Abstract
Aurora B, a member of the Aurora family of serine/threonine protein kinases, is a key player in chromosome segregation. As part of a macromolecular complex known as the chromosome passenger complex, Aurora B concentrates early during mitosis in the proximity of centromeres and kinetochores, the sites of attachment of chromosomes to spindle microtubules. There, it contributes to a number of processes that impart fidelity to cell division, including kinetochore stabilization, kinetochore–microtubule attachment, and the regulation of a surveillance mechanism named the spindle assembly checkpoint. In the regulation of these processes, Aurora B is the fulcrum of a remarkably complex network of interactions that feed back on its localization and activation state. In this review, we discuss the multiple roles of Aurora B during mitosis, focusing in particular on its role at centromeres and kinetochores. Many details of the network of interactions at these locations remain poorly understood, and we focus here on several crucial outstanding questions.
Keywords: centromere, kinetochore, spindle assembly checkpoint, kinase, phosphatase, Aurora B, chromosome passenger complex, bi-orientation
General Remarks
Cells executing mitosis are challenged in ways that can jeopardize their viability and survival (1). The duplicated chromosome pairs (sister chromatids) in the mother cell need to orient on the mitotic spindle so that they can be equally distributed to the two daughter cells after the cohesion that holds them together is dissolved at the metaphase-to-anaphase transition. This process of “bi-orientation” requires that the sister chromatids establish stable “end-on” interactions with microtubules emanating from opposite poles of the mitotic spindle (Figures 1A,B) (2–4). Sister chromatids that fail to bi-orient are mis-segregated into the wrong daughter cell, or separated from the bulk of correctly segregated chromosomes forming the primary nucleus of daughter cells and secluded into extra-nuclear structures called micronuclei. Either fate of mis-oriented chromosomes can have dire consequences for cell physiology (5, 6).
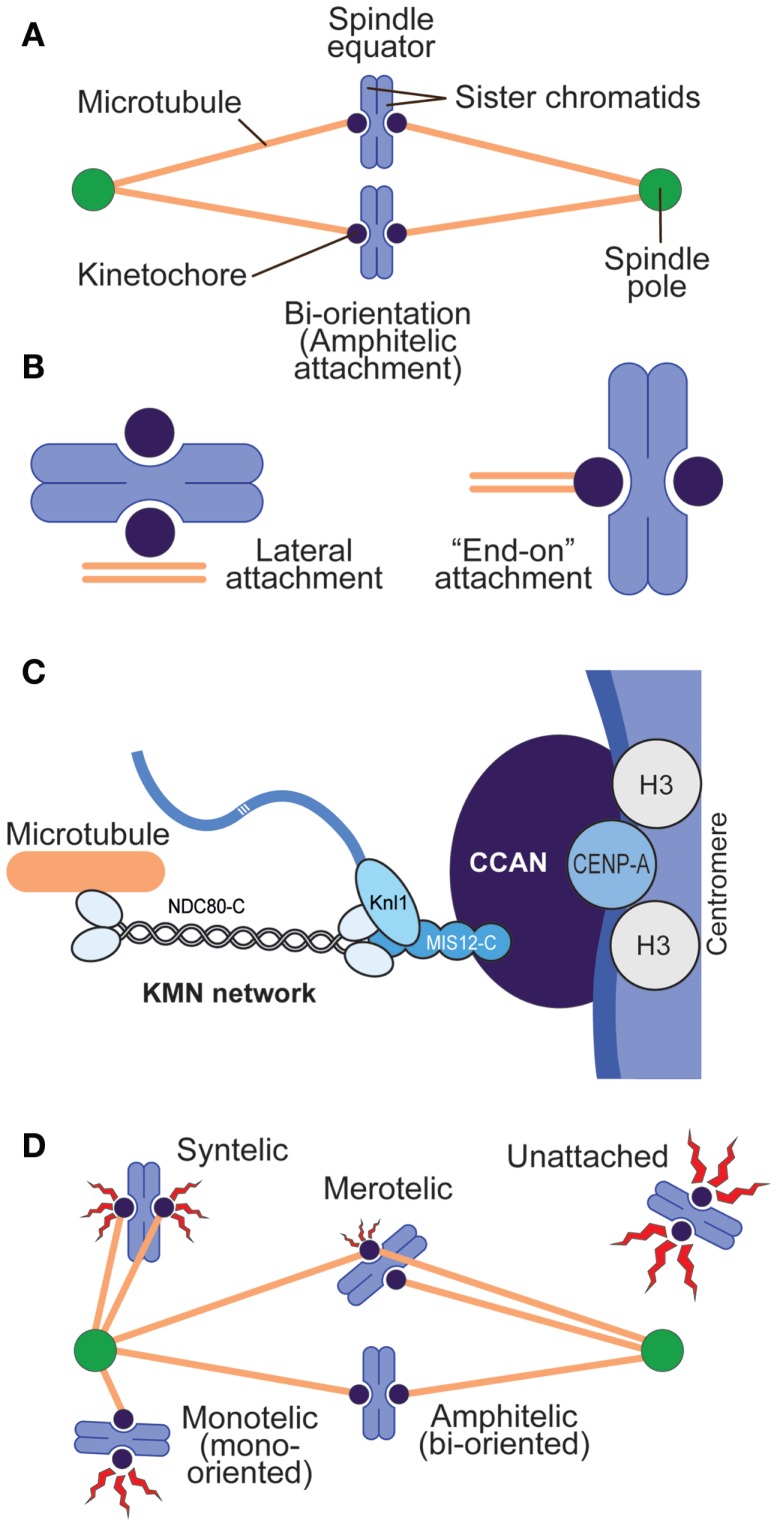
Figure 1.
Chromosome–spindle interactions. (A) A simple spindle with two chromosomes at metaphase. When chromosomes are bi-oriented, the sister chromatids are attached to microtubules, and the microtubules point to opposite spindle poles. (B) Two main modes of kinetochore–microtubule attachment predominate in mitosis. Lateral attachment (left) to the microtubule lattice (as opposed to the microtubule end) is typical of early phases of chromosome congression to the equatorial plane of the mitotic spindle and may not fully engage the core kinetochore machinery devoted to microtubule binding but rather molecular motors (7). “End-on” attachment (right) is typical of the final stages of attachment and involves core kinetochore machinery. (C) Schematic depiction of centromeres and kinetochores. Centromeres host CENP-A, the histone H3 variant, at much higher levels than other segments of the chromosome. CENP-A binds to a subset of 16 or 17 CCAN subunits, collectively represented as a blue oval. The KMN network binds directly to microtubules. (D) Various types of kinetochore–microtubule attachment modes, including erroneous attachments that require correction and that will engage the spindle assembly checkpoint (red flashes). Different “offenses” may provide a graded checkpoint response (variable size of the red flash), with lack of attachment providing a more robust response and merotelic attachment a weak one.
Aurora B is a member of the Aurora family of Serine/Threonine (S/T) protein kinases. Originally discovered as a gene required for maintenance of ploidy in Saccharomyces cerevisiae and named increase in ploidy-1 (IPL1) (8), Aurora B was later found to control several aspects of chromosome segregation in all eukaryotes (9–11). Two additional members of the Aurora family named Aurora A and Aurora C exist in mammals (12, 13). Substrates of these Aurora kinases usually conform to the consensus [RK]-[RK]-X-[TS]-Θ, where X is any residue and Θ is a hydrophobic or aromatic residue. For instance, Aurora B phosphorylates human KNL1 (CASC5) on the RRVSF motif, which, in the non-phosphorylated version, is a recruitment motif for protein phosphatase 1 (PP1) (14–17). Broad analyses of Aurora B substrates and consensus target sequences have been reported (18, 19). Although Aurora kinases share a similar consensus, distinct subcellular localizations ensure that they deliver activity to distinct substrates and regulate different aspects of mitosis (19, 20).
In this review, we discuss the role of Aurora B in the regulation of chromosome segregation, focusing in particular on the roles of Aurora B during prometaphase, the phase of mitosis during which chromosomes attempt to create stable interactions with spindle microtubules. Readers are also referred to comprehensive reviews that discuss the role of Aurora B also in other phases of mitosis (20, 21).
Introductory Concepts I: Centromeres and Kinetochores
Sister chromatids interact with spindle microtubules through specialized and structurally complex protein assemblies known as kinetochores (22, 23). On each chromosome, the kinetochore is established on a unique genetic locus named the centromere (Figure 1C). Centromeres, which may consist of several million base pairs of DNA in metazoans, are specialized chromatin domains whose hallmark is the enrichment of the histone H3 variant CENP-A (also known as CenH3) (24). At centromeres, CENP-A containing nucleosomes are embedded in histone H3-containing chromatin at a ratio that, even if estimated to be as little as 1 CENP-A nucleosomes over 25 H3 nucleosomes, is greatly superior to that in bulk chromatin (25).
CENP-A acts as a platform for the recruitment of kinetochore proteins collectively defined as the constitutive centromere-associated network (CCAN), most of which localize at centromeres during the entire cell cycle (26). These proteins form the so-called “inner kinetochore.” Upon entry into mitosis, an additional protein complex, the Knl1 complex–Mis12 complex–Ndc80 complex (KMN) network, is recruited to the CCAN. The KMN network in the “outer kinetochore” interacts directly with spindle microtubules (27) (Figure 1C).
Introductory Concepts II: Error Correction and the Spindle Assembly Checkpoint
Two feedback mechanisms control the process of kinetochore–microtubule attachment during mitosis, and Aurora B contributes decisively to both of them. These pathways are named error correction (EC) and spindle assembly checkpoint (SAC, also known as mitotic checkpoint, metaphase checkpoint, or “wait anaphase signal”). Error correction is a “local” mechanism that allows kinetochores selectively to stabilize interactions with microtubules that drive chromosome bi-orientation and to weaken those interactions that do not, such as the erroneous configurations known as syntelic and merotelic attachment (Figure 1D) (28). This description of EC summarizes the interpretation of pioneering chromosome micromanipulation experiments carried out over 45 years ago by Bruce Nicklas (29), but is nothing more than a statement of fact, partly because we are still far from a full molecular comprehension of EC. EC is believed to depend on the ability of the kinetochore–centromere system to detect tension, associated with bi-orientation, or lack of tension, associated with lack of bi-orientation. While tension at the bi-oriented chromatids suppresses error correction, lack of tension may not necessarily require error correction (e.g., when lack of tension is due to lack of attachment), but it will require it when kinetochores that are bound to microtubules fail to build tension (as in the case of syntelic or merotelic attachments, Figure 1D).
The KMN complex captures dynamic microtubules to create load-bearing attachments (30, 31). For error correction to occur, kinetochore (KMN)–microtubule interactions need to be sufficiently dynamic to allow the destabilization of erroneous attachments. Aurora B is a key component of the error correction machinery (32, 33). Aurora B inhibition through small-molecule inhibitors or inhibitory antibodies stabilizes incorrect attachments (32–36). Conversely, Aurora B overexpression causes continuous disruption of KT–MT attachments (37), while Aurora B re-activation allows the selective destabilization of incorrect attachments (38–40). Many of the proteins at the interface with microtubules are Aurora B substrates (41).
Similarly to the EC, the SAC also requires kinetochores (42, 43). In contrast to the EC, however, the SAC has the ability to extend into a “global” signal that diffuses away from kinetochores and prevents mitotic exit in the presence of even a single unattached or improperly attached kinetochore (44). The SAC pathway converges on the assembly of a checkpoint effector complex, the mitotic checkpoint complex (MCC), which targets and inhibits the anaphase-promoting complex or cyclosome (APC/C, Figure 2A). The activity of this ubiquitin (Ub) ligase targets Cyclin B and Securin, which are, respectively, the activator of the main mitotic “engine”, the Cdk1 kinase, and a stoichiometric inhibitor of the protease Separase, which is required for dissolution of sister chromatid cohesion. Proteasome-dependent destruction of Cyclin B and Securin upon their ubiquitination by the APC/C inactivates Cdk1 and activates Separase, respectively, triggering mitotic exit and sister chromatid separation (Figure 2A) (4, 45). Cells in which the checkpoint is altered or artificially inactivated undergo precocious mitotic exit in the presence of unattached or incorrectly attached chromosomes and are therefore prone to mis-segregation events (44).
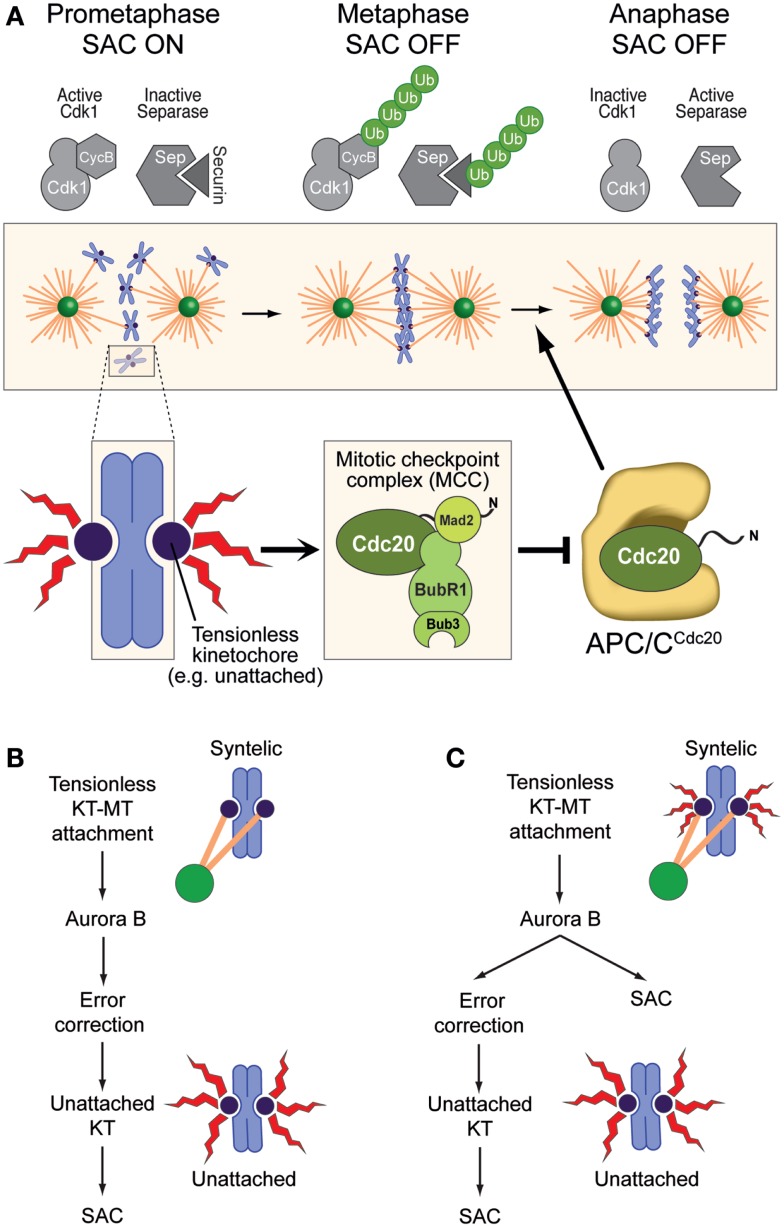
Figure 2.
The spindle assembly checkpoint and error correction. (A) The SAC pathway originates at kinetochores and converges, through several steps, on the assembly of the mitotic checkpoint complex (MCC), which acts as the SAC effector. MCC has been proposed to target the complex of APC/C pre-bound to a second molecule of Cdc20 (which can act both as an APC/C co-activator and as an MCC subunit) (46, 47). APC/CCdc20 promotes poly-ubiquitylation (Ub) of Cyclin B and Securin, promoting mitotic exit and separation of sister chromatids. MCC inhibits this activity of APC/CCdc20 until all chromosomes have achieved bi-orientation, at which point the SAC becomes “satisfied” (it subsides). (B) In this model of Aurora B function, any kinetochore–microtubule interaction, even if erroneous, satisfies the SAC. Aurora B is not a SAC component, but its ability to recognize and correct improper attachment makes it activate the SAC indirectly through generation of unattached kinetochores (as an intermediate in error correction), which are considered the only source of SAC signal. (C) In this alternative model, any tensionless kinetochore is a source of SAC signal, albeit of different signal strengths (size of the red flashes). Aurora B is directly required both for error correction and for the SAC. (B,C) were adapted from Ref. (48).
The general role of Aurora B activity in the EC and the SAC has been widely debated (28, 49). Early models based on experiments with attenuated alleles of Aurora B or at non-saturating doses of small-molecule inhibitors identified in Aurora B an exclusive component of the EC machinery (Figure 2B). According to these models, Aurora B contributed indirectly to SAC activation by generating unattached kinetochores, which became identified as the only structures capable of activating the SAC (32, 50).
This view has been progressively revised, partly because the molecular evidence in favor of a direct role of Aurora B in SAC control has been growing (34–36, 48, 51–54) and partly because there has been a conceptual evolution regarding what the SAC may be monitoring at kinetochores, with a shift from a pure “microtubule occupancy” model to an “intra-kinetochore tension” model (41, 55–58) (Figure 2C). Importantly, results obtained with different experimental approaches have caused the community to oscillate in their preference for a model or the other. However, a full assessment of the virtues and shortcomings of these models remains out of reach, as the molecular understanding of the conditions that lead to EC and SAC activation or silencing remains rudimentary, at least in relation to the considerable complexity of the process. Furthermore, while these two pathways are separable in their downstream components, they may be largely or even completely non-separable in the sensory apparatus that activates or switches them off at the “outer kinetochore,” where they operate. For instance, cyclin-dependent kinase 1 (Cdk1), Aurora B, monopolar spindle 1 (Mps1), and budding uninhibited by benzimidazoles 1 (Bub1), all mitotic protein kinases, are required to promote correct kinetochore–microtubule-binding configurations, as well as for the SAC response (59). They are likely to regulate both phenomena at the same time and from the same place, the kinetochores. In this review, we focus on some of the molecular details that implicate Aurora B in these two pathways.
Aurora B is a Subunit of the CPC
Aurora B kinase is embedded in a multi-protein complex known as the chromosome passenger complex (CPC), whose subunits are codependent for stability and localization (60–64). The three additional CPC subunits are named inner centromeric protein (INCENP, and known as Sli15p in yeast)], Survivin (Bir1p) and Borealin (also known as CSC-1, Dasra, and Nbl1p) (63, 65–69) (Figure 3A). The CPC consists of two functionally distinct modules (20): a module delivering the catalytic activity, composed of Aurora B and a ~50-residue segment at the C-terminal end of INCENP, the so-called IN-box (62, 67, 70–74); and a module mediating localization, consisting of a ~45-residue segment at the N-terminal end of INCENP, Survivin, and Borealin (64, 75–84). The two modules are connected by the central part of INCENP (Figure 3B and see below).
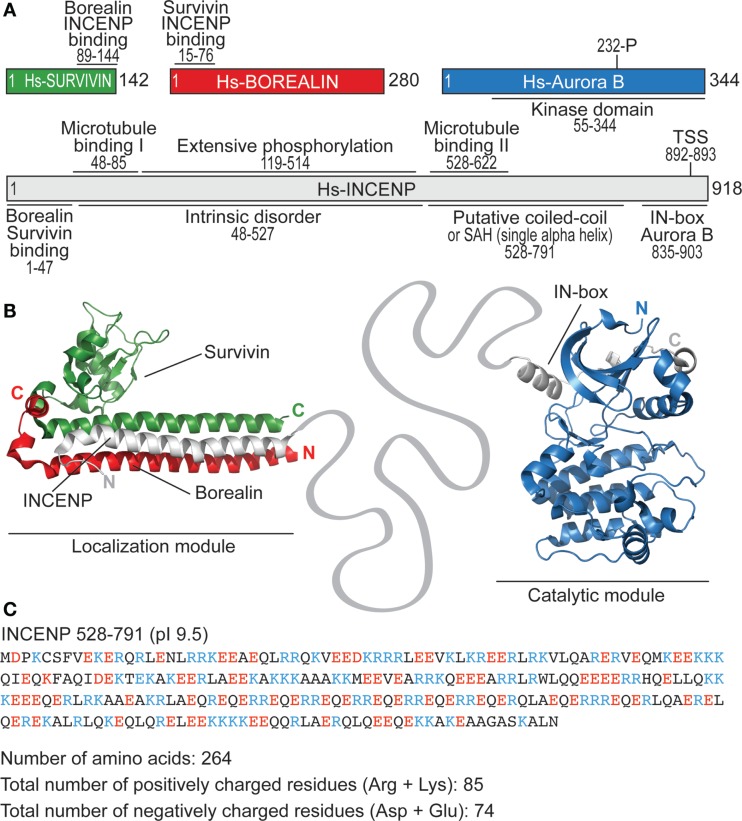
Figure 3.
Structural organization of the CPC. (A) Schematic representation of human CPC subunits with main structural features. (B) Structural organization of the localization module of the CPC (PDB ID 2QFA) (77) and of the catalytic module (PDB ID 2BFX) (70). The linker between the N- and C-terminal regions of INCENP encompasses more than 800 residues. (C) Sequence of the putative coiled-coil region of INCENP shows that its features are hardly compatible with coiled-coil folding (due to insufficient number and irregular spacing of hydrophobic residues). pI defines isoelectric point.
Activation of Aurora B kinase arises from multiple regulatory steps, including the binding of the IN-box of INCENP around the Aurora B active site, the Aurora B-mediated phosphorylation of the IN-box on a Thr-Ser-Ser (TSS) motif, and the auto-phosphorylation of Aurora B at Thr232 (abbreviated as AB-T232-P) in the activation segment (70, 72, 85). Thus, Aurora B activation resembles that of many other kinases, in that it requires interaction with a partner protein and phosphorylation. Phosphorylation at the TSS and at the Aurora B activation segment is likely to occur in trans (70) and may therefore be sensitive to the local concentration of the CPC (86, 87).
Besides the intrinsic mechanisms of regulation described above, Aurora B may also be controlled by extrinsic mechanisms. For instance, phosphorylation of Ser311 of Aurora B by checkpoint kinase 1 (Chk1) may promote catalytic activation of Aurora B near kinetochores (88, 89). In addition, phosphorylation of the CPC targeting subunit Borealin by Mps1 has also been proposed to regulate Aurora B activity (90), but this remains controversial as neither Aurora B nor its activity are grossly perturbed by Mps1 inhibition (91–93). Furthermore, the protein TD-60 has been indicated as an additional CPC subunit required for CPC centromere targeting and Aurora B activation (94, 95). A recent study revealed that TD-60 is as a guanine nucleotide exchange factor (GEF) for the small Ras-like GTPase RalA, and that the latter modulates Aurora B activity and localization (96). While the mechanisms through which the RalA GEF activity of TD-60 influences Aurora B localization and activity requires further investigation, it seems now clear that TD-60 is not part of the CPC.
One of the most exciting chapters in the study of Aurora B kinase has been the development of highly specific and selective chemical inhibitors, spurred by the identification of this kinase as a potential target in oncology (97). Leaving clinical implications aside (98), small-molecule ATP-competitive inhibitors such as Hesperadin and ZM447439 proved invaluable tools for acute mitotic inhibition of Aurora B function and for the investigation of its mitotic functions in basic research laboratories (34, 35).
The Localization Module of the CPC: Centromere Localization and Beyond
The localization module targets the CPC to the centromere, where the bulk of the CPC localizes during mitosis. Crucial for centromere targeting is the CPC subunit Survivin, a member of a family of inhibitor of apoptosis (IAP) proteins containing a BIR domain (99). While Survivin might have lost its function as inhibitor of apoptosis, typical of other IAPs, its BIR domain has retained the ability of recognizing the N-terminus of target proteins. In Survivin, this ability is leveraged to bind a short N-terminal segment of Histone H3 (78, 80, 81) (Figure 4A). In fact, Survivin binds a short N-terminal segment of Histone H3 that must include a phosphorylated version of Thr3 (H3-T3-P) for efficient recognition (76, 78, 80, 81, 83, 84). The kinase responsible for this preeminently mitotic modification of Histone H3 is named Haspin (100). By recognizing H3-T3-P at centromeres, Survivin targets the CPC to the centromere (Figure 4B). Whether this H3-T3-P-dependent mechanism operates in yeast has remained unclear, because deletion of the yeast haspin-like kinases does not result in a growth defect phenotype (101). In yeast, a survivin-dependent mechanism may rely on the interaction of the Survivin homolog Bir1 with Ndc10, a subunit of the CBF3 centromeric complex (102, 103).
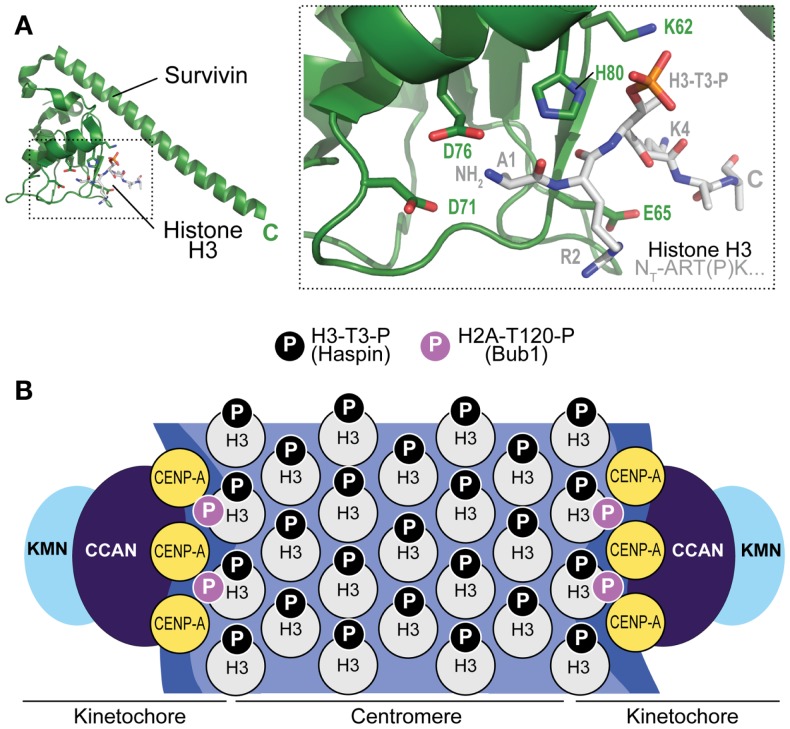
Figure 4.
Mechanism of CPC recruitment to centromeres. (A) Crystal structure of the complex of Survivin with a peptide encompassing the N-terminal region of Histone H3 (PDB ID 4A0J). The peptide has sequence Ala-Arg-Thr(P)-Lys, where (P) indicates that Thr3 is phosphorylated. Asp71 (D71) is implicated in the recognition of the free N-terminus of Ala2 (the N-terminal Met1 is removed by an aminopeptidase). (B) Haspin kinase phosphorylates Thr3 of histone H3 (H3-T3-P) in the centromere region to allow recruitment of the CPC. Bub1 kinase phosphorylates Histone H2A on Thr120 (H2A-T120-P) near kinetochores (i.e., the modification does not extend to centromeres). In principle, both H3-containing and CENP-A containing nucleosomes may contain this modification.
Besides H3-T3-P, also the phosphorylation of Thr120 of Histone H2A by Bub1 kinase (H2A-T120-P, H2A-S121-P in fission yeast) has been implicated in centromere recruitment of the CPC (81, 104, 105) (Figure 4B). The role of this mark, which is detected at kinetochores but not at centromeres, is more elusive. It appears to be crucial to regulate a homeostatic circuit that dynamically controls the activity and abundance of protein kinases, including Plk1 and Aurora B, and protein phosphatases, including members of the protein phosphatase 2A (PP2A) family associated with the B56 regulatory subunit (PP2A-B56), at kinetochores and centromeres. Specifically, H2A-T120-P is believed to promote recruitment of Shugoshin proteins (SGOL1 and SGOL2/TRIPIN in humans) (106, 107). These, in turn, control the recruitment of proteins that play a prominent role in error correction, including kinesin-13 family members such as MCAK, a microtubule depolymerase and Aurora B substrate, and the PP2A-B56 protein phosphatase complex, which balances abundance and activity of Aurora B and Plk1, as well as the phosphorylation of important CPC targets (108–118).
In addition to the recognition of histone marks, other mechanisms have been implicated in CPC centromere recruitment or activation, such as post-translational modifications of Survivin, Borealin, and INCENP, including phosphorylation by Aurora B itself, Cdk1, Mps1, and Plk1 (90, 119–123). Direct binding of Borealin to double-strand DNA has also been reported (64). Finally, there is evidence that oligomerization of the localization module also contributes to its localization (64, 124). How these features may impinge on CPC centromere targeting remains poorly understood.
Incenp, the Bridge Connecting the Two CPC Modules
The functional properties of INCENP that have remained mechanistically obscure are now starting to emerge. INCENP is a rather large protein (918 residues for Isoform 1 in humans; source Uniprot: http://www.uniprot.org) (Figure 3A). Large parts of the INCENP primary sequence are low-complexity and unlikely to adopt a defined three-dimensional tertiary (and even secondary) structure. A predicted coiled-coil between residues 528 and 791 of INCENP is often considered an exception. More careful scrutiny, however, leads to exclude that this segment of INCENP is a genuine coiled-coil. It contains too few hydrophobic residues to support coiled-coil oligomerization and its frequent stretches of positively and negatively charged residues (Figure 3C) may produce false-positive classifications as coiled-coils. Analysis of INCENP residues 528–791 with REPPER, a program that detects short repeats and predicts periodicities in protein sequences (125), suggests that it lacks the regular sequence pattern typical of coiled-coils. In agreement with this idea, a very recent study on avian INCENP showed that this region (residues 503–715, corresponding to residues 528–791 of human INCENP in Figure 3A) folds as a single alpha helix (SAH) domain, rather than as a coiled-coil (126). SAH can unfold reversibly under force, thus extending up to 2.5-fold over their rest length. Because the N-terminal part of the SAH domain contains a second microtubule-binding domain (126), in addition to the one already identified in the N-terminus of INCENP (residues 48–85 of the human protein) (60, 74, 127, 128), it is a potential candidate for regulation by microtubule attachment.
INCENP also contains a large disordered region (residues 48–527, Figure 3A), stuffed with phosphorylation sites. The UNIPROT (http://www.uniprot.org) reports at least 24 phosphorylation sites in residues 119–514 of human INCENP. Phosphorylation of Thr388 in this segment has been implicated in binding and targeting of Plk1 (129). As discussed more thoroughly below, Cdk1-dependent phosphorylation of Thr59 within this segment has important consequences for CPC localization.
The Spatial Separation Model of Aurora B Function
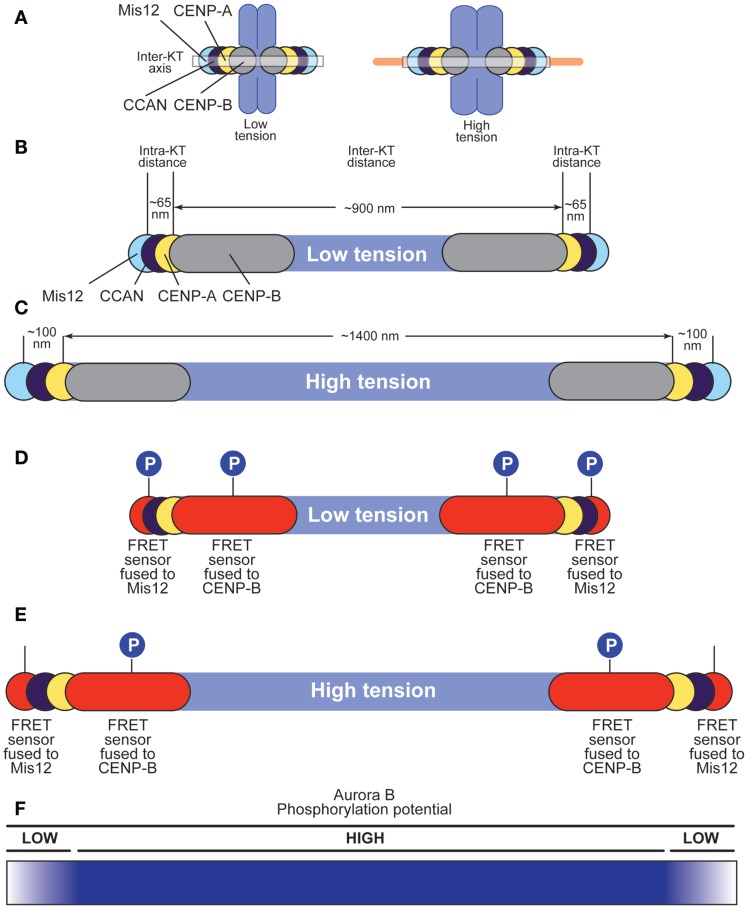
While it is clear that Aurora B substrates at centromeres and kinetochores undergo dynamic changes in their phosphorylation state during the relatively short time it takes kinetochores to attach to the spindle, it is uncertain to which extent these changes reflect the dynamic regulation of Aurora B activity by the intrinsic and extrinsic mechanisms discussed above (41). Rather, current models of Aurora B function focus primarily on the tension-dependent separation of Aurora B from its substrates as the basis of Aurora B regulation in EC and the SAC (41, 58). To appreciate this argument, it is important to explain the geometry of centromeres and kinetochores, their variation during bi-orientation, and how Aurora B may position itself within this system. The size of kinetochores is roughly equivalent to the wavelength of visible light, and in first approximation kinetochores appear as diffraction limited “spots” in the light microscope. In HeLa cells, the distance between the centroids of “spots” corresponding to inner kinetochore proteins in the two sister kinetochores (inter-kinetochore distance) grows from ~0.9 μm in the absence of microtubule binding in prometaphase (i.e., in the absence of tension) to ~1.4 μm upon bi-orientation at metaphase (when chromosomes are end-on-attached and under full tension) (130) (Figure 5A). Similar increases in inter-kinetochore distance have been measured in other cell types: inter-kinetochore distance increases from ~1.1 μm in the absence of microtubule binding to ~2.2 μm upon bi-orientation in newt lung cells (131), and from 0.72 to 0.94 μm in Drosophila S2 cells (57).
Figure 5.
Effects of tension on centromere and kinetochore structure. (A) Chromosome lacking tension (left) or under tension (right). The centroid of the distributions of proteins in the kinetochore, including CENP-B, CENP-A, CCAN subunits, and Mis12 (part of the KMN network) is represented as a circle along the inter-kinetochore axis. Each centroid has a defined coordinate along the axis (57). Microtubules cause changes in the position of the centroids. (B,C) Under low tension (B), inter-kinetochore distance in HeLa cells is ~900 nm (0.9 μm), whereas the distance between the centroids of the distributions of CENP-A and Mis12 is ~65 nm in Drosophila and as little as ~40 nm in human kinetochores (132). CENP-B binds the CENP-B box in alpha-satellite DNA at centromeres, and extends slightly beyond CENP-A toward the centromere (55). Under high tension (C), inter-kinetochore distance grows to 1400 nm (1.4 μm), whereas the distance between the centroids of the CENP-A and Mis12 distributions grows to 100 nm (57). (D,E) A FRET sensor responding to Aurora B phosphorylation was fused either to CENP-B or to Mis12 (55). Under low tension (D), the sensor is phosphorylated regardless of its position, suggesting that Aurora B can reach both positions with similar efficiency. Under high tension (E), the outermost sensor cannot be phosphorylated efficiently (possibly because it becomes dephosphorylated), whereas the innermost sensor continues to be phosphorylated. (F) The phosphorylation potential of Aurora B decays very rapidly after the position defined by the innermost FRET sensor (fused to CENP-B) when chromosomes are under stretch. This rapid decay takes place in ~200 nm or less along the inter-kinetochore axis.
Thus, tension introduces macroscopic changes in the organization of the inter-kinetochore space between sister kinetochores. Importantly, tension also modifies the internal structure of the kinetochore, a condition referred to as intra-kinetochore stretch (Figure 5A). In S2 cells, for instance, the span of the kinetochore [from CENP-A to the centromere-proximal end of the Ndc80 subunit (also known as Hec1), measured along the inter-kinetochore axis] is ~65 nm in the absence of tension, and 102 nm in the presence of tension (57) (Figures 5B,C). Similar tension-driven increases in stretch are observed within human and yeast kinetochores (132, 133). The precise structural changes underlying the establishment of intra-kinetochore tension, however, remain unknown.
The spatial separation model builds on the observation that an Aurora B FRET sensor shows constitutive, tension-independent phosphorylation when positioned close to Aurora B at the interface between the centromere and inner kinetochore, but tension-sensitive phosphorylation when positioned more distantly from the kinase (55) (Figures 5D,E). More specifically, Aurora B phosphorylates a FRET sensor located at the centromere–kinetochore interface (because fused to the CENP-B protein) regardless of attachment status and despite the very significant increase in inter-kinetochore distance upon bi-orientation. Conversely, Aurora B phosphorylates the same FRET sensor now fused to a subunit of the Mis12 complex, in the outer kinetochore, when chromosomes are not under tension, but does so less effectively when tension is present at metaphase (55). Analogous observations have been made with bona fide Aurora B substrates (41, 55–58, 130, 134) and Aurora B substrates located in the outer kinetochore become progressively dephosphorylated during the attachment process (134–136). Furthermore, artificial repositioning of Aurora B to the outer kinetochore prevents dephosphorylation of outer kinetochore substrates (55). The persistence of Aurora B phosphorylation on a sub-class of “proximal” substrates despite full tension suggests that the kinase activity of Aurora B may not per se be force dependent.
Thus, it appears that certain substrates, and in particular substrates in the KMN network that mediates the EC and SAC responses, become physically separated from the kinase as tension arises (41). In the absence of tension, such as in syntelic attachment, substrates remain phosphorylated and attachments intrinsically unstable. In conclusion, the “phosphorylation potential” of Aurora B is dampened with a sharp edge within the very short distance that separates centromeres from kinetochores under tension, whereas it is largely insensitive to the considerable degree of stretching of the inter-kinetochore region (Figure 5F). This clearly suggests that Aurora B is able to read intra-kinetochore tension rather than inter-kinetochore tension, but how it achieves this has remained unclear.
Reading Intra-Kinetochore Tension: From Centromeres?
It was initially hypothesized that the spatial separation that promotes stabilization of kinetochore–microtubule attachment might be linked to the increase in the distance from centromeres, where the bulk of Aurora B is positioned, to kinetochores, where the substrates of Aurora B that mediate microtubule attachment are located (33). With increasing distances, indicative of end-on attachment, the ability of Aurora B at centromeres to reach its substrates would progressively decrease, allowing a progressive stabilization of the kinetochore–microtubule interface. Implicit in this model is the existence of a sharp and separable centromere–kinetochore boundary, but a precise definition of what this boundary looks like is missing. If we consider that CENP-A is embedded in centromeric chromatin containing abundant histone H3 nucleosomes, H3-T3-P is likely to extend to the immediate periphery of kinetochores, and there is no obvious reason why the CPC should not bind to these H3 nucleosomes. It is unknown whether these nucleosomes become separated from CENP-A containing nucleosomes when tension builds up following microtubule end-on attachment to kinetochores.
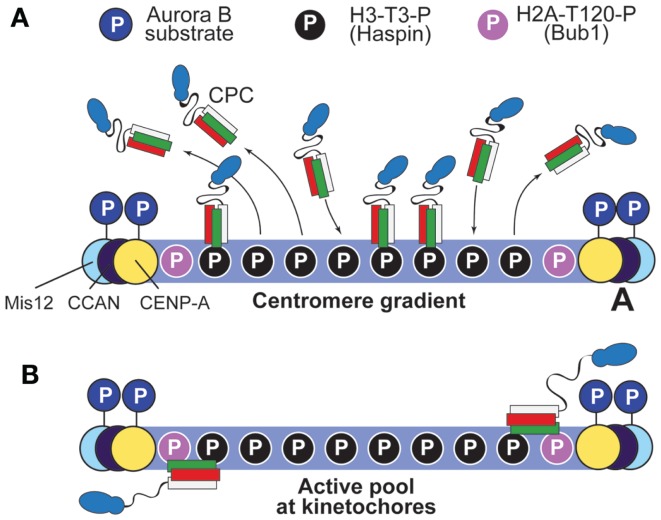
Because the subunits of the CPC turn over at centromeres with halftimes (t1/2) of <1 min (137–139), an alternative hypothesis is that a gradient of Aurora B substrate phosphorylation may be created by initial recruitment of the CPC to H3-T3-P to the centromere and by subsequent release and diffusion from centromeres (41) (Figure 6A). Indeed, this mechanism (“centromere gradient”) can create gradients of Aurora B substrate phosphorylation (140–142). We note, however, that such gradients are relatively flat and form over length scales of several micrometers (140–142). It is therefore unlikely that this diffusible gradient of Aurora B would generate the very sharp edge of activity observed within the ~100 nm (0.1 μm) length scale of the kinetochore.
Figure 6.
Two possible models to account for the gradient of Aurora B phosphorylation. (A) The “centromere gradient” model predicts that the CPC becomes recruited to centromeres as illustrated in Figure 4B, and it then dissociates from them, creating a gradient of CPC concentration (and therefore, by inference, of substrate phosphorylation). We note, however, that it is unlikely that this gradient could account for the sharp transition of phosphorylation potential of Aurora B within the very limited scale length of the kinetochore (see Figure 5). (B) An alternative model posits that an active form of the CPC is anchored near the kinetochore, and that the centromere pool is not strictly required for function (it was therefore omitted from the drawing). Proximity to H2A-T120-P might lead to the activation of this kinetochore pool of the CPC. Interactions with kinetochore subunits are also possible.
The idea that the phosphorylation gradient of Aurora B is created by centromere recruitment and release of the CPC is also at odds with the results from experiments in which centromere enrichment of the CPC was prevented by targeted mutations in CPC subunits. For example, a Survivin mutant impaired in its ability to bind H3-T3-P supports chromosome segregation and long-term viability of DT40 cells deprived of endogenous Survivin (82). Similarly, the deletion of residues 1–228 of Sli1 (Sli15ΔN) rescues the lethality of BIR1 in S. cerevisiae, even if Sli15ΔN does not localize to centromeres (see below) (101). Furthermore, phosphorylation of an inner kinetochore substrate of Aurora B, Ser7 of CENP-A (CENP-A-S7-P) is unaltered after depletion or inhibition of Haspin with 1 μM 5-ITu (5-iodotubercidin), a concentration of the drug that clears centromeres of H3-T3-P (78, 143). Altogether, these observations suggest that a gradient of Aurora B substrate phosphorylation at the centromere–kinetochore interface can be established also in the absence of Aurora B at centromeres.
… Or from Kinetochores?
An alternative hypothesis for Aurora B function is that the functionally relevant pool of Aurora B resides near or at kinetochores, rather than centromeres (22, 59). Strikingly, it was shown that centromeric accumulation of Aurora B is subordinate to kinetochore establishment. An ectopic kinetochore built at a chromosome site containing a Lac-O array, by tethering segments of the kinetochore CCAN subunits CENP-C or CENP-T, promotes accumulation of H3-T3-P and of the CPC in an area comprised between the two ectopic tethering sites, suggesting that the ectopic kinetochore dictates the position of the “centromere” (144). Remarkably, the CENP-C and CENP-T segments used in these experiments do not recruit CENP-A, suggesting that the latter is not required for CPC recruitment at the ectopic “centromere.” Establishment of this ectopic centromere likely involves kinetochore-associated Bub1, which may promote the recruitment of Sgo1. Sgo1, in turn, plays a crucial role in the establishment and protection of centromeric cohesion (104, 107, 115, 145). In S. cerevisiae, the core centromere (a ~125 bp segment on which the kinetochore is built) and two kinetochore proteins, Iml1 and Chl4 (respectively, related to the CCAN subunits CENP-L and CENP-N in humans), are important for the spreading of Sgo1 to pericentromeric regions (146).
Further emphasizing the role of kinetochores in CPC localization is the observation that the abundance of the centromere pool of Aurora B in diploid human cells is controlled dynamically by kinetochore attachment status, with misaligned chromosomes showing an enrichment of Aurora B (135). En passing, this dynamic kinetochore-driven enrichment of Aurora B at centromeres requires Aurora B and Plk1 activity, but not Mps1’s (135). The dispensability of Mps1 is further testified by experiments showing that chemical inhibition of Mps1 activity does not affect the total levels of H3-T3-P, the phosphomark that recruits the CPC to centromeres (124). However, Mps1 may modulate the timing of CPC accumulation at the centromere (147).
The role of kinetochores is further supported by the observation that Knl1, one of the outer kinetochore KMN subunits, is required for Aurora B activation, and that the active form of Aurora B (monitored through AB-T232-P) resides at kinetochores rather than at centromeres (148–150). Identifying the precise reason for this is a crucial question for future analyses. It is plausible that the centromere, contrary to the prometaphase kinetochore, represents a domain of high phosphatase activity that prevents the accumulation of the active form of Aurora B. The kinetochore pool of Aurora B was recently observed under conditions of Haspin inhibition, and was shown to depend on dimerization of Borealin (124).
Sli15ΔN, the Sli15 mutant discussed in the previous section, is also observed at kinetochores in cells depleted of Bir1/Survivin, suggesting that kinetochore localization of this mutant is survivin independent (101). Remarkably, while the Sli15ΔN mutant rescued chromosome segregation and the lethality associated with loss of Bir1/Survivin in S. cerevisiae, it was synthetic lethal with two normally non-essential CCAN subunits at the kinetochore, Mcm21 and Ctf19 (respectively, homologous to CENP-O and CENP-P of higher eukaryotes) (101, 151). These two proteins and their binding partners Okp1 and Ame1 (considered to be homologous to CENP-Q and CENP-U, respectively) have been previously shown to promote kinetochore recruitment of the CPC and to be necessary for the error correction activity of the CPC in S. cerevisiae (152–154).
How Aurora B is recruited to kinetochores is unclear, but a hint comes from the observation that INCENP is phosphorylated on Thr59 (INC-T59-P) by Cdk1 kinase, the master regulator of cell division (64, 129, 155). A phosphomimetic T59E mutant causes INCENP to persist on chromosomes rather than to become relocated on the central spindle at anaphase (155, 156). With the decline in tension upon sister chromatid dissolution in anaphase, the T59E mutant causes the re-activation of typical Aurora B-dependent events at kinetochores, including the recruitment of Mps1, Bub1, and BubR1. Thus, “stripping” of the CPC from centromeres might be required to prevent EC and SAC re-activation during anaphase. However, retention of CPC localization is per se not sufficient for a complete re-activation of these pathways (156–160).
Of note, both H3-T3-P and H2A-T120-P are removed from centromeres at anaphase (81, 161), suggesting that the T59E INCENP mutant may not be retained on chromosomes through these phosphomarks but via a different, currently uncharacterized interaction. Phosphorylation of INC-T59-P may prevent an interaction of INCENP with the MKLP2 kinesin, which is required to relocate the CPC to the central spindle at anaphase (156, 162, 163). Alternatively, it might mediate a direct interaction with one or more kinetochore subunits. This pathway is conserved in S. cerevisiae, where dephosphorylation of Cdk1-dependent sites on Sli15 was shown to be important for CPC relocation at anaphase (123).
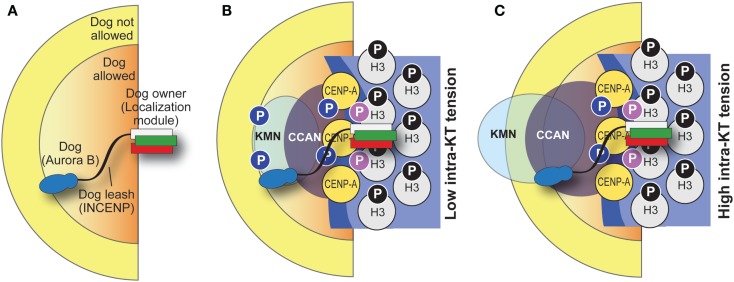
Thus, the active CPC pool that generates the intra-kinetochore phosphorylation gradient discussed above may reside within kinetochores, rather than being delivered there by a diffusible gradient of the kinase (Figure 6B). How does this kinetochore pool of the CPC generate the observed phosphorylation gradient within kinetochores? We have previously proposed that INCENP might act as a flexible arm whose maximal extension limits the reach of Aurora B within kinetochores (22). In this “dog leash” model, intra-kinetochore stretch promoted by microtubule binding might create relative movements of the Aurora B substrates relative to the tethered CPC, until substrates become unreachable by the kinase (Figure 7). As discussed above, the coiled-coil domain of INCENP has been recently shown to contain a SAH domain (126). In agreement with the “dog leash” model, it was shown that the length of the SAH domain modulates the ability of Aurora B to reach its substrates in the centromere and in the outer kinetochore (126).
Figure 7.
“Dog leash” model of CPC function. (A) The idea behind the “dog leash” model is that the localization module of the CPC (the owner) is tethered at the base of kinetochores. INCENP acts as a “dog leash” that allows the “dog,” Aurora B, to phosphorylate substrates only within limits defined by the length of the linker (which may vary, e.g., as a consequence of phosphorylation). This defines a boundary between regions where the dog is allowed and regions where it is not. (B,C) Application of the dog leash model to kinetochores. Under low tension (B), Aurora B can reach out in the kinetochore and phosphorylate substrates there. Under high tension (C), substrates (e.g., in the KMN network) have crossed the boundary defined by the leash and become unreachable. Note that in this drawing the CPC is tethered at the base of the kinetochore and its position is stationary, but this may not be the case and tension might increase its distance from the CENP-A base of the kinetochore. The function of a phosphatase is implicit in the model.
… Or from Microtubules?
Yet, another hypothesis is that Aurora B performs its functions from microtubules (101). This theory builds on previous work characterizing INCENP/Sli15 as a microtubule-binding protein (discussed above). Indeed, Sli15ΔN, the already discussed deletion mutant rescuing the lethality of the bir1 deletion in S. cerevisiae, localizes strongly to microtubules (101). Another recent study suggests that microtubules regulate Aurora B localization and activity in prometaphase (164). Albeit attractive, the hypothesis that microtubule localization is sufficient for CPC function requires further evaluation, not least because CPC function is delivered also in cells lacking microtubules altogether (e.g., because treated with spindle poisons). We reason that because the Sli15ΔN mutant retains kinetochores localization, the most parsimonious interpretation of its ability to suppress the lethality of the bir1 deletion is that it does so from kinetochores.
A Model for CPC Localization and Function
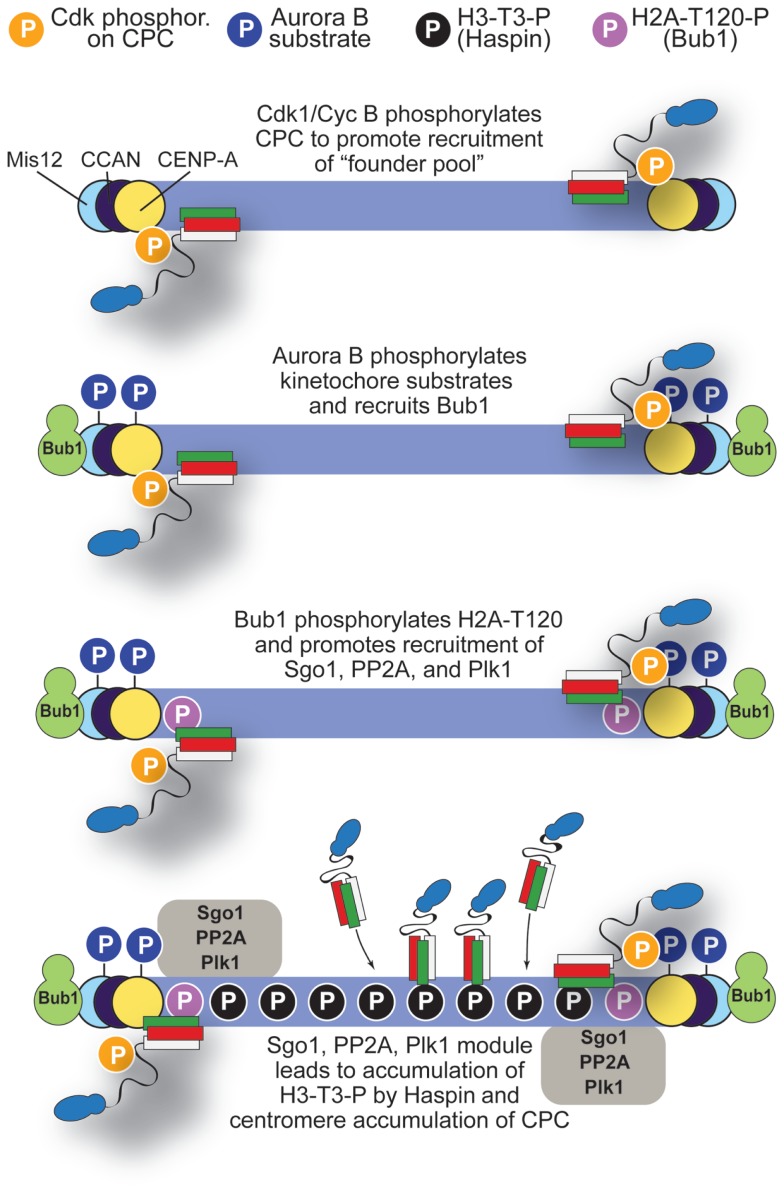
Based on the discussion above, we propose a tentative model for the mechanism of CPC localization (Figure 8). Cdk1-mediated phosphorylation of INCENP may be the initial trigger causing the recruitment of a pool of the CPC to kinetochores through interactions with yet to be identified subunits, possibly within the CCAN at the inner kinetochore. At kinetochores, Aurora B contributes to the recruitment of Bub1 kinase, which creates H2A-T120-P to recruit a kinetochore pool of Sgo1 (107, 115). The latter is responsible for the homeostatic control of phosphorylation at kinetochores through recruitment of PP2A phosphatase and Polo-like kinase 1 (Plk1). How these proteins interact at kinetochores is largely unclear and requires further analysis.
Figure 8.
A model for localization of CPC. The initial step in accumulation of CPC to centromeres may involve Cdk1/Cyclin B, which phosphorylates INCENP-T59 and possibly other sites on CPC. Initial CPC activity at kinetochores recruits Bub1, which in turn, after phosphorylating H2A-T120, promotes recruitment of Sgo1, PP2A, and Plk1. These, in turn, promote activation of Haspin and centromere accumulation of the CPC. See main text for additional detail.
We surmise that execution of this pathway may have two main consequences: (1) limiting the activation of Aurora B to the kinetochore pool and (2) igniting a positive feedback loop that promotes Haspin activation and further CPC accumulation at centromeres via phosphorylation of H3-T3-P in neighboring H3 nucleosomes. Aurora B itself, Plk1, Bub1, and, to a lesser extent, Mps1 may be involved in this positive feedback loop (62, 120, 121, 124, 135, 145, 147, 165). Although Bub1 acts downstream from Mps1 in the SAC pathway (3), there is significant residual Bub1 at kinetochores of cells in which Mps1 activity has been inhibited (91, 92).
Of note, H2A-T120-P is limited to kinetochores (115). Although it has been proposed that the CPC may localize at the intersection of H2A-T120-P and H3-T3-P (81), the overlap between these two marks may be limited to the inner kinetochore, whereas the localization domain of Aurora B is broader and clearly extends to the centromere. However, H2A-T120-P may contribute, by recruiting Shugoshin, to limit the activation of Aurora B to the kinetochore pool (149), although the details of this mechanism remain obscure. Sgo1 may also provide another anchoring point for the CPC at kinetochores, as the BIR domain of Survivin recognizes the N-terminal region of Sgo1 (76).
Mechanisms of Error Correction
A comprehensive picture of the contribution of Aurora B to the establishment of bi-orientation is still missing, but there has been substantial progress in recent years. Importantly, Aurora B has also been shown to regulate the structural stability of the kinetochore. For instance, it phosphorylates the CCAN subunit CENP-C/Mif2 to confer robustness to kinetochore function (166). In addition, phosphorylation of human Dsn1/Mis13 at two closely spaced residues (S100 and S109) increases the binding affinity of the Mis12 complex for CENP-C (42, 134, 166–171).
As already discussed above, anaphase retention of the CPC on kinetochores by expression of the T59E INCENP mutant or by suppression of MKLP2 (see above) leads to loss of tension that re-activates Aurora B-dependent pathways, including re-recruitment of Mps1, Bub1, and BubR1 (156). Nevertheless, kinetochores remain attached to their microtubule fibers under these conditions, indicating that re-activation of Aurora B is not sufficient for error correction. The crucial missing factor is the activity of Cdk1, which declines at anaphase due to degradation of Cyclin B. Artificial retention of Cdk1 activity in cells that have undergone sister chromatid separation leads to extensive destabilization of kinetochore–microtubule attachments (158, 159, 172).
Aurora B contributes at least three, partly related functions, to the process of bi-orientation: (1) the modulation of microtubule-binding affinity of the kinetochore to allow or prevent maturation of attachments; (2) the regulation of microtubule dynamics by controlling the activity and localization of microtubule-associated proteins; (3) the control of the localization of additional proteins involved in the regulation of kinetochore–microtubule attachment, including protein phosphatases that antagonize the phosphorylation of Aurora B substrates (4, 173).
A widely studied example of how Aurora B modulates the affinity of kinetochores for microtubules is the phosphorylation of multiple residues on a disordered and positively charged ~80-residue tail at the N-terminus of Ndc80/Hec1, a subunit of the Ndc80 complex (30, 31, 174–178). This segment of Ndc80 neighbors a calponin-homology (CH) domain that binds directly to microtubules (174, 179). Different models have been proposed for how Ndc80 phosphorylation modulates the binding affinity of the Ndc80 complex for microtubules (174, 175, 178, 180–182). A rigorous recent analysis suggested that each new phosphorylation event on the Ndc80 tail determines a relatively small decrease in microtubule-binding affinity by the Ndc80 complex, regardless of which specific position, among the eight or nine available, becomes phosphorylated (177). In this model, the phosphorylation sites of the Ndc80 tail configure a “rheostat” capable of increasing the microtubule-binding affinity of individual Ndc80 complexes by a factor as small as 20- and as large as 100-fold when transiting from a fully phosphorylated form of the protein to a fully dephosphorylated one (174, 177). Because the degree of phosphorylation of the Ndc80 tail is maximal when tension is low (e.g., in the absence of microtubules) (148), it is plausible that dephosphorylation of the Ndc80 complex is a gradual process that occurs concomitantly with the generation of tension within kinetochores. Consistent with this hypothesis, expression of a non-phosphorylatable mutant of the Ndc80 complex leads to hyper-stretched kinetochore–microtubule attachment and frequent attachment errors (31, 148, 181). Ndc80 has also been shown to have a direct influence on the dynamics of kinetochore microtubules, and Ndc80 phosphorylation may influence this property (183). Importantly, another Aurora family member, Aurora A, has also been very recently implicated in this correction mechanism (184, 185).
In addition to microtubule binding by the KMN network, other Aurora B substrates are important for the stabilization of the kinetochore–microtubule interface. The Dam1 complex in S. cerevisiae and the SKA complex in higher eukaryotes are structurally unrelated but may perform analogous functions as stabilizers of kinetochore–microtubule attachment (186–193). Contrary to the Ndc80 complex, both the Dam1 and the SKA complexes are able to form processive, load-bearing attachments to depolymerizing microtubule in vitro, and both contribute to retaining the Ndc80 complex at depolymerizing microtubule tips, possibly enhancing the overall processivity of microtubule binding (191–197). Importantly, Aurora B phosphorylation negatively regulates the association of the SKA and Dam1 complexes with Ndc80 (194–196, 198–201). An analogous pattern is also observed for the kinetochore recruitment of another microtubule-binding complex, the Astrin–SKAP complex (202). Thus, recruitment of these additional microtubule-binding complexes likely “seals” the kinetochore–microtubule interface of bi-oriented sister chromatids on which the phosphorylation of Aurora B has already faded.
Aurora B also controls kinetochore localization and activity of the non-conventional kinesin-13 family member mitotic centromere-associated kinesin (MCAK, Kif2C), which plays an important role in error correction as a microtubule depolymerase at microtubule ends (39, 203–210). Kinetochore and centromere recruitment of MCAK requires Aurora B phosphorylation of MCAK (203, 206, 210) and the presence of Sgo2 (113, 114, 118, 211).
Also dependent on Aurora B is the recruitment of CENP-E, a kinesin that plays an important role in the initial, lateral attachment of kinetochores to microtubules that precedes end-on attachment (7, 35, 212, 213). Conversion from an initial lateral attachment to end-on attachment occurs also in budding yeast (214). It has been proposed that lateral attachments may be insensitive to Aurora B activity, and therefore may be able to provide a mechanism for establishment of initial kinetochore–microtubule attachments even when Aurora B activity is high (4, 200).
Finally, Aurora B is in an antagonistic relationship with protein phosphatases, most notably of the protein phosphatase 1 (PP1) and PP2A-B56 families (4). These phosphatases counter phosphorylation by Aurora B kinase and other downstream kinases both in the EC and in the SAC (15, 109, 215–218). Many details of the complex molecular mechanisms subtending to the antagonism of Aurora B and PP1 and PP2A phosphatases remain to be elucidated. The following examples illustrate the complexity of this regulation.
Distinct interactions of the B56 regulators with Sgo1, Sgo2, and with the checkpoint component BubR1 recruit the PP2A holoenzyme to centromeres and kinetochores during mitosis (108, 115, 211, 219). The interaction of PP2A-B56 with BubR1 requires the so-called kinetochore attachment regulatory domain (KARD) motif of BubR1, which undergoes multisite phosphorylation (presumably) at kinetochores, partly mediated by Plk1 (219). Interference with the interaction of PP2A with the KARD domain leads to an elevation of Aurora B substrate phosphorylation in the outer kinetochore and prevents the stabilization of kinetochore–microtubule attachment (219).
Repo-Man, a protein scaffold that interacts with the PP1 phosphatase, is responsible for the clearance of the Haspin-mediated phosphorylation of H3-T3-P (161). Aurora B counteracts the chromatin recruitment of Repo-Man by phosphorylating it on Ser893, thus ultimately preventing the dephosphorylation of H3-T3-P. Dephosphorylation of Ser893, which might follow the release of the CPC from its centromeric localization at anaphase, requires an interaction of Repo-Man with PP2A, which is mediated by a motif closely related to the KARD motif of BubR1 (220).
Kinetochore recruitment of PP1 requires interactions with Knl1 and with CENP-E (15, 16, 221). Aurora B prevents kinetochore targeting of PP1 by phosphorylating a PP1-docking motif on Knl1 (15, 16). In S. cerevisiae, a requirement for kinetochore recruitment of PP1 to Knl1 (known as Spc105 in this organism), without which the SAC cannot be silenced, resulting in cell lethality, can be bypassed if Aurora B activity is compromised (16). Both PP1 and PP2A have been implicated as suppressors of the Mps1-dependent phosphorylation of the multiple Met-Glu-Leu-Thr (MELT) repeats of Knl1 that provide a docking site for the Bub1/Bub3 complex at kinetochores (215, 216).
Aurora B in the SAC
The SAC effector MCC consists of three SAC proteins, Mad2 (mitotic arrest deficient 2), Bub3 (budding uninhibited by benzimidazoles 3), BubR1 (Bub1-related 1, the human ortholog of yeast Mad3), and the APC co-activator Cdc20. Additional SAC components are Mad1, the kinases Mps1 (monopolar spindle protein 1), and Bub1 (budding uninhibited by benzimidazoles 1), and, limitedly to metazoans, the components of the Rod-Zwilch-ZW10 complex (RZZ). All SAC components contribute to the formation of the MCC and therefore to APC/C inhibition (3, 45).
The mechanisms through which Aurora B regulates the SAC are likely to be closely interwoven with the mechanisms that trigger error correction. As already pointed out in the previous paragraph, retention of Aurora B activity on chromosomes during anaphase is insufficient to cause error correction, but is sufficient to recruit bona fide SAC components, such as Mps1, Bub1, and BubR1, despite the retention of robust kinetochore fibers (156, 158–160, 172). This observation argues that Aurora B plays a direct role in the recruitment of the SAC components also in the absence of error correction and of unattached kinetochores. Incidentally, the observation that Mps1 can be recruited to anaphase chromosomes that have retained kinetochore fibers needs to be reconciled with the recent proposition that microtubules compete directly with Mps1 localization to kinetochores (222, 223).
Aurora B appears to occupy an upstream position in the pathway of recruitment of SAC components, as its inhibition prevents kinetochore recruitment of all other SAC components (35, 48, 52, 224, 225). Co-inhibition of Aurora B and Mps1 has profound synergistic effects in the impairment of SAC signaling (48, 52). Aurora B inhibition prevents Mps1 recruitment, and artificially tethering Mps1 to the kinetochore bypasses the checkpoint requirement for Aurora B in human cells, suggesting that a primary function of Aurora B in the SAC is the recruitment of Mps1 (51, 52). Conversely, when a downstream SAC component, such as Mad1:Mad2 is tethered to kinetochores, the resulting mitotic arrest depends on Aurora B (51, 226, 227).
Mps1 becomes recruited to the Ndc80 complex of the kinetochore (222, 223, 228, 229). The precise role of Aurora B in the recruitment of Mps1 remains unclear, but a role of Ndc80 phosphorylation has been suggested (223, 229). However, the observation that Aurora B activity becomes at least partly dispensable for kinetochore recruitment of Mps1 when the Mps1 TPR region is deleted suggests that Aurora B does not need to generate a docking site for Mps1 on Ndc80 but rather regulates a conformational transition within Mps1 (230).
After its Aurora B-dependent recruitment to kinetochores, Mps1 promotes the recruitment of downstream SAC component by phosphorylating Knl1 on multiple MELT repeats to dock the Bub1:Bub3 complex (231–234). The latter, in turn, elicits the formation of a comprehensive assembly of SAC protein that may facilitate SAC signaling from kinetochores (235–240).
Conclusion
Aurora B and the CPC are crucial for successful chromosome segregation during cell division. The two pathways Aurora B controls, error correction and the SAC, are tightly interwoven and interdependent. Both appear to rely on spatial control of Aurora B activity, but the precise molecular basis for this spatial control remains unknown. Future analyses will have to rigorously test the implications of the models that have been proposed to explain the spatial regulation of Aurora B activity, including the “centromere gradient” model and the “dog leash” model. It is hoped that global analyses of Aurora B substrate phosphorylation within the framework of predictable alterations of CPC and kinetochore function will finally shed light on the molecular basis of a mechanism that is indispensable for life.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Due to the considerable size of published literature on Aurora B, giving proper account of all contributions in a single review has become difficult. We apologize to the many authors whose work could not be cited due to space restrictions or, in a hopefully small number of cases, to our ignorance. AM would like to acknowledge funding from the EU’s Horizon 2020 European Research Council Advanced Investigator grant RECEPIANCE and from the DFG’s CRC 1093.
References
- 1.Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol (2015) 16:473–85. 10.1038/nrm4025 [DOI] [PubMed] [Google Scholar]
- 2.Duro E, Marston AL. From equator to pole: splitting chromosomes in mitosis and meiosis. Genes Dev (2015) 29:109–22. 10.1101/gad.255554.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.London N, Biggins S. Signalling dynamics in the spindle checkpoint response. Nat Rev Mol Cell Biol (2014) 15:736–47. 10.1038/nrm3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol (2013) 14:25–37. 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet (2012) 13:189–203. 10.1038/nrg3123 [DOI] [PubMed] [Google Scholar]
- 6.Siegel JJ, Amon A. New insights into the troubles of aneuploidy. Annu Rev Cell Dev Biol (2012) 28:189–214. 10.1146/annurev-cellbio-101011-155807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magidson V, Paul R, Yang N, Ault JG, O’Connell CB, Tikhonenko I, et al. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat Cell Biol (2015) 17:1134–44. 10.1038/ncb3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan CS, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics (1993) 135:677–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in Aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell (1995) 81:95–105. 10.1016/0092-8674(95)90374-7 [DOI] [PubMed] [Google Scholar]
- 10.Gopalan G, Chan CS, Donovan PJ. A novel mammalian, mitotic spindle-associated kinase is related to yeast and fly chromosome segregation regulators. J Cell Biol (1997) 138:643–56. 10.1083/jcb.138.3.643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura M, Kotani S, Hattori T, Sumi N, Yoshioka T, Todokoro K, et al. Cell cycle-dependent expression and spindle pole localization of a novel human protein kinase, Aik, related to Aurora of Drosophila and yeast Ipl1. J Biol Chem (1997) 272:13766–71. 10.1074/jbc.272.21.13766 [DOI] [PubMed] [Google Scholar]
- 12.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol (2001) 2:21–32. 10.1038/35048096 [DOI] [PubMed] [Google Scholar]
- 13.Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (Aurora) ABCs of mitosis. Trends Cell Biol (2001) 11:49–54. 10.1016/S0962-8924(00)01880-8 [DOI] [PubMed] [Google Scholar]
- 14.Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, et al. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol (2009) 16:365–71. 10.1016/j.chembiol.2009.02.012 [DOI] [PubMed] [Google Scholar]
- 15.Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol (2010) 188:809–20. 10.1083/jcb.201001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr Biol (2011) 21:942–7. 10.1016/j.cub.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J (1997) 16:1876–87. 10.1093/emboj/16.8.1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander J, Lim D, Joughin BA, Hegemann B, Hutchins JR, Ehrenberger T, et al. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci Signal (2011) 4:ra42. 10.1126/scisignal.2001796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hengeveld RC, Hertz NT, Vromans MJ, Zhang C, Burlingame AL, Shokat KM, et al. Development of a chemical genetic approach for human Aurora B kinase identifies novel substrates of the chromosomal passenger complex. Mol Cell Proteomics (2012) 11:47–59. 10.1074/mcp.M111.013912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol (2012) 13:789–803. 10.1038/nrm3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Horst A, Lens SM. Cell division: control of the chromosomal passenger complex in time and space. Chromosoma (2014) 123:25–42. 10.1007/s00412-013-0437-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J (2009) 28:2511–31. 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westermann S, Schleiffer A. Family matters: structural and functional conservation of centromere-associated proteins from yeast to humans. Trends Cell Biol (2013) 23:260–9. 10.1016/j.tcb.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 24.Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell (2014) 30:496–508. 10.1016/j.devcel.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, et al. The quantitative architecture of centromeric chromatin. Elife (2014) 3:e02137. 10.7554/eLife.02137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma (2011) 120:425–46. 10.1007/s00412-011-0330-0 [DOI] [PubMed] [Google Scholar]
- 27.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol (2008) 9:33–46. 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- 28.Nezi L, Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Curr Opin Cell Biol (2009) 21:785–95. 10.1016/j.ceb.2009.09.007 [DOI] [PubMed] [Google Scholar]
- 29.Nicklas RB, Koch CA. Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J Cell Biol (1969) 43:40–50. 10.1083/jcb.43.1.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell (2006) 127:983–97. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- 31.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell (2006) 127:969–82. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- 32.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev (2001) 15:3118–29. 10.1101/gad.934801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell (2002) 108:317–29. 10.1016/S0092-8674(02)00633-5 [DOI] [PubMed] [Google Scholar]
- 34.Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol (2003) 161:281–94. 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol (2003) 161:267–80. 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kallio MJ, McCleland ML, Stukenberg PT, Gorbsky GJ. Inhibition of Aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol (2002) 12:900–5. 10.1016/S0960-9822(02)00887-4 [DOI] [PubMed] [Google Scholar]
- 37.Munoz-Barrera M, Monje-Casas F. Increased Aurora B activity causes continuous disruption of kinetochore-microtubule attachments and spindle instability. Proc Natl Acad Sci U S A (2014) 111:E3996–4005. 10.1073/pnas.1408017111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol (2004) 6:232–7. 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- 39.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol (2006) 16:1705–10. 10.1016/j.cub.2006.07.057 [DOI] [PubMed] [Google Scholar]
- 40.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol (2006) 16:1711–8. 10.1016/j.cub.2006.07.022 [DOI] [PubMed] [Google Scholar]
- 41.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol (2011) 21:133–40. 10.1016/j.tcb.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim S, Yu H. Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J Cell Biol (2015) 208:181–96. 10.1083/jcb.201407074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, et al. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev (2003) 17:101–14. 10.1101/gad.1040903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol (1995) 130:941–8. 10.1083/jcb.130.4.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol (2012) 22:R966–80. 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- 46.Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature (2015) 517:631–4. 10.1038/nature13911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Primorac I, Musacchio A. Panta rhei: the APC/C at steady state. J Cell Biol (2013) 201:177–89. 10.1083/jcb.201301130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santaguida S, Vernieri C, Villa F, Ciliberto A, Musacchio A. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J (2011) 30:1508–19. 10.1038/emboj.2011.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khodjakov A, Rieder CL. The nature of cell-cycle checkpoints: facts and fallacies. J Biol (2009) 8:88. 10.1186/jbiol195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol (2006) 8:78–83. 10.1038/ncb1341 [DOI] [PubMed] [Google Scholar]
- 51.Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol (2011) 13:475–82. 10.1038/ncb2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saurin AT, van der Waal MS, Medema RH, Lens SM, Kops GJ. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat Commun (2011) 2:316. 10.1038/ncomms1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vader G, Cruijsen CW, van Harn T, Vromans MJ, Medema RH, Lens SM. The chromosomal passenger complex controls spindle checkpoint function independent from its role in correcting microtubule kinetochore interactions. Mol Biol Cell (2007) 18:4553–64. 10.1091/mbc.E07-04-0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petersen J, Hagan IM. S. pombe Aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr Biol (2003) 13:590–7. 10.1016/S0960-9822(03)00205-7 [DOI] [PubMed] [Google Scholar]
- 55.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of Aurora B kinase from kinetochore substrates. Science (2009) 323:1350–3. 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol (2009) 184:383–90. 10.1083/jcb.200811028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol (2009) 184:373–81. 10.1083/jcb.200808130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maresca TJ, Salmon ED. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J Cell Sci (2010) 123:825–35. 10.1242/jcs.064790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Musacchio A. Spindle assembly checkpoint: the third decade. Philos Trans R Soc Lond B Biol Sci (2011) 366:3595–604. 10.1098/rstb.2011.0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and Aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol (2001) 153:865–80. 10.1083/jcb.153.4.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carvalho A, Carmena M, Sambade C, Earnshaw WC, Wheatley SP. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J Cell Sci (2003) 116:2987–98. 10.1242/jcs.00612 [DOI] [PubMed] [Google Scholar]
- 62.Honda R, Korner R, Nigg EA. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol Biol Cell (2003) 14:3325–41. 10.1091/mbc.E02-11-0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gassmann R, Carvalho A, Henzing AJ, Ruchaud S, Hudson DF, Honda R, et al. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J Cell Biol (2004) 166:179–91. 10.1083/jcb.200404001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klein UR, Nigg EA, Gruneberg U. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of borealin, survivin, and the N-terminal domain of INCENP. Mol Biol Cell (2006) 17:2547–58. 10.1091/mbc.E05-12-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Romano A, Guse A, Krascenicova I, Schnabel H, Schnabel R, Glotzer M. CSC-1: a subunit of the Aurora B kinase complex that binds to the survivin-like protein BIR-1 and the incenp-like protein ICP-1. J Cell Biol (2003) 161:229–36. 10.1083/jcb.200207117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooke CA, Heck MM, Earnshaw WC. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J Cell Biol (1987) 105:2053–67. 10.1083/jcb.105.5.2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaitna S, Mendoza M, Jantsch-Plunger V, Glotzer M. Incenp and an Aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr Biol (2000) 10:1172–81. 10.1016/S0960-9822(00)00721-1 [DOI] [PubMed] [Google Scholar]
- 68.Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell (2004) 118:187–202. 10.1016/j.cell.2004.06.026 [DOI] [PubMed] [Google Scholar]
- 69.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med (1997) 3:917–21. 10.1038/nm0897-917 [DOI] [PubMed] [Google Scholar]
- 70.Sessa F, Mapelli M, Ciferri C, Tarricone C, Areces LB, Schneider TR, et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell (2005) 18:379–91. 10.1016/j.molcel.2005.03.031 [DOI] [PubMed] [Google Scholar]
- 71.Adams RR, Wheatley SP, Gouldsworthy AM, Kandels-Lewis SE, Carmena M, Smythe C, et al. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr Biol (2000) 10:1075–8. 10.1016/S0960-9822(00)00673-4 [DOI] [PubMed] [Google Scholar]
- 72.Bishop JD, Schumacher JM. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J Biol Chem (2002) 277:27577–80. 10.1074/jbc.C200307200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bolton MA, Lan W, Powers SE, McCleland ML, Kuang J, Stukenberg PT. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol Biol Cell (2002) 13:3064–77. 10.1091/mbc.E02-02-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kang J, Cheeseman IM, Kallstrom G, Velmurugan S, Barnes G, Chan CS. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J Cell Biol (2001) 155:763–74. 10.1083/jcb.200105029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ainsztein AM, Kandels-Lewis SE, Mackay AM, Earnshaw WC. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J Cell Biol (1998) 143:1763–74. 10.1083/jcb.143.7.1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeyaprakash AA, Basquin C, Jayachandran U, Conti E. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure (2011) 19:1625–34. 10.1016/j.str.2011.09.002 [DOI] [PubMed] [Google Scholar]
- 77.Jeyaprakash AA, Klein UR, Lindner D, Ebert J, Nigg EA, Conti E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell (2007) 131:271–85. 10.1016/j.cell.2007.07.045 [DOI] [PubMed] [Google Scholar]
- 78.Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, et al. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science (2010) 330:231–5. 10.1126/science.1189435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vader G, Kauw JJ, Medema RH, Lens SM. Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep (2006) 7:85–92. 10.1038/sj.embor.7400562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science (2010) 330:235–9. 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science (2010) 330:239–43. 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- 82.Yue Z, Carvalho A, Xu Z, Yuan X, Cardinale S, Ribeiro S, et al. Deconstructing survivin: comprehensive genetic analysis of survivin function by conditional knockout in a vertebrate cell line. J Cell Biol (2008) 183:279–96. 10.1083/jcb.200806118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Du J, Kelly AE, Funabiki H, Patel DJ. Structural basis for recognition of H3T3ph and Smac/DIABLO N-terminal peptides by human survivin. Structure (2012) 20:185–95. 10.1016/j.str.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Niedzialkowska E, Wang F, Porebski PJ, Minor W, Higgins JM, Stukenberg PT. Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres. Mol Biol Cell (2012) 23:1457–66. 10.1091/mbc.E11-11-0904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bayliss R, Fry A, Haq T, Yeoh S. On the molecular mechanisms of mitotic kinase activation. Open Biol (2012) 2:120136. 10.1098/rsob.120136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT, Funabiki H. Chromosomal enrichment and activation of the Aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell (2007) 12:31–43. 10.1016/j.devcel.2006.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tseng BS, Tan L, Kapoor TM, Funabiki H. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev Cell (2010) 18:903–12. 10.1016/j.devcel.2010.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC, et al. Chk1 is required for spindle checkpoint function. Dev Cell (2007) 12:247–60. 10.1016/j.devcel.2007.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Petsalaki E, Akoumianaki T, Black EJ, Gillespie DA, Zachos G. Phosphorylation at serine 331 is required for Aurora B activation. J Cell Biol (2011) 195:449–66. 10.1083/jcb.201104023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jelluma N, Brenkman AB, van den Broek NJ, Cruijsen CW, van Osch MH, Lens SM, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell (2008) 132:233–46. 10.1016/j.cell.2007.11.046 [DOI] [PubMed] [Google Scholar]
- 91.Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol (2010) 190:25–34. 10.1083/jcb.201002133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol (2010) 190:73–87. 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maciejowski J, George KA, Terret ME, Zhang C, Shokat KM, Jallepalli PV. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol (2010) 190:89–100. 10.1083/jcb.201001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mollinari C, Reynaud C, Martineau-Thuillier S, Monier S, Kieffer S, Garin J, et al. The mammalian passenger protein TD-60 is an RCC1 family member with an essential role in prometaphase to metaphase progression. Dev Cell (2003) 5:295–307. 10.1016/S1534-5807(03)00205-3 [DOI] [PubMed] [Google Scholar]
- 95.Rosasco-Nitcher SE, Lan W, Khorasanizadeh S, Stukenberg PT. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science (2008) 319:469–72. 10.1126/science.1148980 [DOI] [PubMed] [Google Scholar]
- 96.Papini D, Langemeyer L, Abad MA, Kerr A, Samejima I, Eyers PA, et al. TD-60 links RalA GTPase function to the CPC in mitosis. Nat Commun (2015) 6:7678. 10.1038/ncomms8678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and Aurora kinases in cancer. Nat Rev Cancer (2010) 10:825–41. 10.1038/nrc2964 [DOI] [PubMed] [Google Scholar]
- 98.Malumbres M, Perez de Castro I. Aurora kinase A inhibitors: promising agents in antitumoral therapy. Expert Opin Ther Targets (2014) 18:1377–93. 10.1517/14728222.2014.956085 [DOI] [PubMed] [Google Scholar]
- 99.Silke J, Vaux DL. IAP gene deletion and conditional knockout models. Semin Cell Dev Biol (2015) 39:97–105. 10.1016/j.semcdb.2014.12.004 [DOI] [PubMed] [Google Scholar]
- 100.Higgins JM. Haspin: a newly discovered regulator of mitotic chromosome behavior. Chromosoma (2010) 119:137–47. 10.1007/s00412-009-0250-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Campbell CS, Desai A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature (2013) 497:118–21. 10.1038/nature12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cho US, Harrison SC. Ndc10 is a platform for inner kinetochore assembly in budding yeast. Nat Struct Mol Biol (2012) 19:48–55. 10.1038/nsmb.2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yoon HJ, Carbon J. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc Natl Acad Sci U S A (1999) 96:13208–13. 10.1073/pnas.96.23.13208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting shugoshin localization. Curr Biol (2005) 15:353–9. 10.1016/j.cub.2004.12.044 [DOI] [PubMed] [Google Scholar]
- 105.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science (2010) 327:172–7. 10.1126/science.1180189 [DOI] [PubMed] [Google Scholar]
- 106.Marston AL. Shugoshins: tension-sensitive pericentromeric adaptors safeguarding chromosome segregation. Mol Cell Biol (2015) 35:634–48. 10.1128/MCB.01176-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu H, Qu Q, Warrington R, Rice A, Cheng N, Yu H. Mitotic transcription installs Sgo1 at centromeres to coordinate chromosome segregation. Mol Cell (2015) 59:426–36. 10.1016/j.molcel.2015.06.018 [DOI] [PubMed] [Google Scholar]
- 108.Meppelink A, Kabeche L, Vromans MJ, Compton DA, Lens SM. Shugoshin-1 balances Aurora B kinase activity via PP2A to promote chromosome bi-orientation. Cell Rep (2015) 11:508–15. 10.1016/j.celrep.2015.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol (2011) 13:1265–71. 10.1038/ncb2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peplowska K, Wallek AU, Storchova Z. Sgo1 regulates both condensin and Ipl1/Aurora B to promote chromosome biorientation. PLoS Genet (2014) 10:e1004411. 10.1371/journal.pgen.1004411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nerusheva OO, Galander S, Fernius J, Kelly D, Marston AL. Tension-dependent removal of pericentromeric shugoshin is an indicator of sister chromosome biorientation. Genes Dev (2014) 28:1291–309. 10.1101/gad.240291.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Verzijlbergen KF, Nerusheva OO, Kelly D, Kerr A, Clift D, de Lima Alves F, et al. Shugoshin biases chromosomes for biorientation through condensin recruitment to the pericentromere. Elife (2014) 3:e01374. 10.7554/eLife.01374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rattani A, Wolna M, Ploquin M, Helmhart W, Morrone S, Mayer B, et al. Sgol2 provides a regulatory platform that coordinates essential cell cycle processes during meiosis I in oocytes. Elife (2013) 2:e01133. 10.7554/eLife.01133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rivera T, Ghenoiu C, Rodriguez-Corsino M, Mochida S, Funabiki H, Losada A. Xenopus Shugoshin 2 regulates the spindle assembly pathway mediated by the chromosomal passenger complex. EMBO J (2012) 31:1467–79. 10.1038/emboj.2012.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu H, Jia L, Yu H. Phospho-H2A and cohesin specify distinct tension-regulated Sgo1 pools at kinetochores and inner centromeres. Curr Biol (2013) 23:1927–33. 10.1016/j.cub.2013.07.078 [DOI] [PubMed] [Google Scholar]
- 116.Eshleman HD, Morgan DO. Sgo1 recruits PP2A to chromosomes to ensure sister chromatid bi-orientation during mitosis. J Cell Sci (2014) 127:4974–83. 10.1242/jcs.161273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Parra MT, Gomez R, Viera A, Llano E, Pendas AM, Rufas JS, et al. Sequential assembly of centromeric proteins in male mouse meiosis. PLoS Genet (2009) 5:e1000417. 10.1371/journal.pgen.1000417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang H, Feng J, Famulski J, Rattner JB, Liu ST, Kao GD, et al. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J Cell Biol (2007) 177:413–24. 10.1083/jcb.200701122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tsukahara T, Tanno Y, Watanabe Y. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature (2010) 467:719–23. 10.1038/nature09390 [DOI] [PubMed] [Google Scholar]
- 120.Colnaghi R, Wheatley SP. Liaisons between survivin and Plk1 during cell division and cell death. J Biol Chem (2010) 285:22592–604. 10.1074/jbc.M109.065003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chu Y, Yao PY, Wang W, Wang D, Wang Z, Zhang L, et al. Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J Mol Cell Biol (2011) 3:260–7. 10.1093/jmcb/mjq037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nakajima Y, Cormier A, Tyers RG, Pigula A, Peng Y, Drubin DG, et al. Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP regulates CPC-spindle interaction to ensure proper microtubule dynamics. J Cell Biol (2011) 194:137–53. 10.1083/jcb.201009137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pereira G, Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science (2003) 302:2120–4. 10.1126/science.1091936 [DOI] [PubMed] [Google Scholar]
- 124.Bekier ME, Mazur T, Rashid MS, Taylor WR. Borealin dimerization mediates optimal CPC checkpoint function by enhancing localization to centromeres and kinetochores. Nat Commun (2015) 6:6775. 10.1038/ncomms7775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gruber M, Soding J, Lupas AN. REPPER – repeats and their periodicities in fibrous proteins. Nucleic Acids Res (2005) 33:W239–43. 10.1093/nar/gki405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Samejima K, Platani M, Wolny M, Ogawa H, Vargiu G, Knight PJ, et al. The INCENP coil is a single alpha helical (SAH) domain that binds directly to microtubules and is important for CPC localization and function in mitosis. J Biol Chem (2015) 290:21460–72. 10.1074/jbc.M115.645317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wheatley SP, Kandels-Lewis SE, Adams RR, Ainsztein AM, Earnshaw WC. INCENP binds directly to tubulin and requires dynamic microtubules to target to the cleavage furrow. Exp Cell Res (2001) 262:122–7. 10.1006/excr.2000.5088 [DOI] [PubMed] [Google Scholar]
- 128.Sandall S, Severin F, McLeod IX, Yates JR, III, Oegema K, Hyman A, et al. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell (2006) 127:1179–91. 10.1016/j.cell.2006.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Goto H, Kiyono T, Tomono Y, Kawajiri A, Urano T, Furukawa K, et al. Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat Cell Biol (2006) 8:180–7. 10.1038/ncb1350 [DOI] [PubMed] [Google Scholar]
- 130.Liu D, Davydenko O, Lampson MA. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. J Cell Biol (2012) 198:491–9. 10.1083/jcb.201205090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Waters JC, Skibbens RV, Salmon ED. Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. J Cell Sci (1996) 109(Pt 12):2823–31. [DOI] [PubMed] [Google Scholar]
- 132.Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell (2009) 137:672–84. 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol (2009) 19:694–9. 10.1016/j.cub.2009.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Welburn JP, Vleugel M, Liu D, Yates JR, III, Lampson MA, Fukagawa T, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell (2010) 38:383–92. 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Salimian KJ, Ballister ER, Smoak EM, Wood S, Panchenko T, Lampson MA, et al. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr Biol (2011) 21:1158–65. 10.1016/j.cub.2011.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Keating P, Rachidi N, Tanaka TU, Stark MJ. Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores. J Cell Sci (2009) 122:4375–82. 10.1242/jcs.055566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Beardmore VA, Ahonen LJ, Gorbsky GJ, Kallio MJ. Survivin dynamics increases at centromeres during G2/M phase transition and is regulated by microtubule-attachment and Aurora B kinase activity. J Cell Sci (2004) 117:4033–42. 10.1242/jcs.01242 [DOI] [PubMed] [Google Scholar]
- 138.Murata-Hori M, Wang YL. Both midzone and astral microtubules are involved in the delivery of cytokinesis signals: insights from the mobility of Aurora B. J Cell Biol (2002) 159:45–53. 10.1083/jcb.200207014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ahonen LJ, Kukkonen AM, Pouwels J, Bolton MA, Jingle CD, Stukenberg PT, et al. Perturbation of Incenp function impedes anaphase chromatid movements and chromosomal passenger protein flux at centromeres. Chromosoma (2009) 118:71–84. 10.1007/s00412-008-0178-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, Tobelmann P, et al. Midzone activation of Aurora B in anaphase produces an intracellular phosphorylation gradient. Nature (2008) 453:1132–6. 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tan L, Kapoor TM. Examining the dynamics of chromosomal passenger complex (CPC)-dependent phosphorylation during cell division. Proc Natl Acad Sci U S A (2011) 108:16675–80. 10.1073/pnas.1106748108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang E, Ballister ER, Lampson MA. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J Cell Biol (2011) 194:539–49. 10.1083/jcb.201103044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.De Antoni A, Maffini S, Knapp S, Musacchio A, Santaguida S. A small-molecule inhibitor of Haspin alters the kinetochore functions of Aurora B. J Cell Biol (2012) 199:269–84. 10.1083/jcb.201205119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hori T, Shang WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol (2013) 200:45–60. 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Boyarchuk Y, Salic A, Dasso M, Arnaoutov A. Bub1 is essential for assembly of the functional inner centromere. J Cell Biol (2007) 176:919–28. 10.1083/jcb.200609044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kiburz BM, Reynolds DB, Megee PC, Marston AL, Lee BH, Lee TI, et al. The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev (2005) 19:3017–30. 10.1101/gad.1373005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.van der Waal MS, Saurin AT, Vromans MJ, Vleugel M, Wurzenberger C, Gerlich DW, et al. Mps1 promotes rapid centromere accumulation of Aurora B. EMBO Rep (2012) 13:847–54. 10.1038/embor.2012.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.DeLuca KF, Lens SM, DeLuca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci (2011) 124:622–34. 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Caldas GV, DeLuca KF, DeLuca JG. KNL1 facilitates phosphorylation of outer kinetochore proteins by promoting Aurora B kinase activity. J Cell Biol (2013) 203:957–69. 10.1083/jcb.201306054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Posch M, Khoudoli GA, Swift S, King EM, Deluca JG, Swedlow JR. Sds22 regulates Aurora B activity and microtubule-kinetochore interactions at mitosis. J Cell Biol (2010) 191:61–74. 10.1083/jcb.200912046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ng TM, Waples WG, Lavoie BD, Biggins S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell (2009) 20:3818–27. 10.1091/mbc.E09-04-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev (2003) 17:2902–21. 10.1101/gad.1144403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Knockleby J, Vogel J. The COMA complex is required for Sli15/INCENP-mediated correction of defective kinetochore attachments. Cell Cycle (2009) 8:2570–7. 10.4161/cc.8.16.9267 [DOI] [PubMed] [Google Scholar]
- 154.Pot I, Knockleby J, Aneliunas V, Nguyen T, Ah-Kye S, Liszt G, et al. Spindle checkpoint maintenance requires Ame1 and Okp1. Cell Cycle (2005) 4:1448–56. 10.4161/cc.4.10.2106 [DOI] [PubMed] [Google Scholar]
- 155.Hummer S, Mayer TU. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr Biol (2009) 19:607–12. 10.1016/j.cub.2009.02.046 [DOI] [PubMed] [Google Scholar]
- 156.Vazquez-Novelle MD, Petronczki M. Relocation of the chromosomal passenger complex prevents mitotic checkpoint engagement at anaphase. Curr Biol (2010) 20:1402–7. 10.1016/j.cub.2010.06.036 [DOI] [PubMed] [Google Scholar]
- 157.Mirchenko L, Uhlmann F. Sli15(INCENP) dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr Biol (2010) 20:1396–401. 10.1016/j.cub.2010.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rattani A, Vinod PK, Godwin J, Tachibana-Konwalski K, Wolna M, Malumbres M, et al. Dependency of the spindle assembly checkpoint on Cdk1 renders the anaphase transition irreversible. Curr Biol (2014) 24:630–7. 10.1016/j.cub.2014.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vazquez-Novelle MD, Sansregret L, Dick AE, Smith CA, McAinsh AD, Gerlich DW, et al. Cdk1 inactivation terminates mitotic checkpoint surveillance and stabilizes kinetochore attachments in anaphase. Curr Biol (2014) 24:638–45. 10.1016/j.cub.2014.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kamenz J, Hauf S. Slow checkpoint activation kinetics as a safety device in anaphase. Curr Biol (2014) 24:646–51. 10.1016/j.cub.2014.02.005 [DOI] [PubMed] [Google Scholar]
- 161.Qian J, Lesage B, Beullens M, Van Eynde A, Bollen M. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal Aurora B targeting. Curr Biol (2011) 21:766–73. 10.1016/j.cub.2011.03.047 [DOI] [PubMed] [Google Scholar]
- 162.Gruneberg U, Neef R, Honda R, Nigg EA, Barr FA. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J Cell Biol (2004) 166:167–72. 10.1083/jcb.200403084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kitagawa M, Fung SY, Hameed UF, Goto H, Inagaki M, Lee SH. Cdk1 coordinates timely activation of MKlp2 kinesin with relocation of the chromosome passenger complex for cytokinesis. Cell Rep (2014) 7:166–79. 10.1016/j.celrep.2014.02.034 [DOI] [PubMed] [Google Scholar]
- 164.Banerjee B, Kestner CA, Stukenberg PT. EB1 enables spindle microtubules to regulate centromeric recruitment of Aurora B. J Cell Biol (2014) 204:947–63. 10.1083/jcb.201307119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang F, Ulyanova NP, van der Waal MS, Patnaik D, Lens SM, Higgins JM. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr Biol (2011) 21:1061–9. 10.1016/j.cub.2011.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Westermann S, Cheeseman IM, Anderson S, Yates JR, III, Drubin DG, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol (2003) 163:215–22. 10.1083/jcb.200305100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang Y, Wu F, Ward T, Yan F, Wu Q, Wang Z, et al. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J Biol Chem (2008) 283:26726–36. 10.1074/jbc.M804207200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rago F, Gascoigne KE, Cheeseman IM. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr Biol (2015) 25:671–7. 10.1016/j.cub.2015.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Akiyoshi B, Nelson CR, Biggins S. The Aurora B kinase promotes inner and outer kinetochore interactions in budding yeast. Genetics (2013) 194:785–9. 10.1534/genetics.113.150839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Emanuele MJ, Lan W, Jwa M, Miller SA, Chan CS, Stukenberg PT. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol (2008) 181:241–54. 10.1083/jcb.200710019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Matson DR, Stukenberg PT. CENP-I and Aurora B act as a molecular switch that ties RZZ/Mad1 recruitment to kinetochore attachment status. J Cell Biol (2014) 205:541–54. 10.1083/jcb.201307137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat Cell Biol (2010) 12:185–92. 10.1038/ncb2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Funabiki H, Wynne DJ. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma (2013) 122:135–58. 10.1007/s00412-013-0401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell (2008) 133:427–39. 10.1016/j.cell.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature (2010) 467:805–10. 10.1038/nature09423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tooley JG, Miller SA, Stukenberg PT. The Ndc80 complex uses a tripartite attachment point to couple microtubule depolymerization to chromosome movement. Mol Biol Cell (2011) 22:1217–26. 10.1091/mbc.E10-07-0626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zaytsev AV, Mick JE, Maslennikov E, Nikashin B, DeLuca JG, Grishchuk EL. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol Biol Cell (2015) 26:1829–44. 10.1091/mbc.E14-11-1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zaytsev AV, Sundin LJ, DeLuca KF, Grishchuk EL, DeLuca JG. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J Cell Biol (2014) 206:45–59. 10.1083/jcb.201312107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol (2007) 14:54–9. 10.1038/nsmb1186 [DOI] [PubMed] [Google Scholar]
- 180.Alushin GM, Musinipally V, Matson D, Tooley J, Stukenberg PT, Nogales E. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat Struct Mol Biol (2012) 19:1161–7. 10.1038/nsmb.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol (2008) 18:1778–84. 10.1016/j.cub.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1). Curr Biol (2008) 18:1785–91. 10.1016/j.cub.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Umbreit NT, Gestaut DR, Tien JF, Vollmar BS, Gonen T, Asbury CL, et al. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc Natl Acad Sci U S A (2012) 109:16113–8. 10.1073/pnas.1209615109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chmatal L, Yang K, Schultz RM, Lampson MA. Spatial regulation of kinetochore microtubule attachments by destabilization at spindle poles in meiosis I. Curr Biol (2015) 25:1835–41. 10.1016/j.cub.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ye AA, Deretic J, Hoel CM, Hinman AW, Cimini D, Welburn JP, et al. Aurora A kinase contributes to a pole-based error correction pathway. Curr Biol (2015) 25:1842–51. 10.1016/j.cub.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, III, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell (2002) 111:163–72. 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- 187.Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J (2009) 28:1442–52. 10.1038/emboj.2009.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J (2006) 25:5504–15. 10.1038/sj.emboj.7601426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci (2009) 122:2436–45. 10.1242/jcs.051912 [DOI] [PubMed] [Google Scholar]
- 190.Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol (2007) 178:269–81. 10.1083/jcb.200702141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, III, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell (2009) 16:374–85. 10.1016/j.devcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Westermann S, Wang HW, vila-Sakar AA, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature (2006) 440:565–9. 10.1038/nature04409 [DOI] [PubMed] [Google Scholar]
- 193.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A (2006) 103:9873–8. 10.1073/pnas.0602249103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol (2010) 189:641–9. 10.1083/jcb.200912021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell (2012) 23:968–80. 10.1016/j.devcel.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by Aurora B. J Cell Biol (2010) 189:713–23. 10.1083/jcb.200910142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, Westermann S, Drubin D, et al. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci U S A (2008) 105:6918–23. 10.1073/pnas.0801811105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Abad MA, Medina B, Santamaria A, Zou J, Plasberg-Hill C, Madhumalar A, et al. Structural basis for microtubule recognition by the human kinetochore Ska complex. Nat Commun (2014) 5:2964. 10.1038/ncomms3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol (2012) 196:563–71. 10.1083/jcb.201109001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kalantzaki M, Kitamura E, Zhang T, Mino A, Novak B, Tanaka TU. Kinetochore-microtubule error correction is driven by differentially regulated interaction modes. Nat Cell Biol (2015) 17:421–33. 10.1038/ncb3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Maure JF, Komoto S, Oku Y, Mino A, Pasqualato S, Natsume K, et al. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr Biol (2011) 21:207–13. 10.1016/j.cub.2010.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Schmidt JC, Kiyomitsu T, Hori T, Backer CB, Fukagawa T, Cheeseman IM. Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. J Cell Biol (2010) 191:269–80. 10.1083/jcb.201006129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, et al. Aurora B regulates MCAK at the mitotic centromere. Dev Cell (2004) 6:253–68. 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- 204.Ems-McClung SC, Hainline SG, Devare J, Zong H, Cai S, Carnes SK, et al. Aurora B inhibits MCAK activity through a phosphoconformational switch that reduces microtubule association. Curr Biol (2013) 23:2491–9. 10.1016/j.cub.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell (2004) 15:1146–59. 10.1091/mbc.E03-08-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol (2004) 14:273–86. 10.1016/j.cub.2004.01.055 [DOI] [PubMed] [Google Scholar]
- 207.Ohi R, Sapra T, Howard J, Mitchison TJ. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol Biol Cell (2004) 15:2895–906. 10.1091/mbc.E04-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tanenbaum ME, Medema RH, Akhmanova A. Regulation of localization and activity of the microtubule depolymerase MCAK. Bioarchitecture (2011) 1:80–7. 10.4161/bioa.1.2.15807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol (2007) 179:869–79. 10.1083/jcb.200707120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhang X, Lan W, Ems-McClung SC, Stukenberg PT, Walczak CE. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol Biol Cell (2007) 18:3264–76. 10.1091/mbc.E07-01-0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tanno Y, Kitajima TS, Honda T, Ando Y, Ishiguro K, Watanabe Y. Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev (2010) 24:2169–79. 10.1101/gad.1945310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, et al. Chromosomes can congress to the metaphase plate before biorientation. Science (2006) 311:388–91. 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Magidson V, O’Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell (2011) 146:555–67. 10.1016/j.cell.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature (2005) 434:987–94. 10.1038/nature03483 [DOI] [PubMed] [Google Scholar]
- 215.Espert A, Uluocak P, Bastos RN, Mangat D, Graab P, Gruneberg U. PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing. J Cell Biol (2014) 206:833–42. 10.1083/jcb.201406109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nijenhuis W, Vallardi G, Teixeira A, Kops GJ, Saurin AT. Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat Cell Biol (2014) 16:1257–64. 10.1038/ncb3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Vanoosthuyse V, Hardwick KG. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr Biol (2009) 19:1176–81. 10.1016/j.cub.2009.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Pinsky BA, Nelson CR, Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol (2009) 19:1182–7. 10.1016/j.cub.2009.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Suijkerbuijk SJ, Vleugel M, Teixeira A, Kops GJ. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev Cell (2012) 23:745–55. 10.1016/j.devcel.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 220.Qian J, Beullens M, Lesage B, Bollen M. Aurora B defines its own chromosomal targeting by opposing the recruitment of the phosphatase scaffold repo-man. Curr Biol (2013) 23:1136–43. 10.1016/j.cub.2013.05.017 [DOI] [PubMed] [Google Scholar]
- 221.Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell (2010) 142:444–55. 10.1016/j.cell.2010.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hiruma Y, Sacristan C, Pachis ST, Adamopoulos A, Kuijt T, Ubbink M, et al. Cell division cycle. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science (2015) 348:1264–7. 10.1126/science.aaa4055 [DOI] [PubMed] [Google Scholar]
- 223.Ji Z, Gao H, Yu H. Cell division cycle. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science (2015) 348:1260–4. 10.1126/science.aaa4029 [DOI] [PubMed] [Google Scholar]
- 224.Famulski JK, Chan GK. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr Biol (2007) 17:2143–9. 10.1016/j.cub.2007.11.037 [DOI] [PubMed] [Google Scholar]
- 225.Vigneron S, Prieto S, Bernis C, Labbe JC, Castro A, Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell (2004) 15:4584–96. 10.1091/mbc.E04-01-0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ballister ER, Riegman M, Lampson MA. Recruitment of Mad1 to metaphase kinetochores is sufficient to reactivate the mitotic checkpoint. J Cell Biol (2014) 204:901–8. 10.1083/jcb.201311113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kuijt TE, Omerzu M, Saurin AT, Kops GJ. Conditional targeting of MAD1 to kinetochores is sufficient to reactivate the spindle assembly checkpoint in metaphase. Chromosoma (2014) 123:471–80. 10.1007/s00412-014-0458-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, et al. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J (2009) 28:1099–110. 10.1038/emboj.2009.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zhu T, Dou Z, Qin B, Jin C, Wang X, Xu L, et al. Phosphorylation of microtubule-binding protein Hec1 by mitotic kinase Aurora B specifies spindle checkpoint kinase Mps1 signaling at the kinetochore. J Biol Chem (2013) 288:36149–59. 10.1074/jbc.M113.507970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Nijenhuis W, von Castelmur E, Littler D, De Marco V, Tromer E, Vleugel M, et al. module of MPS1 is required for its kinetochore localization by Aurora B. J Cell Biol (2013) 201:217–31. 10.1083/jcb.201210033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Yamagishi Y, Yang CH, Tanno Y, Watanabe Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol (2012) 14:746–52. 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- 232.Shepperd LA, Meadows JC, Sochaj AM, Lancaster TC, Zou J, Buttrick GJ, et al. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr Biol (2012) 22:891–9. 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol (2012) 22:900–6. 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Primorac I, Weir JR, Chiroli E, Gross F, Hoffmann I, van Gerwen S, et al. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. Elife (2013) 2:e01030. 10.7554/eLife.01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Krenn V, Overlack K, Primorac I, van Gerwen S, Musacchio A. KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT repeats. Curr Biol (2014) 24:29–39. 10.1016/j.cub.2013.11.046 [DOI] [PubMed] [Google Scholar]
- 236.Overlack K, Primorac I, Vleugel M, Krenn V, Maffini S, Hoffmann I, et al. A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. Elife (2015) 4:e05269. 10.7554/eLife.05269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Vleugel M, Omerzu M, Groenewold V, Hadders MA, Lens SM, Kops GJ. Sequential multisite phospho-regulation of KNL1-BUB3 interfaces at mitotic kinetochores. Mol Cell (2015) 57:824–35. 10.1016/j.molcel.2014.12.036 [DOI] [PubMed] [Google Scholar]
- 238.Vleugel M, Tromer E, Omerzu M, Groenewold V, Nijenhuis W, Snel B, et al. Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. J Cell Biol (2013) 203:943–55. 10.1083/jcb.201307016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.London N, Biggins S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev (2014) 28:140–52. 10.1101/gad.233700.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Moyle MW, Kim T, Hattersley N, Espeut J, Cheerambathur DK, Oegema K, et al. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J Cell Biol (2014) 204:647–57. 10.1083/jcb.201311015 [DOI] [PMC free article] [PubMed] [Google Scholar]