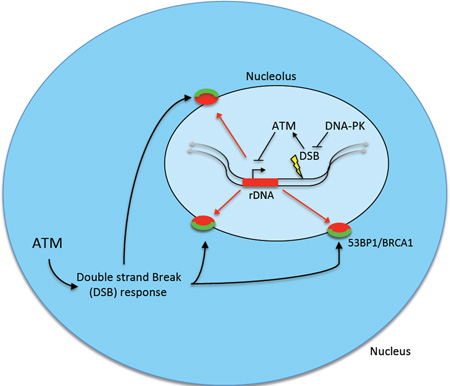
Summary
Resolution of DNA double-strand breaks (DSBs) is essential for the suppression of genome instability. DSB repair in transcriptionally active genomic regions represents a unique challenge that is associated with an Ataxia telangiectasia mutated (ATM) kinase-mediated transcriptional silencing. Despite emerging insights into the underlying mechanisms, how DSB silencing connects to DNA repair remains undefined. We observe that silencing within the rDNA depends on persistent DSBs. Non-homologous end-joining (NHEJ) was the predominant mode of DSB repair allowing transcription to resume. ATM-dependent rDNA silencing in the presence of persistent DSBs led to the large-scale reorganization of nucleolar architecture, with movement of damaged chromatin to nucleolar cap regions. These findings identify ATM-dependent temporal and spatial control of DNA repair and provide insights into how communication between DSB signaling and ongoing transcription promotes genome integrity.
Graphical abstract
Introduction
DNA double strand breaks (DSBs) occur naturally as byproducts of cellular metabolism and in response to environmental and therapeutic stresses. In response to DSBs, cells activate a kinase directed molecular cascade to activate a network of subsequent responses. Central to this process is the phosphatidylinositol kinase-like-kinase (PIKK), Mutated in Ataxia Telangiectasia (ATM), which phosphorylates thousands of targets to affect a myriad of cellular processes including cell-cycle checkpoints and DSB repair (Matsuoka et al., 2013). Accurate DSB repair is fundamental for the suppression of sequence alterations and translocations that cause genomic instability and cancer. Cells have evolved two complementary pathways, non-homologous end-joining (NHEJ) and homologous recombination (HR), that repair the majority of DSBs. NHEJ mediated DSB repair requires the activation of the PIKK, DNA-dependent protein kinase (DNA-PK) that promotes end processing and ligation by a complex of several proteins including the Ku70/80 heterodimer, XRCC4 and DNA ligase 4 (Williams et al., 2014). Conversely, HR is considered a restorative process that uses a homologous template for synthesis-driven repair and the BRCA1/2 proteins to nucleate Rad51 filaments that initiate synapsis between single stranded regions of the break and homologous regions of the genome, usually residing on a sister chromatid (Helleday, 2010).
The DSB response is required to cope with ongoing cellular processes on chromatin, such as transcription. A striking example of this interplay occurs during spermatogenesis, where unsynapsed sex chromosomes replete with programmed DSBs activate ATR dependent DSB responses to silence transcription in a process known as meiotic sex chromosome inactivation (Turner, 2007). Somatic cells also silence RNA Pol I and RNA Pol II mediated transcription in response to DSBs. Nucleolar DSBs generated by ionizing radiation (IR) or UV-microbeams caused ATM-dependent silencing of RNA Polymerase I (Pol I) transcription (Kruhlak et al., 2007). In this instance an ATM kinase dependent interaction between NBS1 and Treacle contributed to Pol I silencing (Ciccia et al., 2014; Larsen et al., 2014). Using a cellular reporter system, we found that an ATM- and ubiquitin- driven chromatin modification caused silencing of RNA polymerase II (Pol II) transcription in cis to DSBs (Shanbhag et al., 2010). This ATM-driven transcriptional silencing is mediated in part by recruitment of polycomb repressive and SWI/SNF complexes to DSBs (Kakarougkas et al., 2014a; Ui et al., 2015). Despite accumulating mechanistic insight, how silencing transcription impacts the complex process of DSB recognition and repair is still unclear.
Ribosomal DNA (rDNA) is the most actively transcribed region of the human genome and occurs within a defined nuclear compartment, the nucleolus (Grummt, 2013). Hundreds of 43kb repeats of rDNA are located on the short arms of the acrocentric chromosomes in human cells to facilitate the rapid production of rRNA molecules required for ribosome biogenesis (Huang et al., 2006; Russell and Zomerdijk, 2006). These rDNA loci organize into nucleoli following mitosis where Pol I and rRNA processing machineries concentrate (Prieto and McStay, 2008). The nucleolus acts as a stress sensor and many types of cellular stresses lead to marked changes in its organization (Boulon et al., 2010). Electron microscopy studies found that inhibition of rDNA transcription by Actinomycin D (ActD) lead to dense “caps” surrounding the nucleolus (Reynolds et al., 1964). Subsequent studies have determined that these nucleolar caps are formed of Pol I components and the rRNA processing machinery that redistribute following transcriptional silencing (Shav-Tal et al., 2005). As cellular stress sensors, the nucleoli and rDNA represent a unique compartmentalized system to examine the impact of DSBs and ATM-dependent transcriptional silencing on nuclear architecture.
Here, we explore how DSBs generated within the rDNA repeats are sensed and repaired. Persistent breaks are required to fully induce transcriptional silencing by ATM. Rapid DNA repair by NHEJ mitigates the impact of DNA damage on transcription, thus preserving ribosomal RNA synthesis. Remarkably, ATM-dependent transcriptional silencing induced nucleolar reorganization and the recognition of rDNA DSBs at the nucleolar periphery by DNA damage response (DDR) factors. These findings uncover a role for ATM-dependent silencing in connecting nuclear organization to the DNA damage response.
Results
NHEJ mitigates ATM-Dependent DSB Silencing of the rDNA
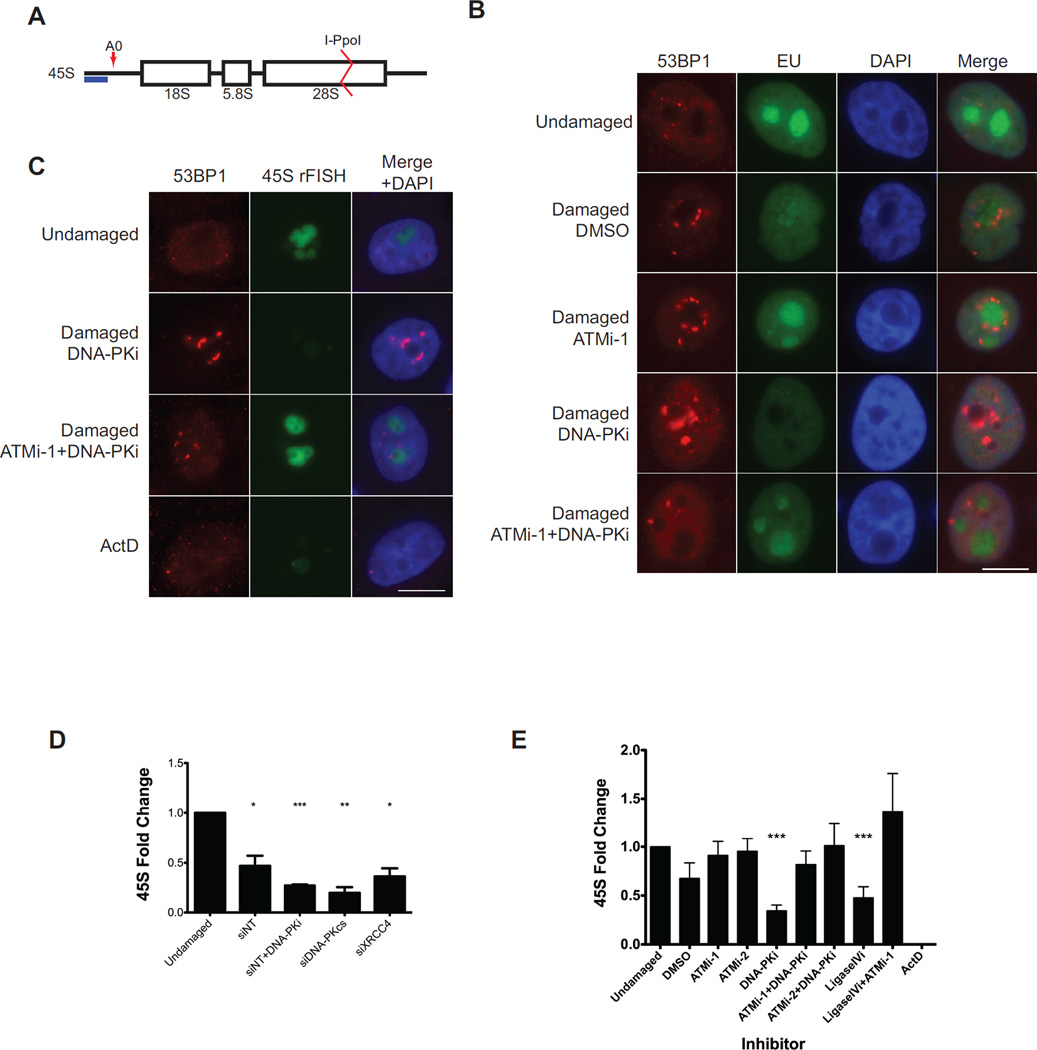
To generate DSBs within the rDNA repeats we introduced an inducible I-PpoI endonuclease into immortalized, non-transformed MCF10A mammary epithelial cells. I-PpoI recognizes a sequence within the 28S portion of the rDNA repeat (Figure 1A) and has been previously developed by the Kastan group to enable site specific DSB formation in mammalian cells (Berkovich et al., 2007). To control I-PpoI induction we included an N-terminal estradiol receptor (ER) and a C-terminal destabilization domain. Upon nuclease induction, the DSB-response protein 53BP1 localized to the perinucleolar region and an associated reduction in incorporation of the ribonucleoside analog 5-Ethynyl uridine (EU) was observed in nucleoli indicating rDNA silencing had occurred during the ensuing DSB response (Figure 1B) as previously described (Kruhlak et al., 2007; Larsen et al., 2014). This phenotype was reversed by ATM inhibition (ATMi) and exacerbated by inhibition of DNA-PK (DNA-PKi). Remarkably, ATMi overcame DSB-silencing even in the presence of DNA-PKi. Similar results were observed in HeLa S3 cells and in colon cancer DLD-1 cells following induction of I-PpoI (Data not shown).
Figure 1. Inhibition of NHEJ exacerbates ATM-dependent silencing of rDNA transcription.
(A) Schematic drawing of a nascent 45S rRNA transcript. The 45S transcript is rapidly cleaved at A0 as indicated by the red arrow. The blue bar indicates the amplicon monitored by RT-qPCR. The I-PpoI cut site within the 28S region is indicated.
(B) Immunofluorescence of 53BP1 (red) and nascent transcription measured by 5-Ethynyl uridine (EU) incorporation following I-PpoI induction in the presence of the indicated inhibitors. A merge overlaid with DAPI is shown. Scale bar is 10µm.
(C) Combined immunofluorescence of 53BP1 (red) and nascent rRNA measured by RNA-FISH. Scale bar is 10µm.
(D) MCF10A cells were treated with the indicated siRNA for 72h before I-PpoI induction and nascent transcription was monitored 5 hr later. Error bars represent SEM. *p<0.05. **p<0.01, ***p<0.001
(E) RT-qPCR measurement of nascent 45S transcript following induction of I-PpoI endonuclease for 5h in the presence of the indicated inhibitors in MCF10A cells. Please see supplemental experimental procedures for inhibitor details. Error bars represent SEM of at least 3 biological replicates. ***p<0.001
Transcription of the rDNA results in a long nascent transcript that is rapidly processed by a series of nucleases to produce mature RNAs for incorporation into ribosomes. One of the earliest steps in rRNA maturation involves cleavage of the precursor at site A0 (Figure 1A). We exploited this early processing to measure nascent transcription following DSBs using two methods. First, we observed the ATM-kinase dependent loss of nascent RNA-FISH signal following DSB induction (Figure 1C). As a control for silencing, ActD inhibition of RNA polymerase I caused loss of RNA-FISH signal. We next monitored 45S transcript levels by RT-qPCR. Although decreased in vehicle treated cells, transcriptional silencing was increased when components of NHEJ were inhibited pharmacologically or by siRNA (Figures 1D–E and S1A). Simultaneous inhibition of ATM by either of 2 different inhibitors led to reversal of DSB silencing (Figure 1E), consistent with previous studies at both RNA polymerase I and II loci (Kakarougkas et al., 2014b; Kruhlak et al., 2007; Larsen et al., 2014; Shanbhag et al., 2010; Ui et al., 2015). Together these results show that DSBs generated by I-PpoI endonuclease cause robust ATM-dependent silencing of rDNA transcription and that inhibition of NHEJ exacerbates this phenotype.
Persistent DSBs are Required for ATM Dependent rDNA-silencing
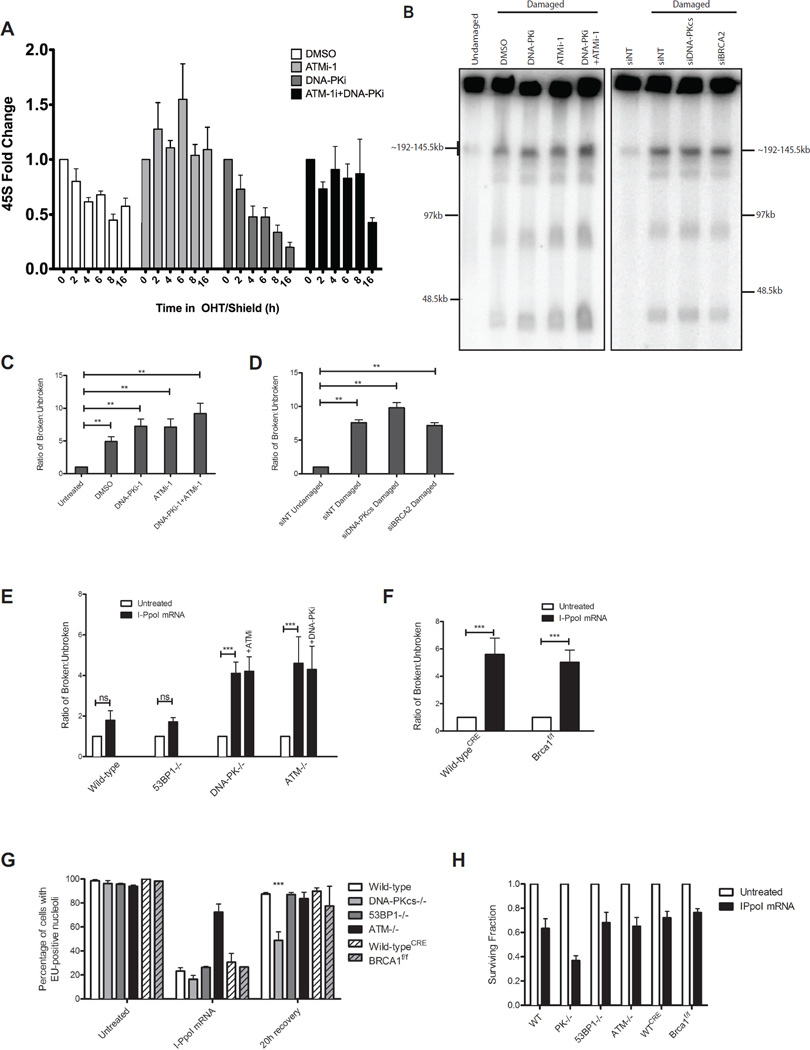
The observation that inhibition of NHEJ increased DSB-silencing suggested that ongoing DNA repair suppresses silencing. To examine this possibility, I-PpoI was induced and transcription of 45S was monitored over 16h. In vehicle treated cells, transcriptional inhibition reached a plateau between 4–8h and was reversible with ATMi (Figure 2A). When simultaneously treated with DNA-PKi to block NHEJ, transcriptional silencing continued to decrease over 16h that was again reversible by ATMi. The degree of silencing correlated with the increased fraction of DSBs generated as monitored by southern blotting (Figure S1B). To further examine the fraction of broken rDNA loci we adapted a classical method for DSB monitoring based on pulsed-field gel electrophoresis (PFGE). Broken rDNA loci are released into the gel forming a ladder of products, the smallest of which corresponds to a single rDNA repeat unit (~43kb; Figure 2B). Quantification of the released fragments as a ratio to the signal retained in the well indicated that DNA-PKi or ATMi alone increase the fraction of broken loci and that combined inhibition modestly increased this effect (Figure 2C). Concordant results were obtained using neutral comet assays (S1C–D). Importantly, despite the increase in DSBs when both ATM and DNA-PK are inhibited, silencing is still reversed (Figure 1E), consistent with the dependence on ATM signaling of this phenomenon. Interestingly, siRNA directed to DNA-PK increased rDNA damage using the PFGE assay, whereas that directed to BRCA2 had no effect (Figure 2D). We verified these results using neutral comet assays and found that siRNA to multiple HR factors (BRCA1, BRCA2, RAD51) did not increase DSBs whereas siRNA to NHEJ factors (DNA-PKcs, XRCC4) caused a statistically significant increase (Figure S1D).
Figure 2. Persistent DSBs are required for ATM-dependent rDNA silencing.
(A) Nascent 45S transcription was measured as in Figure 1B at the indicated times following I-PpoI induction. Error bars represent SEM of 3 biological replicates
(B) PFGE and hybridization of rDNA probes following I-PpoI induction with indicated treatments. Approximate sizes are indicated based on DNA markers.
(C) Quantification of PFGE hybridization signal for inhibitor treated MCF10A cells as indicated.
(D) Quantification of PFGE hybridization signal for siRNA treated MCF10A cells as indicated.
(E) Quantification of PFGE hybridization signal for MEFs either untreated or transfected with I-PpoI mRNA.
(F) Quantification as in (E), except CreERT2-expressing wild-type and conditionally deleted Brca1f/f were assayed.
(G) Fraction of EU-positive cells before and 20h following I-PpoI mRNA transfection. ***p<0.001.
(H) Cellular viability was measured by the MTS assay at 40h following washout of transfected I-PpoI mRNA.
All error bars represent SEM.
To extend these assays to alternative cellular systems we transfected I-PpoI mRNA into genetically defined MEFs (Van Sluis and McStay, 2015). DNA-PK−/− and ATM−/− MEFs showed a significant increase in DSBs in comparison to WT or 53BP1−/− MEFs, a finding confirmed using comet assays (Figure 2E and S1F–H). Conditional knockout of mouse BRCA1 did not increase DSBs beyond controls (Figure 2F and S1H). Consistent with a role of NHEJ in restoring transcription following DSB silencing, only DNA-PK−/− cells maintained a significant fraction of cells with EU-negative nucleoli 20h following I-PpoI washout (Figure 2G and S2). We also monitored survival of these cells using the MTS assay and found DNA-PK−/− cells to be less viable than the other genotypes examined (Figure 2H). Although 53BP1 is essential for NHEJ in certain contexts, such as class switch recombination at immunoglobulin loci, it is more dispensable for DNA repair than canonical NHEJ factors (Panier and Boulton, 2014), consistent with results in the rDNA. Together these results confirm that persistent DSBs drive silencing and that NHEJ is the predominant pathway for repair of I-PpoI generated DSBs in the rDNA.
ATM-Dependent DSB Silencing Induces Dynamic Nucleolar Reorganization
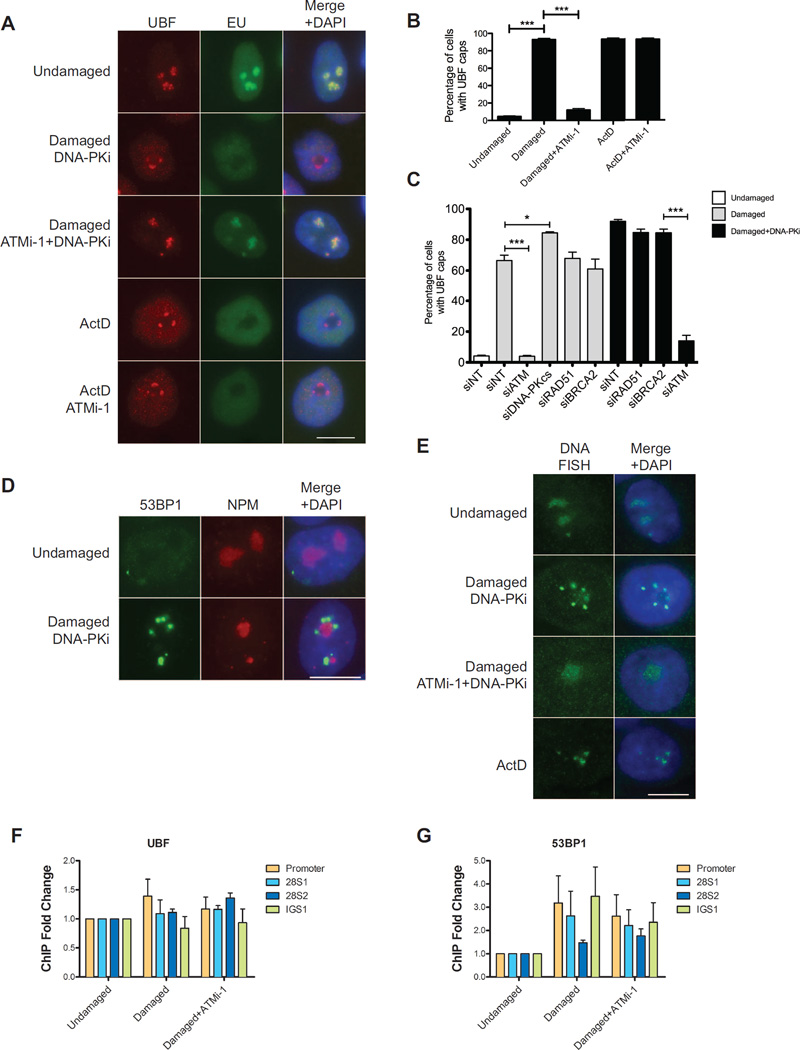
To determine how ATM-dependent silencing influences nucleolar structure we examined the localization of several Pol I components and associated factors after DSB induction. Following I-PpoI induction we observe robust redistribution of nucleolar proteins UBF, Nop58, RPA135 and PAF49 into caps localizing around DAPI-sparse nucleoli (Figure 3A and S3). When steady state DSB levels are maximized by DNA-PKi, 93% of cells form UBF nucleolar caps, this was reversible by ATMi (Figure 3B). Inhibition of Pol I transcription by ActD induced nucleolar cap formation but this was not reversed by ATMi (Figures 3A and 3B). Therefore, ATM-driven nucleolar reorganization is a specific consequence of its actions to silence DSBs, predicting that silencing precedes nucleolar cap formation. In accordance with the role of NHEJ in suppressing DSB silencing in rDNA we observe an increase in the fraction of cells displaying nucleolar UBF caps when NHEJ was blocked by siRNA but not when HR was inhibited (Figure 3C). Interestingly another component of nucleoli, nucleophosmin (NPM) was not observed to redistribute following DSB generation (Figure 3D). This suggests that the structure of nucleoli is at least partially maintained and is consistent with previous findings after ActD induced silencing (Shav-Tal et al., 2005).
Figure 3. ATM-dependent DSB silencing leads to nucleolar reorganization.
(A) Immunofluorescence of UBF (red) and nascent transcription as measured in Figure 1D. Actinomycin D treatment serves as a control for cap formation in the absence of I-PpoI induction. Scale bar is 10µm.
(B) Quantification of the percentage of cells with UBF nucleolar caps for the experiment described in (A). Error bar represents SEM of at least 3 biological replicates. ***p<0.001
(C) Percentage of cells with UBF nucleolar caps following I-PpoI induction 72h after siRNA transfection. *p<0.05, ***p<0.001
(D) Immunofluorescent staining of 53BP1 (green) and Nucleophosmin (NPM). Scale bar is 10µm.
(E) DNA-FISH of rDNA following I-PpoI induction. Scale bar is 10µm.
(F) Fold change of ChIP-qPCR signal for UBF as indicated following induction of I-PpoI in the presence of indicated inhibitors. Primer sets were targeted to the promoter, 28S coding region or the intergenic spacer (IGS) between rDNA repeats. Error bars represent SEM of 3 biological replicates.
(G) Fold change of ChIP-qPCR signal for 53BP1 as in Figure 3F. Error bars represent SEM of 3 biological replicates.
We next sought to determine how the rDNA itself is organized following DSBs. Using rDNA FISH we observe the redistribution of rDNA repeats into nucleolar cap-like structures that was dependent on DSBs and on ATM activity (Figure 3E). ChIP-qPCR showed that the UBF was distributed throughout the rDNA repeat as previously described (O'Sullivan et al., 2002), and the distribution pattern of UBF was largely unchanged in the presence of DSBs with or without ATM inhibition (Figure 3F). Conversely, 53BP1 localization was induced by DSBs, a response that was mildly reduced by inhibition of ATM (Figure 3G). Together these data demonstrate large-scale alterations in rDNA chromatin occur following DSBs and that this dynamic is controlled by ATM kinase activity.
Redistribution of Nucleolar DSBs into Nucleolar Caps Facilitates DSB-Recognition
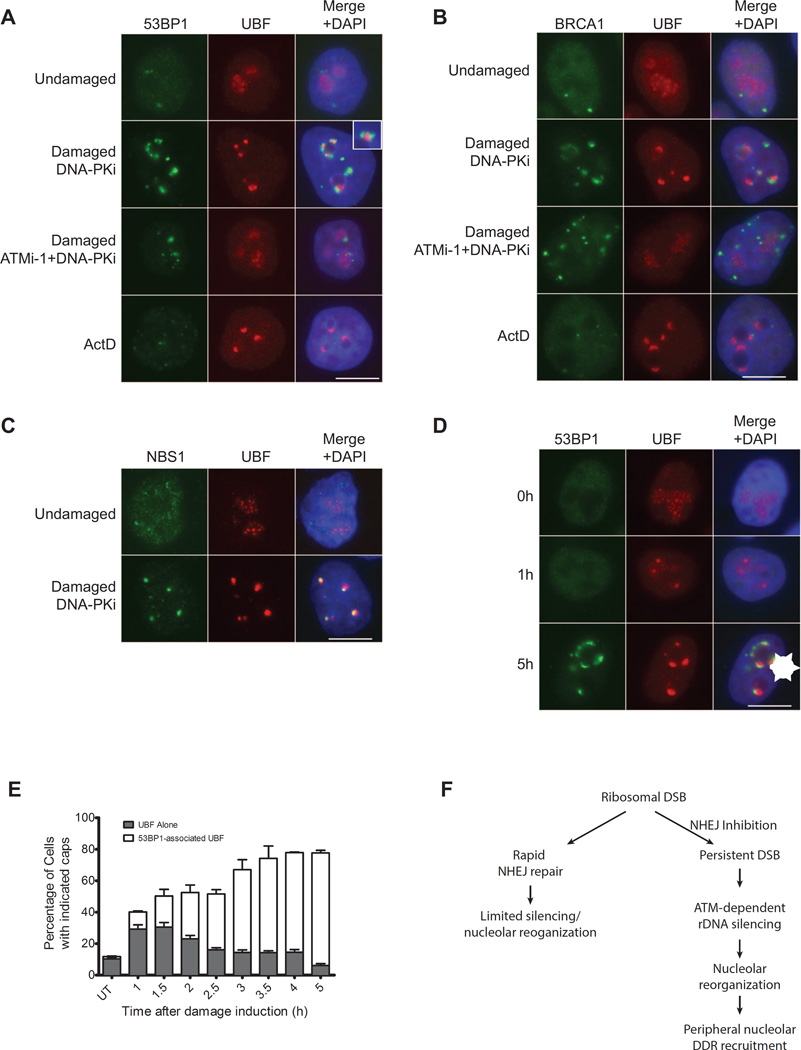
The redistribution of nucleolar DSBs into caps in response to ATM dependent silencing suggested that this movement facilitates recognition of the breaks by the DDR. Indeed, initial observations indicate that 53BP1 does not localize to the interior of the nucleolus but instead only reaches a detectable level when the breaks are relocated to the nucleolar periphery (see Figures 1B and S4). Co-staining of damaged cells with 53BP1 and UBF shows that they visibly juxtapose only in the nucleolar periphery (Figure 4A and inset). We also observe BRCA1 and Rad51 associate with UBF caps (Figure 4B and S4A). Unlike 53BP1 and BRCA1, NBS1 colocalized with UBF, suggesting that the DSB response distributes unevenly through the rDNA repeats at nucleolar caps (Figure 4C). This is not unlike the response to single DSBs outside of rDNA where the DNA damage machinery distributes in a non-uniform manner across DSB-flanking chromatin (Berkovich et al., 2007; Goldstein and Kastan, 2015).
Figure 4. Nucleolar DSBs are recognized following ATM-dependent nucleolar cap formation.
(A) Immunofluorescence staining of 53BP1 (green) and UBF (red). Merged images were overlaid with DAPI-stained nuclei. Inset depicts a 2× digital zoom of 53BP1 juxtaposition to UBF at nucleolar caps. Representative images of 3 biological replicates are shown. Scale bar is 10µm.
(B) Immunofluorecence of BRCA1 (green) and UBF (red) as in Figure 4A.
(C) Immunofluorecence of NBS1 (green) and UBF (red) as in Figure 4A.
(D) Immunofluorescence of 53BP1 (green) and UBF (red) at 0hr, 1 hr and 5 hr following I-PpoI induction in the presence of DNA-PK inhibitor. An arrowhead indicates coalesced UBF. An asterisk indicates 53BP1-associated UBF nucleolar caps. Scale bar is 10µm.
(E) Quantification of Figure 4E. Cells displaying punctate non-53BP1-associated UBF (Arrowhead in Figure 4D) and 53BP1-associated UBF nucleolar caps (Asterisk in Figure 4D) were quantified at indicated times following I-PpoI induction in the presence of DNA-PK inhibitor. Error bars represent SEM for 3 biological replicates.
(F) Schematic of rDNA DSB silencing. As described in the text, persistent DSBs exact ATM dependent nucleolar reorganization and DSB recognition subsequent to transcriptional silencing of rDNA loci.
To determine the temporal relationship between nucleolar reorganization and DSB recognition, we performed a timecourse for UBF cap localization and 53BP1 association. At early times following break induction we observed concentrated UBF structures that were generally internal to the nucleolus and did not associate with 53BP1 (Figure 4D). By 5 hours the majority of cells exhibited UBF caps that associated with 53BP1 at the nucleolar periphery (Figure 4D). We did not observe 53BP1 association with UBF inside the nucleolus throughout the course of the experiment. Quantification of this phenomenon indicates a redistribution of UBF before movement to the periphery where DNA damage response factors can recognize the lesions (Figure 4E). Time-lapse microscopy allowed dynamic visualization of this initial coalescing of UBF followed by movement to the nucleolar periphery (Figure S4B). These data demonstrate that large-scale alterations in nucleolar architecture accompany DSB silencing and that this facilitates ATM dependent temporal and spatial control of the DDR.
Together these results suggest a model where rDNA silencing is most efficient when rapid NHEJ repair is inhibited (Figure 4F). In this context persistent DSBs lead to ATM-dependent transcriptional silencing and reorganization of nucleolar proteins and chromatin into caps where the DNA damage machinery is efficiently recruited.
Discussion
The process of DSB-induced transcriptional silencing has been examined in a variety of cellular contexts. Here, we illustrate complex and multidirectional relationships between the DDR and transcription. Rapid repair preserves transcriptional output, while ATM kinase dependent signaling silences transcription in the face of prolonged DSBs (Fig 4). The DSB response is reversed by multiple phosphatases, deubiquitylating enzymes and the proteasome (Panier and Durocher, 2013). This reversibility fits well with the prior observation of a rapid recovery of transcription following the termination of nuclease activity (Shanbhag et al., 2010). We propose that rapid breakage-repair cycles do not provide sufficient time for the damage response to fully activate DSB silencing at a given locus. Perhaps a threshold of steady state breaks is required to efficiently silence repetitive loci such as the rDNA. This cumulative DSB signaling may be akin to checkpoint activation, which is estimated to require ~20 DSBs to efficiently prevent entry into mitosis (Deckbar et al., 2007).
DSB repair predominantly occurs through a combination of HR and NHEJ mechanisms. A recent elegant study reported similar findings on the ATM-driven DSB silencing of rDNA and the redistribution of nucleolar components into caps (van Sluis and McStay, 2015). The authors observe localization of HR factors and DNA synthesis at nucleolar caps suggesting that HR repairs these breaks. We also observe similar phenomena of HR protein localization at all cell cycle stages (data not shown). However, using several assays to directly measure DSBs, only inhibition of NHEJ appreciably increases steady state DSB levels (Figures 2 and S2). Additionally, deficiency in NHEJ, but not HR, led to an increase in the fraction of cells with nucelolar caps (Figure 3B). We do not exclude a role for HR in rDNA repair; rather our data supports the conclusion that NHEJ is the predominant repair mechanism, as it is for DSB repair elsewhere in the genome (Rothkamm, et al., 2003).
ATM dependent DSB silencing has been reported by numerous independent studies at PolI and PolII dependent loci (Ciccia et al., 2014; Kruhlak et al., 2007; Kakarougkas et al., 2014b; Larsen et al., 2014; Shanbhag et al., 2010; van Sluis and McStay, 2015; Ui et al., 2015). The related kinase ATR is required for the physiologic DSB silencing phenomena meiotic sex chromosome inactivation (Turner, 2007) as well as silencing of stalled replication forks (Im et al., 2014). Interestingly, a singular report suggests that DNA-PK directly induces transcriptional silencing at I-PpoI induced DSBs (Pankotai et al., 2012). In contrast to ATM and ATR, which prevent silencing distal to DSBs, DNA-PK was stated to prevent traversal of RNA-PolII transcription across a DSB by undefined mechanisms. Our results reveal the opposite at I-PpoI breaks within the rDNA. DNA-PK loss enhanced rather than mitigated transcriptional silencing in the rDNA locus, consistent with other reports of ATM dependent silencing (Kruhlak et al., 2007). Collectively, these results raise the possibility of differential usage of PIKKs to silence transcription under various scenarios. It will therefore be important to reexamine the relative usage of each PIKK using genome wide approaches.
Together with a recent report (Van sluis and McStay, 2015), this study implicates ATM in large-scale nuclear architecture changes in response to DSBs in mammalian cells. Previously, dynamic chromatin movements following DSB induction have been described, some of which are ATM-dependent (Dion and Gasser, 2013). In mammalian cells that utilize alternative lengthening of telomeres, a meiotic-like HR mechanism drives telomere movement (Cho et al., 2014). The primary mode of DSB driven rDNA reorganization in yeast is also HR (Torres-Rosell et al., 2007). Surprisingly, loss of HR did not influence nucelolar reorganization and cap formation following I-PpoI induction. This may be due in part to the propensity of these loci to be repaired by NHEJ. Our data also suggest that some fraction of breaks is rapidly repaired by NHEJ before relocalization is induced. The relocalization to the periphery of persistent DSBs may serve to activate other aspects of the DDR such as cell cycle checkpoints or HR. Inhibition of canonical DSB repair mechanisms did not recapitulate ATM loss with respect to silencing, highlighting the unique and multifaceted roles of ATM in DNA damage response coordination. The cellular targets of ATM are broad and encompass proteins involved in not only DSB recognition and repair, but also in protein trafficking, RNA processing and transcription (Matsuoka et al., 2007). How these events influence genome integrity remains an open question and may provide unique insights into physiologic function of ATM.
Experimental Procedures
Cell culture, immunofluorescence, RT-qPCR and Southern blotting were performed using standard methods; antibodies, primers and probe sequences are found in Supplemental Experimental Procedures. Comet assays were performed under neutral conditions using the Trevigen CometAssay Kit according to manufacturers directions and analyzed using OpenComet software on ImageJ (Gyori et al., 2014). ChIP-qPCR assays were performed as described previously (Tang et al., 2013). RNA-FISH was performed essentially as described (Fannucchi et al., 2013). Detailed protocols can be found in the Supplemental Experimental Procedures.
PFGE in MCF10A and MEFs
Plugs were prepared in 1% low melting agarose/PBS from 0.5–1×106 cells. After overnight digestion in digestion buffer (100mM EDTA, 0.2% Sodium deoxycholate, 1% Sodium laurel sarcosine, 1mg/mL Proteinase K) at 50°C plugs were washed 3×30 minutes in TE then 1×30 minutes in 0.5× TBE. Plugs were run on a 1% agarose/0.5× TBE gels for 18 hours at 14°C with 6V/cm and 5s switch times on a CHEF DRII apparatus (Bio-Rad). The Low Range PFG Marker (New England Biolabs) was used as a size standard. Gels were dried, denatured and hybridized overnight at 42°C using oligonucleotide probes indicated in Supplementary Table 1 that were end-labeled with PNK and 32P-ATP. Gels were imaged as described for southern blots (see supplemental procedures). Using the gel analysis tools in ImageJ the quantity of DNA released into the gel and retained in the wells was quantified and a ratio was calculated as unbroken:broken rDNA repeats.
Supplementary Material
List of Changes.
The reviewers indicated that no further changes were necessary.
We have removed all priority claims as recommended by the editor.
We corrected minor grammatical errors in the text that do not impact the conclusions of the manuscript.
We combined Supplemental Figures 1 and 2 to conform to the requirement that no more supplemental figures be submitted than the number of main figures.
Acknowledgements
We thank S. Franco (Johns Hopkins Medical School), A. Nussenzweig (NCI), S. Zha (Columbia Medical School) for providing MEF cell lines, and H. Winters for technical support. This work was supported by NIH grants GM101149, CA138835, and CA17494, (to RAG), who is also supported by funds from the Abramson Family Cancer Research Institute and Basser Research Center for BRCA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
S.M.H. designed and performed experiments and wrote the manuscript. J.A.B. performed and analyzed DNA FISH experiments. R.A.G. designed and supervised experiments and wrote the manuscript.
References
- Berkovich E, Monnat RJ, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert F-M, Lamond AI. The Nucleolus under Stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NW, Dilley RL, Lampson MA, Greenberg RA. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell. 2014;159:108–121. doi: 10.1016/j.cell.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Huang J-W, Izhar L, Sowa ME, Harper JW, Elledge SJ. Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proc. Natl. Acad. Sci. U.S.a. 2014;111:18631–18636. doi: 10.1073/pnas.1422488112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckbar D, Birraux J, Krempler A, Tchouandong L, Beucher A, Walker S, Stiff T, Jeggo P, Löbrich M. Chromosome breakage after G2 checkpoint release. J. Cell Biol. 2007;176:749–755. doi: 10.1083/jcb.200612047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V, Gasser SM. Chromatin Movement in the Maintenance of Genome Stability. Cell. 2013;152:1355–1364. doi: 10.1016/j.cell.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Goldstein M, Kastan MB. Repair versus checkpoint functions of Brca1 are differentially regulated by site of chromatin binding. Cancer Res. canres. 2015 doi: 10.1158/0008-5472.CAN-15-0400. 0400.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannucchi S, Shibayama Y, Burd S, Weinberg MS, Mhalanga M. Chromosomal contact permits transcription between coregulated genes. Cell. 2013;155:606–620. doi: 10.1016/j.cell.2013.09.051. [DOI] [PubMed] [Google Scholar]
- Grummt I. The nucleolus—guardian of cellular homeostasis and genome integrity. Chromosoma. 2013;122:487–497. doi: 10.1007/s00412-013-0430-0. [DOI] [PubMed] [Google Scholar]
- Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, Clement M-V. OpenComet: An automated tool for comet assay image analysis. Redox Biology. 2014;2:457–465. doi: 10.1016/j.redox.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis. 2010;31:955–960. doi: 10.1093/carcin/bgq064. [DOI] [PubMed] [Google Scholar]
- Huang S, Rothblum LI, Chen D. Ribosomal chromatin organization. Biochem. Cell Biol. 2006;84:444–439. doi: 10.1139/o06-089. [DOI] [PubMed] [Google Scholar]
- Im J-S, Keaton M, Lee KY, Kumar P, Park J, Dutta A. ATR checkpoint kinase and CRL1βTRCP collaborate to degrade ASF1a and thus repress genes overlapping with clusters of stalled replication forks. Genes and Development. 2014;28:875–887. doi: 10.1101/gad.239194.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Künzel J, Löbrich M, Jeggo PA, Downs JA. Requirement for PBAF in Transcriptional Repression and Repair at DNA Breaks in Actively Transcribed Regions of Chromatin. Mol. Cell. 2014b;55:723–732. doi: 10.1016/j.molcel.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montaño C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nat Cell Biol. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- Larsen DH, Hari F, Clapperton JA, Gwerder M, Gutsche K, Altmeyer M, Jungmichel S, Toledo LI, Fink D, Rask M-B, et al. The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat Cell Biol. 2014;16:792–803. doi: 10.1038/ncb3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- O'Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Boulton SJ. Double strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Panier S, Durocher D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol. 2013;14:661–672. doi: 10.1038/nrm3659. [DOI] [PubMed] [Google Scholar]
- Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- Prieto J-L, McStay B. Pseudo-NORs: a novel model for studying nucleoli. Biochim. Biophys. Acta. 2008;1783:2116–2123. doi: 10.1016/j.bbamcr.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds RC, Montgomery PO, Hughes B. Nucleolar “Caps” Produced by Actinomycin D. Cancer Res. 1964;24:1269–1277. [PubMed] [Google Scholar]
- Russell J, Zomerdijk JCBM. The RNA polymerase I transcription machinery. Biochem. Soc. Symp. 2006:203–216. doi: 10.1042/bss0730203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragón L, Lisby M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- Turner JMA. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Ui A, Nagaura Y, Yasui A. Transcriptional Elongation Factor ENL Phosphorylated by ATM Recruits Polycomb and Switches Off Transcription for DSB Repair. Mol. Cell. 2015;58:468–482. doi: 10.1016/j.molcel.2015.03.023. [DOI] [PubMed] [Google Scholar]
- Williams GJ, Hammel M, Radhakrishnan SK, Ramsden D, Lees-Miller SP, Tainer JA. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst.) 2014;17:110–120. doi: 10.1016/j.dnarep.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.