SUMMARY
Let-7 microRNAs (miRNAs) are critical regulators of animal development, stem cell differentiation, glucose metabolism, and tumorigenesis. Mammalian genomes contain twelve let-7 isoforms that suppress expression of a common set of target mRNAs. LIN28 proteins selectively block let-7 biogenesis in undifferentiated cells and in cancer. The current model for coordinate let-7 repression involves the LIN28 cold shock domain (CSD) binding the terminal loop and the two CCHC-type zinc fingers recognizing a GGAG sequence motif in precursor let-7 (pre-let-7) RNAs. Here we perform a systematic analysis of all let-7 miRNAs and find that a single let-7 family member, human let-7a-3 (and its murine ortholog let-7c-2), escapes LIN28-mediated regulation. Mechanistically, we find that the pre-let-7c-2 loop precludes LIN28A binding and regulation. These findings refine the current model of let-7 regulation by LIN28 proteins and have important implications for understanding the LIN28/let-7 axis in development and disease.
Keywords: microRNA, let-7a-3, let-7c-2, LIN28A, LIN28B, embryonic stem cells, cancer
Graphical abstract
INTRODUCTION
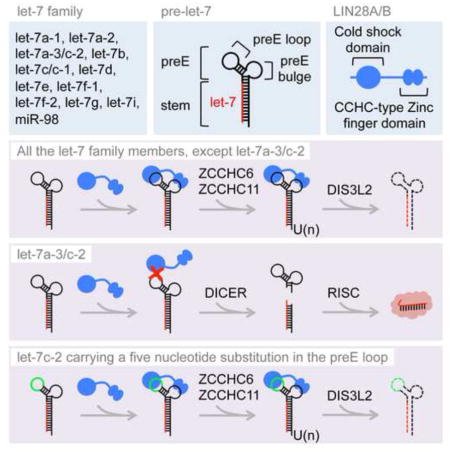
Let-7 is one of the most highly conserved microRNA (miRNAs) in animals(Pasquinelli et al., 2000; Reinhart et al., 2000). The human let-7 family comprises twelve members that are expressed from eight different loci (let-7a-1, let-7a-2, let-7a-3, let-7b, let-7c, let-7d, let-7e, let-7f-1, let-7f-2, let-7g, let-7i, miR-98)(Roush and Slack, 2008). Each member is embedded in a let-7 primary miRNA hairpin (let-7 pri-miRNA or pri-let-7) that is processed by the DROSHA-DGCR8 containing Microprocessor complex(Denli et al., 2004; Gregory et al., 2004). This processing generates 67–80 nucleotide (nt) long let-7 precursors miRNA (let-7 pre-miRNAs or pre-let-7) that can be classified into two groups: group I let-7 pre-miRNAs (let-7a-2, let-7c, let-7e) that are directly processed by the Dicer complex in the cytoplasm; and group II let-7 pre-miRNAs (all the other let-7 members) that undergo 3′ mono-uridylation by terminal uridyl transferases (TUTases) ZCCHC6, ZCCHC11 and GLD2, in order to be efficiently matured by DICER(Heo et al., 2012). Nearly identical 22 nt long mature let-7 miRNAs (namely let-7-5p) are generated by DICER processing and associate with Argonaute proteins in the miRNA-induced silencing complex (miRISC) where they function by repressing a broad array of genes involved in the control of development, cell proliferation, cell growth, metabolism and inflammation(Bussing et al., 2008). These functions of let-7 miRNAs are primarily accomplished in differentiated cells where they are abundantly expressed.
Let-7 pri- and pre-miRNAs harbor a typical hairpin structure with a stem containing the let-7-5p miRNA sequence base-paired extensively with the partially complementary let-7-3p miRNA sequence, connected by a so-called terminal loop region of variable lengths and structures among different let-7 family members, a region referred to as pre-element (preE)(Nam et al., 2011). Let-7 preE serves as a platform to recruit RNA-binding proteins such as LIN28, KHSRP (also known as KSRP), hnRNPA1 and TRIM25, in order to posttranscriptionally regulate let-7 biogenesis(Heo et al., 2008; Michlewski and Caceres, 2010; Newman et al., 2008; Rybak et al., 2008; Trabucchi et al., 2009; Viswanathan et al., 2008; Zhang et al., 2015). LIN28 proteins play pervasive roles during animal development and are often dysregulated in cancer(Ambros and Horvitz, 1984; Moss et al., 1997; Moss and Tang, 2003; Shyh-Chang and Daley, 2013; Thornton and Gregory, 2012). The paralogous LIN28A and LIN28B genes are predominantly expressed in undifferentiated cells such as embryonic stem cells (ESCs) and primarily function to repress let-7 miRNA expression thereby relieving repression of let-7 target mRNAs, and possibly also function to regulate mRNA translation and/or splicing by unknown mechanisms(Polesskaya et al., 2007; Wilbert et al., 2012). LIN28 proteins contain a cold shock domain and two CCHC-type zinc finger domains that bind respectively to the GNGAY consensus sequence (Y = pyrimidine; N = any base) in the let-7 preE loop and a conserved GGAG motif in the let-7 preE bulge(Ali et al., 2012; Heo et al., 2009; Loughlin et al., 2011; Mayr et al., 2012; Nam et al., 2011; Piskounova et al., 2008). LIN28A and LIN28B are predominantly cytoplasmic and nuclear, respectively, and repress let-7 biogenesis by two distinct mechanisms. LIN28A recruits ZCCHC6 and ZCCHC11, which catalyze 3′ oligo-uridylation of pre-let-7 miRNA(Hagan et al., 2009; Heo et al., 2009; Thornton et al., 2012). This modification inhibits Dicer processing and induces 3′-5′ degradation of let-7 precursors by the DIS3L2 exonuclease(Chang et al., 2013; Faehnle et al., 2014; Ustianenko et al., 2013). LIN28B binds let-7 preE in the nucleus and blocks Microprocessor-mediated cleavage of pri-let-7 miRNA by an unknown mechanism(Piskounova et al., 2011).
The LIN28/let-7 axis profoundly impacts diverse biological processes in mammals including stem cell pluripotency, development, glucose metabolism, tissue regeneration, organismal growth, and the age of onset of menopause and puberty in humans(Shyh-Chang and Daley, 2013; Shyh-Chang et al., 2013; Thornton and Gregory, 2012; Viswanathan and Daley, 2010). This embryonic pathway is frequently reactivated in human cancers, correlates with poor patient survival, and is sufficient to drive tumorigenesis in cell and mouse cancer models(Diskin et al., 2012; Madison et al., 2013; Molenaar et al., 2012; Nguyen et al., 2014; Thornton and Gregory, 2012; Urbach et al., 2014; Viswanathan et al., 2009). Importantly, where examined, the regulation of let-7 plays a central role in these numerous biological functions of LIN28A and LIN28B. While it has been widely appreciated that these RNA-binding proteins coordinately inhibit let-7 miRNA biogenesis, a systematic analysis and comparison of all let-7 miRNAs has not yet been performed. Here we find that although LIN28A and LIN28B exert a similar degree of repression on different let-7 miRNAs, a specific let-7 member - human let-7a-3 (and its murine ortholog let-7c-2) bypasses LIN28-mediated blockade both in vitro and in human cancer cells and mouse ESCs (mESCs). We furthermore provide mechanistic insight into this escape from LIN28-mediated regulation.
RESULTS
Analysis of let-7 regulation by LIN28 proteins
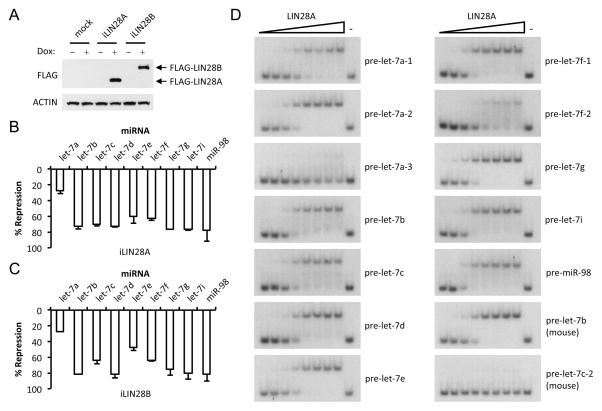
To compare the repressive activity of LIN28 proteins on let-7, we generated doxycycline-inducible Hela cell lines, which do not have detectable expression of endogenous LIN28A and LIN28B and conversely express high levels of all let-7 isoforms(Heo et al., 2012; Piskounova et al., 2011; Thornton et al., 2014). Isogenic clones expressing either FLAG-LIN28A (iLIN28A) or FLAG-LIN28B (iLIN28B) were obtained. Upon doxycycline treatment, each protein was expressed to a comparable level by Western blot and displayed a typical localization pattern as reported before (Figure 1A and S1A) (Hafner et al., 2010; Piskounova et al., 2011). Of note, this system achieved respectively lower and higher expression of LIN28A and LIN28B compared to human ESCs, and similar expression level for LIN28B compared to a panel of LIN28B-expressing human cancer cell lines, indicating that these isogenic clones express LIN28A/B at physiologically relevant levels (Figure S1B and S1C). We next examined the relative expression of different let-7 miRNAs upon LIN28A or LIN28B induction by quantitative reverse transcription PCR (q.RT-PCR). Examination of let-7 levels showed a specific reduction of each let-7 isoform two days after doxycycline treatment (Figure 1B, 1C and S1D). Although LIN28 proteins inhibit let-7 biogenesis by different mechanisms(Piskounova et al., 2011), a comparable degree of let-7 repression was observed between LIN28A and LIN28B. Therefore, LIN28 proteins achieve the same role in regulating let-7 expression. We also noticed that different let-7 isoforms were down regulated with variable degrees after LIN28 protein induction, ranging from <30% for let-7a to >70% for most other isoforms (Figure 1B and 1C). While this q.RT-PCR analysis cannot distinguish between the three let-7a miRNAs expressed from different loci (let-7a-1, let-7a-2, and let-7a-3), it suggests that one or more of let-7a miRNA isoforms might be differentially sensitive to LIN28 repression. We hypothesized that LIN28 proteins might bind these isoforms with different affinities and chose to systematically investigate LIN28A binding to each of the twelve human let-7 precursors by electrophoretic mobility shift assays (EMSA). Our analysis revealed that one pre-let-7 isoform, namely pre-let-7a-3, binds LIN28A and LIN28B with a much lower affinity than the other pre-let-7 members. Mouse pre-let-7c-2, which is orthologous to human pre-let-7a-3, also displayed strikingly lower affinity for LIN28 binding (Figure 1D, S1E and S1F).
Figure 1. Systematic analysis of let-7 regulation by LIN28 proteins.
(A) Western blot analyses showing expression of FLAG-LIN28A and FLAG-LIN28B upon doxycyline treatment. (B, C) q.RT-PCR analyses of let-7 expression in iLIN28A (B) and iLIN28B Hela cells (C). Results are the average of two biological replicate experiments represented as percentage of repression relative to mock treated cells. Error bars represent SD. (D) Electrophoretic mobility shift assay (EMSA) showing relative LIN28A binding to individual pre-let-7 RNAs. A twofold dilution series was used for the LIN28A titrations. Human precursors were used in these assays unless stated otherwise. See also Figure S1.
A single let-7 isoform bypasses LIN28A regulation
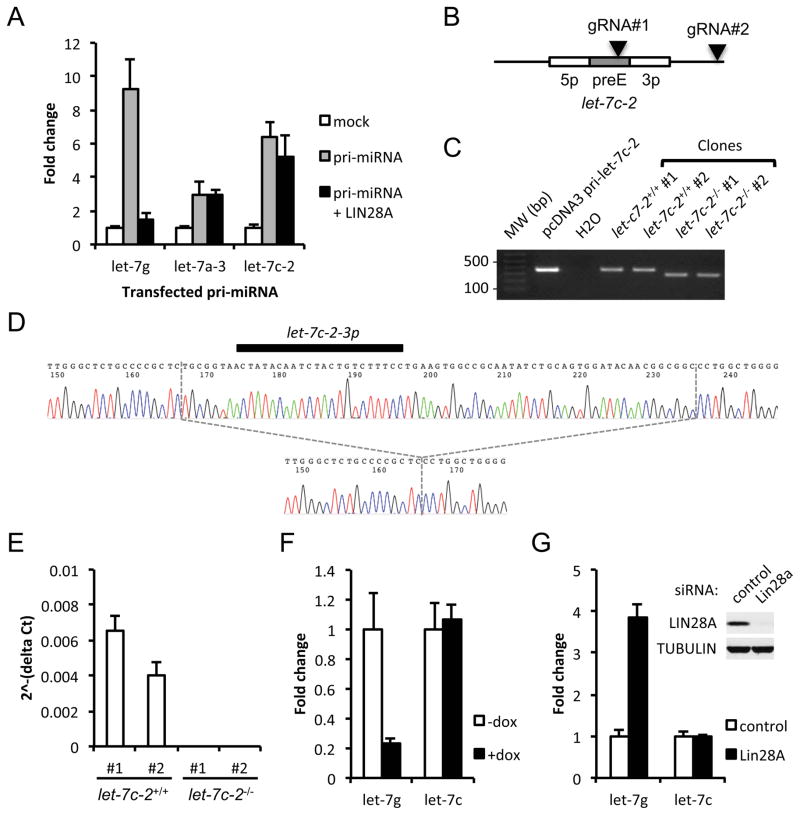
Considering the in vitro binding data, we next asked whether these let-7 isoforms could be repressed by LIN28 in cells. We individually overexpressed pri-let-7g, pri-let-7a-3, or pri-let-7c-2 in Hela cells in the presence or absence of LIN28A and measured the corresponding mature let-7 miRNA levels by qRT-PCR (Figure 2A). While let-7g was strongly repressed (more than 80%) by LIN28A, we did not detect any significant change in the expression of let-7a and let-7c upon LIN28A overexpression, thereby indicating that let-7a-3 and let-7c-2 isoforms can bypass LIN28A-mediated repression. We also evaluated this bypass mechanism in mESCs by examining endogenous let-7c expression. The mouse genome encodes for two let-7c isoforms, namely let-7c-1 and let-7c-2, which cannot be distinguished by qRT-PCR analyses since the mature let-7 miRNA sequences are identical. Upon deletion of the let-7c-2 locus using the CRISPR/Cas9 genome-editing system, we found that let-7c expression was lost in let-7c-2−/− mouse embryonic stem cells, suggesting that let-7c-2 is the most abundant let-7c isoform expressed in these cells (Figure 2B–E). We found that, while let-7g levels were substantially suppressed by LIN28A overexpression in mESCs, the levels of let-7c were unaffected (Figure 2F). Similarly, let-7g levels accumulated upon LIN28A knockdown whereas let-7c levels remained unchanged (Figure 2G). Therefore, physiological expression of LIN28A in cells does not impact endogenous let-7c-2 expression. Altogether, these results show that unlike all other let-7 miRNAs, a single member (mouse let-7c-2, and human let-7a-3) escapes LIN28 binding and regulation.
Figure 2. Human let-7a-3 and mouse let-7c-2 bypass LIN28A-mediated repression.
(A) q.RT-PCR analyses of let-7g, let-7a and let-7c expression in Hela cells upon transfection of empty vectors (mock), let-7 pri-miRNA only or let-7 pri-miRNA and FLAG-LIN28A as indicated. (B) Schematic representation of the let-7c-2 locus. Arrowheads indicate guide RNA (gRNA) target sites used for CRISPR/Cas9 DNA editing. (C) PCR for genotyping analysis of let-7c-2+/+ and let-7c-2−/− TC1 mESC clones. (D) Sequencing analyses showing the deletion in the let-7c-2 locus. The upper track and the lower track correspond to the sequencing results obtained from the let-7c-2+/+ clone #1 and the let-7c-2−/− clone #1 respectively. (E) q.RT-PCR analyses for let-7c expression in let-7c-2+/+ and let-7c-2−/− clones. (F) q.RT-PCR analyses for let-7g and let-7c expression in KH2-iLIN28A mESCs treated with or without doxycycline (dox). (G) RT-qPCR analyses for let-7g and let-7c expression in TC1 mESCs transfected with control or Lin28a siRNA. Upper right panels represent Western blot results validating LIN28A knockdown. (A), (E-G) Results are the average of two independent experiments. Error bars represent SD.
The preE loop in pre-let-7c-2 precludes LIN28A binding
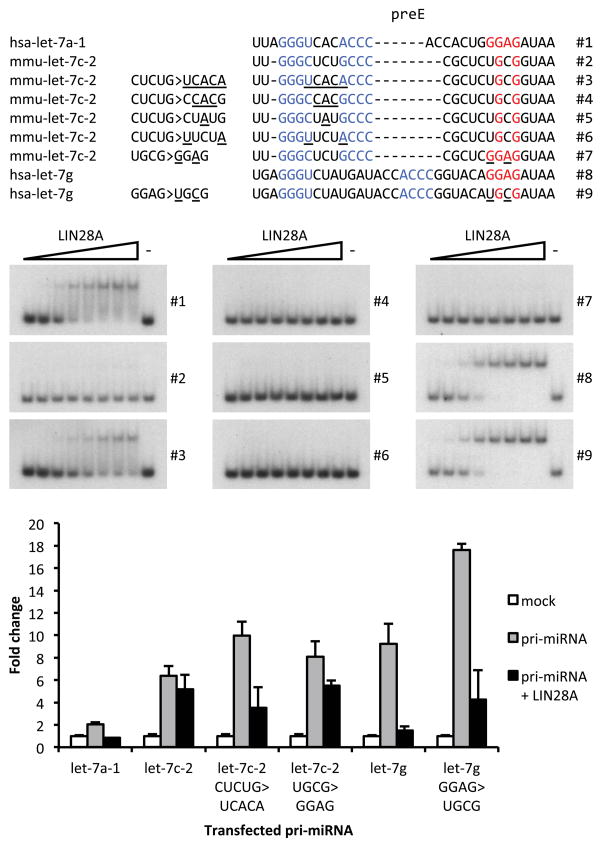
We sought to address at the molecular level how pre-let-7c-2 might escape the LIN28 blockade. We noticed that let-7c-2 precursor lacks the canonical GGAG motif in the preE bulge (Figure 3A). We postulated that restoring this motif would be sufficient to confer LIN28A binding and regulation. We performed mutagenesis to substitute the atypical UGCG motif within the pre-let-7c-2 preE bulge with a canonical GGAG motif found in most other pre-let-7 family members. To our surprise we observed that addition of the GGAG sequence was not sufficient to restore LIN28A binding to the mutated pre-let-7c-2 (Figure 3A and 3B). Accordingly, pre-let-7c-2 containing GGAG was not appreciably suppressed by LIN28A expression in transfected cells (Figure 3C). Considering these results we also replaced the GGAG motif in let-7g preE with UGCG (i.e. the sequence from pre-let-7c-2) and did not observe any dramatic differences in LIN28A binding to or repression of this let-7g mutant. These data suggest that the UGCG motif is functional and that other sequence and/or structural features of pre-let-7c-2 are responsible for the escape from LIN28 regulation (Figure 3A–C). We considered that the terminal loop of the preE of pre-let-7c-2 might inhibit LIN28 binding. To explore this, we generated a mutant form of pre-let-7c-2 by substituting nucleotides in the preE loop with corresponding nucleotides from let-7a-1 preE loop, which shares very comparable structure and yet interacts with and is regulated by LIN28A. A five-nucleotide substitution comprising the pre-let-7c-2 preE loop with the stem-loop junction (CUCUG>UCACA) was sufficient to rescue LIN28A binding in EMSA (Figure 3A and 3B). EMSA performed with additional mutations within this region revealed that that the five-nucleotide change is necessary and sufficient to confer LIN28 binding in the context of pre-let-7c-2 (Figure 3B). LIN28A-mediated repression was also established by this terminal loop replacement (Figure 3C). Therefore these results strongly suggest that let-7c-2 bypasses LIN28A repression primarily because the interaction between LIN28A CSD and let-7c-2 preE loop is compromised.
Figure 3. The preE loop of pre-let-7c-2 precludes LIN28A binding and regulation.
(A) Representation of different wild type and mutant preE sequences of let-7 constructs used in EMSA and transfection experiments. Mutations are underlined, preE stem is in blue and GGAG motif is in red. (B) EMSA showing specific binding of LIN28A to individual let-7 wild type and mutant pre-miRNA depicted in (A). (C) q.RT-PCR analyses of let-7a, let-7c and let-7g expression in Hela cells upon transfection of empty vectors (mock), let-7 pri-miRNA only or let-7 pri-miRNA and FLAG-LIN28A as indicated. Results are the average of two independent experiments. Error bars represent SD.
DISCUSSION
By performing a systematic analysis of all twelve let-7 miRNAs, we surprisingly found that a single let-7 family member, namely let-7a-3 in human and let-7c-2 in mouse, escapes LIN28-mediated regulation. Using both EMSA to measure relative LIN28 binding affinities to different pre-let-7 family members, as well as measuring let-7 miRNA levels in response to LIN28A or LIN28B overexpression and LIN28A knockdown in cells, we conclude that these two miRNA precursors are refractory to LIN28 binding and regulation. Although previous analyses have examined the global effects of LIN28 gain- or loss-of-function on miRNA expression, these studies failed to appreciate that not all let-7 miRNAs are regulated by these proteins(Hagan et al., 2009; Newman et al., 2008), and contributed to the current dogma that LIN28 proteins coordinately represses let-7 expression. This finding likely remained obscure because both let-7c in mouse and let-7a in human are expressed from multiple different loci, i.e. let-7c-1, and let-7c-2 (mouse) and let-7a-1, let-7a-2, and let-7a-3 (human). Since the mature miRNA sequence within each subfamily is identical, it is not possible to distinguish them by q.RT-PCR, microarray, or RNA sequencing, thereby confounding the interpretation of miRNA expression studies.
Mechanistically we found that the five-nucleotide long sequence forming the short apical stem-loop of let-7c-2 preE precludes LIN28A binding in vitro and LIN28A-mediated repression in cells. In fact, replacing this sequence with the corresponding nucleotides from pre-let-7a-1 preE loop substantially restored LIN28A interaction. This interaction might result from the combined effect of: 1) creating a more favorable CSD binding motif; 2) weakening the base pairing in the stem by changing a G-C pair to an A-U pair, thus allowing the CSD to protrude through the terminal loop and remodel the base pairing in the stem. Our results also suggest that the lack of a canonical GGAG motif in let-7c-2 preE bulge was not responsible for the bypass. Indeed, swapping the respective GGAG and UGCG motifs in pre-let-7g and pre-let-7c-2 did not have a significant impact on the interaction or lack of interaction with LIN28A. These results are in accordance with a previous study where it was proposed that LIN28 CSD binds first to let-7 preE, and remodels its structure to allow for subsequent binding of the GGAG motif by LIN28 CCHC-type zinc fingers in order to anchor the protein to the precursor RNA(Mayr et al., 2012).
It will be interesting to explore the full range of physiological contexts where that bypass is relevant. Mouse models with let-7c-2 deletion will help dissect the importance of this mechanism. Since LIN28A and LIN28B mRNAs both contain let-7 target sites and are repressed in let-7 expressing cells, it is tempting to speculate that this LIN28 bypass might be important for resetting the bistable switch to alter cell fate decisions. Related to this, the efficiency of reprogramming mouse embryonic fibroblasts (MEFs) to induced pluripotent stem cells (iPSCs) was more substantially enhanced by let-7 antisense oligonucleotides (that should antagonize all let-7 members) than by ectopic LIN28 expression(Worringer et al., 2014). It will be important to understand which cell-types and developmental stages express high levels of these individual let-7 miRNAs and the upstream mechanisms controlling their transcription(Patterson et al., 2014). Given also the strong links between let-7 repression and tumorigenesis, it will also be interesting to explore the epigenetic/genetic loss of let-7a-3. In this regard, deletions containing the let-7a-3~let-7b locus have been reported in human tumors (Wang et al., 2012). Our results also predict that epigenetic/genetic loss of let-7a-3 would cooperate with the oncogenic effects of LIN28A and LIN28B activation in human tumorigenesis. Intriguingly, it was recently demonstrated in a mouse model that conditional deletion of the let-7c-2~let-7b locus functionally cooperates with transgenic Lin28b overexpression to drive hyperplasia of the intestinal epithelium(Madison et al., 2015; Madison et al., 2013). Our mechanistic studies can help explain this result and will likely provoke future studies that address the relative contribution of this newly identified mechanism in the many different contexts where the LIN28/let-7 axis is known to be important.
EXPERIMENTAL PROCEDURES
DNA cloning and mutagenesis
Human LIN28A and LIN28B cDNA were PCR amplified from pFLAG-CMV2 LIN28A and pFLAG-CMV2 LIN28B respectively(Piskounova et al., 2011), and cloned into pcDNA5/FRT/TO vector (Invitrogen). Human and mouse pri-let-7 cDNA were PCR amplified from Hela cells and J1 embryonic stem cells genomic DNA respectively and cloned into pcDNA3 vector. Pri-let-7 mutant plasmids were generated by site-directed mutagenesis using QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). For CRISPR/Cas9-mediated Let-7c-2 gene editing, oligonucleotides were designed on line (http://crispr.mit.edu/), annealed and cloned into BbsI site of pX330 vector (see Table S1 for oligonucleotide sequences).
Transfection
Plasmids and siRNA transfections were performed with Lipofectamine 2000 (Invitrogen). Mouse embryonic stem cells were transfected by reverse transfection with the following synthetic RNA (all from Dharmacon): ON-TARGETplus Non-targeting Pool (D-001810-10-20) and ON-TARGETplus Mouse Lin28a (83557) siRNA – SMARTpool (L-051530-01-0020).
Cell culture
Hela, HEK293T, HepG2 and H1299 cell lines were maintained in DMEM (Gibco) supplemented with penicillin/streptomycin (Gibco) and 10% fetal bovine serum (Gemini Bio Products). K562 cells were maintained in RPMI (Gibco) supplemented with penicillin/streptomycin and 10% fetal bovine serum. Flp-In T-REx Hela cells (a kind gift of Stephen Taylor, University of Manchester) were maintained in DMEM supplemented with penicillin/streptomycin, 4 μg/ml blasticidin (Gibco), 50 μg/ml zeocin (Gibco) and 10% fetal bovine serum. Flp-In T-REx iLIN28A and iLIN28B Hela cells were generated by cotransfecting pcDNA5/FRT/TO FLAG-LIN28A or pcDNA5/FRT/TO FLAG-LIN28B vector with the Flp recombinase encoding plasmid pOG44 (Invitrogen). Isogenic clones were isolated and maintained in DMEM supplemented with penicillin/streptomycin, 4 μg/ml blasticidin, 200 μg/ml hygromycin (Sigma) and 10% Tet System Approved fetal bovine serum (Clontech). Transgene expression was assayed upon treatment with 1 μg/ml doxycycline (Sigma) for 48 hours. TC1 mESCs were cultured feeder-free on gelatin-coated dishes with KO-DMEM medium (Gibco) supplemented with 15% fetal bovine serum (Gemini Bio Products), L-glutamine (Gibco), sodium pyruvate (Gibco), non-essential amino acids (Gibco), 2-mercaptoethanol, penicillin/streptomycin, 1000 U/ml LIF (Gemini Bio Products) that is referred as LIF/serum condition. For CRIPSR/Cas9-mediated gene manipulation of TC1 cells and for KH2 iFLAG-Lin28A mESCs culture, cells were propagated in LIF/2i condition, consisting of N2B27 medium supplemented with LIF, as well as 3 μM CHIR99021 (Stemgent) and 1 μM PD0325901 (Stemgent).
CRISPR/Cas9-mediated gene editing
TC1 mESCs were cotransfected by nucleofection (Lonza) with 1 μg of each pX330 vector encoding for let-7c-2-specific guide RNA #1 and #2 and 0.1 μg of vector for expression of puromycin resistance gene. One day later, 2.5 μg/ml puromycin (Sigma) was added to the media for 24 hrs. Individual mESC clones were isolated and genotyped by PCR and Sanger sequencing.
EMSA
PCR amplification of pri-let-7 expression plasmids was used to generate DNA templates for in vitro transcription of radiolabeled pre-let-7 RNA (see table for oligonucleotide sequences). These DNA templates were gel-purified and in vitro transcription was performed according to Riboprobe in vitro Transcription Systems (Promega) using [a-32P] rUTP and T7 RNA polymerase. Radiolabeled pre-let-7 RNAs were subsequently treated with RQ1 DNase and purified using illustra MicroSpin G-25 Columns (GE Healthcare Life Sciences). For EMSA, reactions were set up in binding buffer (50 mM Tris, pH 7.6, 100 mM NaCl, 10 mM 2-mercaptoethanol, 1 U/μl rRNasin [Promega]) with radiolabelled RNA and varying amounts of immunopurified FLAG-LIN28A or FLAG-LIN28B proteins and were incubated for 30 min at room temperature. Protein-RNA complexes were resolved on native 5% polyacrylamide gels and visualized by autoradiography.
Real-time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) and reverse transcribed using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was performed using individual TaqMan Assays (Applied Biosystems) with TaqMan universal PCR master mix, no AmpErase UNG (Applied Biosystems). Gene expression was normalized to U6 snRNA.
Antibodies
For western blot and immunocytochemistry analyses, the following antibodies were used: anti-FLAG (Sigma, A8592), anti-LIN28A (A177)(Cell Signaling, 3978), anti-LIN28B (Cell Signaling, 4196), anti-OCT4 (Abcam, ab19857), anti-ACTIN (Sigma, A2066), anti-beta TUBULIN (Abcam, ab6046).
Supplementary Material
Highlights.
Human let-7a-3 and mouse let-7c-2 miRNA escape LIN28 regulation
Human let-7a-3 and mouse let-7c-2 pre-miRNA are refractory to LIN28 binding
LIN28 cold-shock domain interaction with let-7c-2 pre-miRNA loop is compromised
A five-nucleotide substitution in pre-let-7c-2 loop restores LIN28 repression
Acknowledgments
We thank Stephen Taylor for Flp-In T-REx Hela cells. This work was supported by a grant to R.I.G. from the US National Institute of General Medical Sciences (NIGMS) (R01GM086386).
Footnotes
AUTHOR CONTRIBUTIONS
R.T. performed all experiments. M.P. helped generate let-7c-2−/− mESCs using CRISPR/Cas9 technology. R.T., and R.I.G. designed all experiments, analyzed data, and wrote the manuscript with helpful discussion with M.P.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali PS, Ghoshdastider U, Hoffmann J, Brutschy B, Filipek S. Recognition of the let-7g miRNA precursor by human Lin28B. FEBS Lett. 2012;586:3986–3990. doi: 10.1016/j.febslet.2012.09.034. [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Diskin SJ, Capasso M, Schnepp RW, Cole KA, Attiyeh EF, Hou C, Diamond M, Carpenter EL, Winter C, Lee H, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44:1126–1130. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle CR, Walleshauser J, Joshua-Tor L. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature. 2014 doi: 10.1038/nature13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Monouridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Loughlin FE, Gebert LF, Towbin H, Brunschweiger A, Hall J, Allain FH. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2202. [DOI] [PubMed] [Google Scholar]
- Madison BB, Jeganathan AN, Mizuno R, Winslow MM, Castells A, Cuatrecasas M, Rustgi AK. Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2. PLoS Genet. 2015;11:e1005408. doi: 10.1371/journal.pgen.1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Liu Q, Zhong X, Hahn CM, Lin N, Emmett MJ, Stanger BZ, Lee JS, Rustgi AK. LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let-7. Genes Dev. 2013;27:2233–2245. doi: 10.1101/gad.224659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr F, Schutz A, Doge N, Heinemann U. The Lin28 cold-shock domain remodels pre-let-7 microRNA. Nucleic acids research. 2012;40:7492–7506. doi: 10.1093/nar/gks355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar JJ, Domingo-Fernandez R, Ebus ME, Lindner S, Koster J, Drabek K, Mestdagh P, van Sluis P, Valentijn LJ, van Nes J, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012;44:1199–1206. doi: 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular Basis for Interaction of let-7 MicroRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LH, Robinton DA, Seligson MT, Wu L, Li L, Rakheja D, Comerford SA, Ramezani S, Sun X, Parikh MS, et al. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell. 2014;26:248–261. doi: 10.1016/j.ccr.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Patterson M, Gaeta X, Loo K, Edwards M, Smale S, Cinkornpumin J, Xie Y, Listgarten J, Azghadi S, Douglass SM, et al. let-7 miRNAs can act through notch to regulate human gliogenesis. Stem Cell Reports. 2014;3:758–773. doi: 10.1016/j.stemcr.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, Lapierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B Inhibit let-7 MicroRNA Biogenesis by Distinct Mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNAbinding protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, Daley GQ. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 2013;155:778–792. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Du P, Jing L, Sjekloca L, Lin S, Grossi E, Sliz P, Zon LI, Gregory RI. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4) Nucleic acids research. 2014 doi: 10.1093/nar/gku805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22:474–482. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A, Yermalovich A, Zhang J, Spina CS, Zhu H, Perez-Atayde AR, Shukrun R, Charlton J, Sebire N, Mifsud W, et al. Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev. 2014;28:971–982. doi: 10.1101/gad.237149.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013;19:1632–1638. doi: 10.1261/rna.040055.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hu X, Greshock J, Shen L, Yang X, Shao Z, Liang S, Tanyi JL, Sood AK, Zhang L. Genomic DNA copy-number alterations of the let-7 family in human cancers. PLoS One. 2012;7:e44399. doi: 10.1371/journal.pone.0044399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX, Yan BY, Nathanson JL, Hutt KR, Lovci MT, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worringer KA, Rand TA, Hayashi Y, Sami S, Takahashi K, Tanabe K, Narita M, Srivastava D, Yamanaka S. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell. 2014;14:40–52. doi: 10.1016/j.stem.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Elabd S, Hammer S, Solozobova V, Yan H, Bartel F, Inoue S, Henrich T, Wittbrodt J, Loosli F, et al. TRIM25 has a dual function in the p53/Mdm2 circuit. Oncogene. 2015 doi: 10.1038/onc.2015.21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.