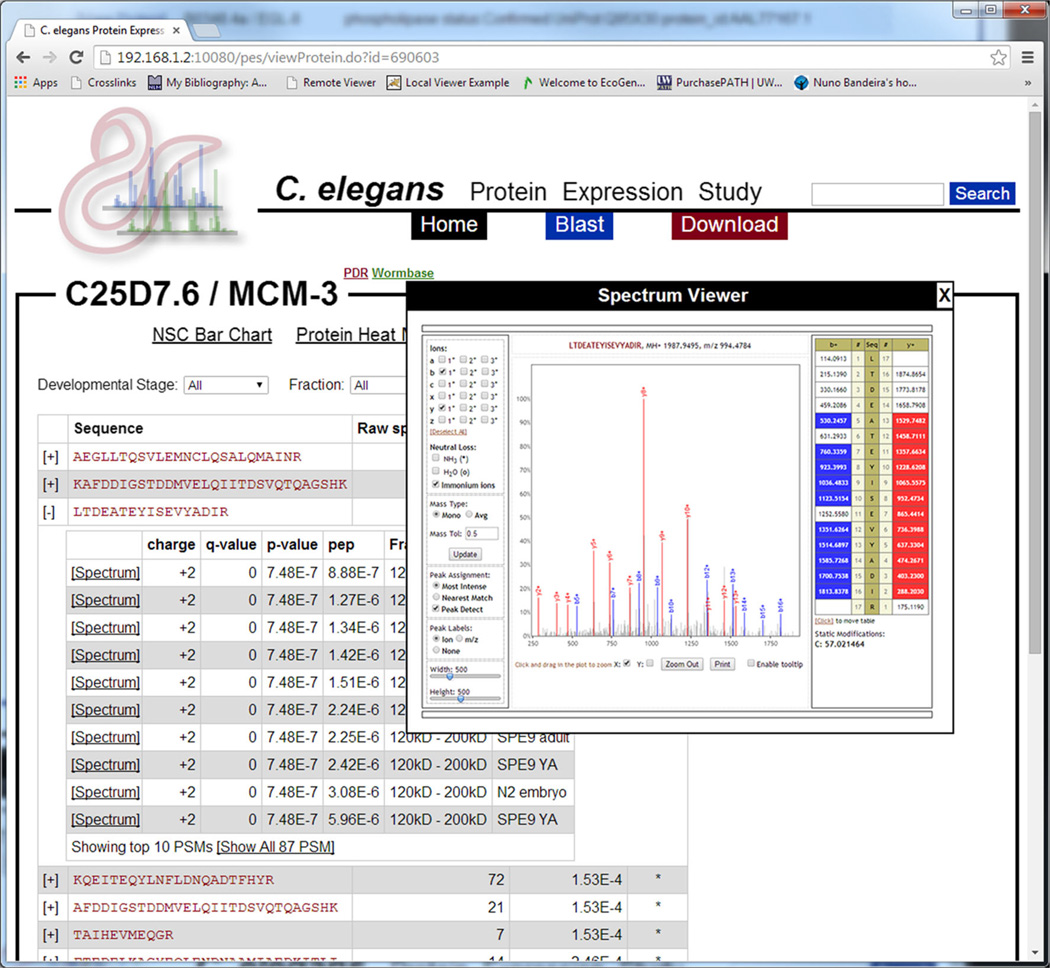
As previously stated, the underlying MS/MS data (peptide sequences, PSMs, and spectra) are available from all data visualization pages (Figure 6). Additionally, users may click the “View Spectra” tab to view a list of all peptides identified that mapped to the current protein. For each peptide, users may view all PSMs as well as in which developmental stage and mass fraction those PSMs were identified. For each PSM, users may view the underlying MS/MS spectrum using the built-in Lorikeet spectrum viewer (https://code.google.com/p/lorikeet/). Additionally, the list of peptides may be filtered by developmental stage, mass fraction, or both.
Figure 6.
A screenshot illustrating the view of the underlying proteomics data in the resource. For a given protein, all peptides that mapped to that protein are listed in the order of peptide q-value. The sequence, raw spectrum count, q-value, and whether or not that peptide uniquely maps to this protein are presented. Each peptide’s row may be expanded to view the underlying PSMs in order of q-value. For each PSM, the charge, q-value, P-value, posterior error probability, mass fraction, and developmental stage are listed. The spectra associated with each PSM may also be viewed using the built-in Lorikeet spectrum viewer. Additionally, the list of peptides may be filtered by developmental stage, fraction, or both, using the form at the top of the page
Conclusions
We have presented a web application and data resource designed to search, visualize, and interpret data generated by SEQUEST when applied to multiple mass fractions from multiple developmental stages of C. elegans. The application has been designed to not only illustrate how proteins may change between developmental stages but also to deduce whether proteoforms are present, the character of those proteoforms, and how they may be affecting the estimation of abundance for a given protein. The web application is freely accessible at http://www.yeastrc.org/wormpes/. All the instrument raw files and minimally-processed MS/MS data are available for download at the site.
Acknowledgments
The authors acknowledge support for this work by grants P41 GM103533, R01 DK069386, and U01 HG004263 from the National Institutes of Health, and the University of Washington Proteomics Resource (UWPR95794).
References
- 1.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 2.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 6.Vogel C, Marcotte EM. Label-free protein quantitation using weighted spectral counting. Methods Mol. Biol. 2012;893:321–341. doi: 10.1007/978-1-61779-885-6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harshman SW, Canella A, Ciarlariello PD, Rocci A, Agarwal K, Smith EM, Talabere T, Efebera YA, Hofmeister CC, Benson DM, Jr, Paulaitis ME, Freitas MA, Pichiorri F. Characterization of multiple myeloma vesicles by label-free relative quantitation. Proteomics. 2013;13:3013–3029. doi: 10.1002/pmic.201300142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodiger A, Agne B, Baerenfaller K, Baginsky S. Arabidopsis proteomics: a simple and standardizable workflow for quantitative proteome characterization. Methods Mol. Biol. 2014;1072:275–288. doi: 10.1007/978-1-62703-631-3_20. [DOI] [PubMed] [Google Scholar]
- 9.de Wit M, Kant H, Piersma SR, Pham TV, Mongera S, van Berkel MP, Boven E, Pontén F, Meijer GA, Jimenez CR, Fijneman RJ. Colorectal cancer candidate biomarkers identified by tissue secretome proteome profiling. J. Proteome. 2014;99:26–39. doi: 10.1016/j.jprot.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, Alves P, Chateigner A, Perry M, Morris M, Auerbach RK, Feng X, Leng J, Vielle A, Niu W, Rhrissorrakrai K, Agarwal A, Alexander RP, Barber G, Brdlik CM, Brennan J, Brouillet JJ, Carr A, Cheung MS, Clawson H, Contrino S, Dannenberg LO, Dernburg AF, Desai A, Dick L, Dosé AC, Du J, Egelhofer T, Ercan S, Euskirchen G, Ewing B, Feingold EA, Gassmann R, Good PJ, Green P, Gullier F, Gutwein M, Guyer MS, Habegger L, Han T, Henikoff JG, Henz SR, Hinrichs A, Holster H, Hyman T, Iniguez AL, Janette J, Jensen M, Kato M, Kent WJ, Kephart E, Khivansara V, Khurana E, Kim JK, Kolasinska-Zwierz P, Lai EC, Latorre I, Leahey A, Lewis S, Lloyd P, Lochovsky L, Lowdon RF, Lubling Y, Lyne R, MacCoss M, Mackowiak SD, Mangone M, McKay S, Mecenas D, Merrihew G, Miller DM, 3rd, Muroyama A, Murray JI, Ooi SL, Pham H, Phippen T, Preston EA, Rajewsky N, Rätsch G, Rosenbaum H, Rozowsky J, Rutherford K, Ruzanov P, Sarov M, Sasidharan R, Sboner A, Scheid P, Segal E, Shin H, Shou C, Slack FJ, Slightam C, Smith R, Spencer WC, Stinson EO, Taing S, Takasaki T, Vafeados D, Voronina K, Wang G, Washington NL, Whittle CM, Wu B, Yan KK, Zeller G, Zha Z, Zhong M, Zhou X, modENCODE Consortium. Ahringer J, Strome S, Gunsalus KC, Micklem G, Liu XS, Reinke V, Kim SK, Hillier LW, Henikoff S, Piano F, Snyder M, Stein L, Lieb JD, Waterston RH. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran JC, Doucette AA. Multiplexed size separation of intact proteins in solution phase for mass spectrometry. Anal. Chem. 2009;81:6201–6209. doi: 10.1021/ac900729r. [DOI] [PubMed] [Google Scholar]
- 12.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh EJ, Hoopmann MR, MacLean B, MacCoss MJ. Comparison of database search strategies for high precursor mass accuracy MS/MS data. J. Proteome Res. 2010;9:1138–1143. doi: 10.1021/pr900816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein L, Sternberg P, Durbin R, Thierry-Mieg J, Spieth J. WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 2001;29:82–86. doi: 10.1093/nar/29.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hillier LW, Reinke V, Green P, Hirst M, Marra MA, Waterston RH. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res. 2009;19:657–666. doi: 10.1101/gr.088112.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merrihew GE, Davis C, Ewing B, Williams G, Kall L, Frewen BE, Noble WS, Green P, Thomas JH, MacCoss MJ. Use of shotgun proteomics for the identification, confirmation, and correction of C. elegans gene annotations. Genome Res. 2008;18:1660–1669. doi: 10.1101/gr.077644.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 18.Kall L, Storey JD, Noble WS. QVALITY: nonparametric estimation of q-values and posterior error probabilities. Bioinformatics. 2009;25:964–966. doi: 10.1093/bioinformatics/btp021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Jaschob D, Riffle M. JobCenter: an open source, cross-platform, and distributed job queue management system optimized for scalability and versatility. Source Code Biol. Med. 2012;7:8. doi: 10.1186/1751-0473-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaschob D, Davis TN, Riffle M. Mason: a JavaScript web site widget for visualizing and comparing annotated features in nucleotide or protein sequences. BMC Res. Notes. 2015;8:70. doi: 10.1186/s13104-015-1009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piano F, Schetter AJ, Morton DG, Gunsalus KC, Reinke V, Kim SK, Kemphues KJ. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 2002;12:1959–1964. doi: 10.1016/s0960-9822(02)01301-5. [DOI] [PubMed] [Google Scholar]
- 23.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]