Abstract
Yersinia pestis, the causative agent of plague, is a highly virulent bacterium responsible for millions of human deaths throughout history. In the last decade, two natural plague foci have been described in the Republic of Georgia from which dozens of Y. pestis strains have been isolated. Analyses indicate that there are genetic differences between these strains, but it is not known if these differences are also reflected in protein expression. We chose four strains of Y. pestis (1390, 1853, 2944, and 8787) from the National Center for Disease Control and Public Health collection for proteomic studies based on neighbor-joining tree genetic analysis and geographical loci of strain origin. Proteomic expression was analyzed using two-dimensional gel electrophoresis and mass spectrometry. Select Y. pestis strains were grown under different physiological conditions and their proteomes were compared: (1) 28°C without calcium; (2) 28°C with calcium; (3) 37°C without calcium; and (4) 37°C with calcium. Candidate proteins were identified and the differences in expression of F1 antigen, tellurium-resistance protein, and outer membrane protein C, porin were validated by Western blotting. The in vitro cytotoxicity activity of these strains was also compared. The results indicate that protein expression and cytotoxic activities differ significantly among the studied strains; these differences could contribute to variations in essential physiological functions in these strains.
Keywords: Yersinia pestis, proteome, 2-D gel electrophoresis, virulence, Republic of Georgia
Introduction
Yersinia pestis is a Gram-negative, rod-shaped bacterium that is the causative agent of plague, a high-mortality disease recorded throughout history. Most human Y. pestis infections are flea-borne and manifest as bubonic plague; the other two forms of plague, septicemic and pneumonic, are rare. Public health experts recognize plague as a re-emerging infectious disease. In addition, Y. pestis carries the potential for use as a biological weapon because it can be mass-produced, is easily aerosolized, and may result in highly fatal pneumonic plague, which can spread person-to-person via inhalation of infectious air-borne droplets (1, 2).
Strains of Y. pestis isolated from different natural foci exhibit distinct biochemical and virulent phenotypes (1); laboratory passaging is also known to influence the accumulation of genetic polymorphisms in plague vaccine strains (3). In the event of infectious disease outbreaks, detailed knowledge of the molecular characteristics of Y. pestis strains is important to enable epidemiological traceability.
Several factors are thought to contribute to the persistence of Y. pestis in the environment. The stability of the rodent-flea infection cycle allows for constant circulation of the organism within susceptible host species (4). In this repeating cycle of rodent-flea infection, the organism is capable of causing fatal disease in Muridae and Sciuridae populations, and fleas must continually infect new hosts to find new blood meals (5). Y. pestis is therefore dependent on vectors and reservoirs for continued enzootic replication.
The longevity of fleas infected with Y. pestis also contributes to the duration of the enzootic cycle of infection; these insect reservoirs can sustain enzootic foci over long periods before their death, even when the host rodents are asymptomatic (6). In addition, the longevity of certain species of rodents post-infection can play a role in enzootic cycles of infection: certain rodent species, including some within the Microtus (7–10) and Meriones (11–14) genera, exhibit moderate- to high-resistance to plague and can carry Y. pestis without quickly dying (15). Some isolates of Y. pestis are less virulent than others, and this decreased virulence is thought to contribute to the stable enzootic circulation of Y. pestis because host and reservoir live longer and therefore have more opportunities to transmit the organism (16). In ancient plague foci, such as those in the Caucasus, vector–host cycles with different transmission and disease dynamics may co-exist (17) in which Y. pestis variants are highly virulent in certain rodents but avirulent in man (16).
Plague re-emergence in areas where the disease has been absent for many decades is not well understood (18–20). Two different mechanisms have been proposed: either Y. pestis continues circulating in animal populations at undetectable levels, or it is reintroduced to the animal population (21, 22). Although Y. pestis evolved from the enteric bacterium Yersinia pseudotuberculosis, which can survive for long periods in soil and water, selective pressure exerted by vector-borne transmission has resulted in the loss of such functionality (18, 23, 24). Y. pestis lacks the survival traits of other yersiniae, and long-term persistence outside of a host or vector is unlikely.
Plague epidemics have been recorded throughout the last century in the Republic of Georgia. During the last 50 years, two natural foci have been described from which multiple Y. pestis strains have been isolated (3). These foci are located in the Transcaucasian highland (encompassing the Ninotsminda and Akhalkalaki regions on the Javakheti plateau) and the Iori region (encompassing eastern Georgia and the border between Georgia and Azerbaijan) (25).
Yersinia pestis was isolated for the first time in Georgia in the spring of 1966, coinciding with a rodent epizootic of plague in several foci within the country. The cultures were isolated from the carcasses of Meriones libycus (red gerbils) and their fleas, Xenopsylla conformis and Ceratophyllus laeviceps. Strains were isolated from Eldari, Nazarlebi, and Taribani steppes, and the existence of a natural focus of plague among a Georgian population of M. libycus was confirmed for the first time. Later studies showed that M. libycus is the plague reservoir in this focus and the primary vectors are X. conformis and C. laeviceps fleas (26, 27).
The Transcaucasian desert plague foci are spread across approximately 500,000 ha, with each focus covering 40,000–200,000 ha. The foci are in a semi-desert zone near the Iori and Alazani rivers. Epizootics are severe in this area and Y. pestis strains that are isolated tend to be highly virulent; most of these strains were isolated from dead rodents. The epizootic process is spread across several individual foci with the same geographic features.
Georgian Anti-Plague Stations have conducted active surveillance in these foci since the early 1900s and have collected over 120 Y. pestis strains from these regions (28). The National Center for Disease Control and Public Health (NCDCPH) collection currently contains 46 strains, 40 of which were isolated from Georgian plague foci between 1966 and 1997 (25).
The differences between Y. pestis strains, including Georgian strains, on a genomic level have been extensively studied (25, 29–32). It is likely that some of these strain variations are also reflected at the proteomic level as the final products of gene expression. At the time of publication, no comparative studies of trans-Caucasian Y. pestis strains at the proteomic level were available in published literature.
A neighbor-joining tree was generated using multilocus variable number tandem repeat analysis for the 46 unique Y. pestis strains in the NCDCPH collection in Tbilisi, Georgia (30). Four strains of Y. pestis located at different positions on this tree were selected for comparative proteomic analysis.
Early studies of Yersinia physiology uncovered the low calcium response, whereby bacterial cultures grown in rich medium at an elevated temperature (37°C) exhibit a growth defect upon chelation of calcium ions. This defect was shown to be a result of a type III secretion system in Y. pestis, the Ysc TTSS, and is responsible for the secretion of virulence factors known as Yersinia outer proteins (YOPS) (33). This secretion system can be activated in vitro and virulence factors can be released into the medium when Y. pestis is grown at 37°C in the presence of millimolar concentrations of calcium (34). The Y. pestis proteome was previously examined using two-dimensional (2-D) gel electrophoresis (35, 36). These studies showed that virulence factors were induced at 26 or 37°C in the presence of 2.5 mM Ca2+ (a concentration similar to that in mammalian plasma). The Y. pestis proteome varies as a function of temperature and calcium, and expression of virulence factors clearly depends on these physiological conditions (37); thus, differences between the proteomes of Y. pestis strains could be masked under one set of physiological conditions, but expressed under another.
In this study, we examined and characterized proteomes of Y. pestis strains from the trans-Caucasian area as a function of temperature and calcium, which were used to affect induction of virulence. Y. pestis strains were grown at different temperatures in the presence and absence of calcium ions (Ca2+) and their proteomes were compared by 2-D gel electrophoresis and mass spectrometry (MS) and confirmed by Western blotting.
Materials and Methods
Yersinia pestis Strains
The following strains of Y. pestis were chosen for comparative proteomic studies: 1390, 1853, 8787, and 2944. These strains were provided by the NCDCPH in Tbilisi, Georgia. Strains 1390, 1853, and 8787 were isolated in Ninotsminda, Georgia, in 1979, 1980, and 1992, respectively; strain 2944 was isolated in Kabardino-Balkaria, Russia, in 1975.
Yersinia pestis Growth with Temperature and Calcium Concentration Changes
The following procedure was carried out in quadruplicate for each Y. pestis strain included in the study. Cultures were grown overnight in Mueller-Hilton broth at 28°C with continuous shaking. After incubation, 0.1 mL of each culture was transferred into 15 mL of fresh broth. Two aliquots were incubated at 37°C, one of which was adjusted with 0.4 M CaCl2 (15 μL) to a final concentration of 4 mM; 15 μL of sterile distilled water was added to the second aliquot. The remaining two aliquots were treated as described above but incubated at 28°C. All aliquots were incubated for another 4 h at the specified temperatures. Cells were harvested during the exponential phase of growth. Aliquots were centrifuged at 3,000 × g for 10 min, and duplicate bacterial pellets from strains under each set of physiological conditions were prepared for one-dimensional (1-D) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and 2-D gel electrophoresis.
Preparation for 1-D SDS-PAGE
Bacterial pellets were resuspended in 5% SDS and incubated at 95°C for 10 min. Lysates were centrifuged at 15,000 × g for 15 min, and the supernatant was collected and tested for sterility; inactivated samples were inoculated and grown in tryptic soy broth with 0.0125% phenol red for 72 h at 28°C. After incubation, samples were assessed for color change or turbidity; samples were considered sterile if neither color change nor turbidity were observed.
Preparation for 2-D Gel Electrophoresis
Bacterial pellets were resuspended in buffer (25 mM Tris-acetate, pH 7.8, 5 mM EDTA, 150 μg/mL lysozyme, 2 mM PMSF, 0.05% Triton X-100, and 1 mM benzamidine). Samples were incubated for 30 min at 20°C with intermittent vortexing. Lysates were centrifuged at 20,000 × g for 60 min at 4°C. The supernatant was collected and 20% CHAPS stock solution was added to a final concentration of 2% (w/v), followed by incubation at 95°C for 10 min. After incubation, the supernatant was tested for sterility as previously described.
Protein Quantification
A micro-BCA kit (Pierce, Thermo Scientific) was used to quantify the protein concentration in supernatants. Protein quantification was performed in quadruplicate for each sample with the appropriate buffer controls, according to the manufacturer’s instructions.
2-D Gel Electrophoresis
Sample Preparation and Isoelectrofocusing
Isoelectrofocusing (IEF) strips (linear pH 3–10) were rehydrated in 8M urea, 0.5% Triton X-100, 0.5% Pharmalyte 3–10, and 30 mM DTT overnight. Protein samples (30 μg) were loaded onto rehydrated strips in buffer containing 7M urea, 2M thiourea, 2% CHAPS, 2% Triton X-100, 0.1% ASB-14, 2-mercaptoethanol, 2% Pharmalyte 3–10, and bromophenol blue. IEF was carried out at 500 V for 3 h and 3,500 V for 18 h ± 30 min.
Equilibration
Strips were equilibrated for 15 min in a buffer containing 0.05M Tris-HCl (pH 6.8), 6M urea, 30% glycerol, 3% SDS, and 1% DTT, followed by equilibration in the same buffer with 2.5% DTT iodoacetamide instead of 1% DTT for 15 min.
SDS Electrophoresis
SDS electrophoresis was run on a 1-mm thick, 12.5% polyacrylamide gel at 25°C, first at 10 mA per gel at 80 V for 1 h, and then at 12 mA per gel at 150 V for 17 h.
Staining, Scanning, and Analysis
The gels were stained with a silver stain kit (GE-Healthcare), omitting the glutaraldehyde step. Silver-stained gels were scanned with an image scanner (Labscan 6.0). Images were digitalized and processed using Image Master 2D platinum 7.0.
In each series of experiments, the supernatants of the four Y. pestis strains were analyzed concurrently for each physiological condition. Proteins that exhibited at least 1.5-fold difference between the strains were selected. The relative intensities of protein spots coinciding by location (isoelectric point and molecular weight) from different experiments were compared by t-test. The significantly differentially expressed protein spots (p < 0.05) were excised, destained, and stored at −20°C until MS analysis.
In-Gel Digestion and MS Analysis
Excised proteins were reduced with TCEP and alkylated with iodoacetamide. Samples were then treated with acetonitrile, dried, and rehydrated in activated trypsin (Thermo Scientific, Pierce) to begin digestion; proteins were digested at 37°C for 4 h. Digested samples were processed using nanospray ionization tandem HPLC-MS/MS CID performed with helium (Finnigan LTQ, Thermo Scientific). MS/MS spectra data was analyzed using SEQUEST (Proteome Discoverer 2.0), searching against UniProt UniRef100 databases.
Antibodies
Antibodies against F1 antigen were obtained from Life Science-Meridian (cat. # C86308M). Polyclonal antibodies against DNA-binding protein H-NS, tellurium-resistance protein, and outer membrane protein C, porin, were raised in rabbits against the following peptide sequences:
H-NS: EMLEKLEVVVN (amino acids 28–38)
Tellurium-resistance protein: PADVDKIVFVVT (amino acids 99–110)
Outer membrane protein C, porin: NTDDIVAVGMVYQ (amino acids 332–344)
Antibodies against H-NS and tellurium-resistance protein were affinity-purified on corresponding peptide columns; antibodies against outer membrane protein C, porin were purified on protein-A Sepharose columns. The specificities of antibodies were tested by inhibition of binding with immunizing peptides using Western blotting.
1-D SDS-PAGE and Western Blotting
Laemli SDS sample buffer was added to protein aliquots (30 μg). Samples were then analyzed by SDS-PAGE, followed by Western blotting. After electrophoretic transfer onto nitrocellulose membranes, protein bands were detected by staining with Ponceau S to confirm the uniform loading of the gel. Nitrocellulose membranes were sectioned according to the molecular weight standard and were then probed with primary antibodies against the following antigens: F1; tellurium-resistance protein; outer membrane protein C, porin; and DNA-binding protein H-NS. Because F1 and outer membrane protein C, porins are of a similar molecular weight, separate electrophoresis were carried out for each antigen.
Standard immunochemical procedures were carried out, and bands were visualized using peroxidase-labeled secondary antibodies (anti-rabbit) and a SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Pierce). Intensifying screens were used to expose the blots to X-ray films pre-flashed with Sensitize (Amersham). The densities of the protein bands were measured using LabWorks 4.0 software (UVP).
The autoradiographs were calibrated using combined protein from the four strains (15, 30, 45, and 60 μg total protein). Standard curves were prepared for each protein by plotting the optical density of the immunostained band against the protein concentration. The least squares regression showed a significant fit to a straight line for all standards. In experimental samples, to calculate protein amount, the optical density of each band was divided by the optical density of the 30-μg total protein standard; this value was considered to be the “relative amount” of protein (e.g., F1, Tellurium-resistance protein, and H-NS) (38).
In vitro Cytotoxicity Assay
In vitro cytotoxicity of Y. pestis strains and the vaccine strain EV was evaluated based on strain ability to induce apoptosis in macrophage cultures. Apoptosis was assayed using a colorimetric caspase-3 assay kit (Sigma-Aldrich, cat. #CASP-3-C) according to the manufacturer’s instructions. This kit measures the activity of caspase-3, one of the critical enzymes of apoptosis, and includes a specific inhibitor for precise measurement of caspase-3 activity.
Macrophages were cultivated as previously described (39, 40). Briefly, J774A.1 mouse macrophages were grown in DMEM supplemented with 10% heat-inactivated FBS. Cultures were incubated for 72 h at 37°C in a 5% CO2 atmosphere, after which the wells were washed to remove non-adherent cells.
To each well, 100 μL of Y. pestis (3 × 108 cfu/mL) was added and incubated at 37°C for 3 h. Plates were centrifuged at 600 × g for 5 min at 4°C, washed once with 1 mL of PBS per well, and 1× lysis buffer was added (100 μL/105cells). After 15 min of incubation on ice, the lysate was centrifuged at 3,000 × g for 5 minutes at 4°C, and the supernatant was stored at −70°C. Supernatants were filtered, divided into aliquots, and tested for sterility as previously described. Cell lysates and positive controls were brought to the required volume with 1× assay buffer and incubated for 2 h at 37°C with caspase-3 substrate. For each aliquot, caspase-3 inhibitor was included in addition to the peptide substrate, and parallel measurements were taken. The amount of p-nitroaniline (pNA) released in the assay was measured using a spectrophotometer (OD405 nm) and the concentration was determined by the standard curve. These values were subtracted from the values obtained without the inhibitor. The protein amount was determined in cell lysates, and enzyme activity expressed as nanomoles of pNA released per minute per 1 mg of cell lysate protein.
Statistical Analysis
The relative amounts of proteins were subjected to analysis of variance (ANOVA) by strain (1390, 1853, 2944, and 8787) and physiological condition (28°C, 28°C with Ca2+, 37°C, 37°C with Ca2+). Results from different strains grown under the same physiological conditions were compared by t-test.
Caspase-3 activity data was analyzed by strain using one-way ANOVA. Planned comparisons were done by t-test. All t-tests were two-tailed unless otherwise indicated.
Results
2-D Gel Electrophoresis
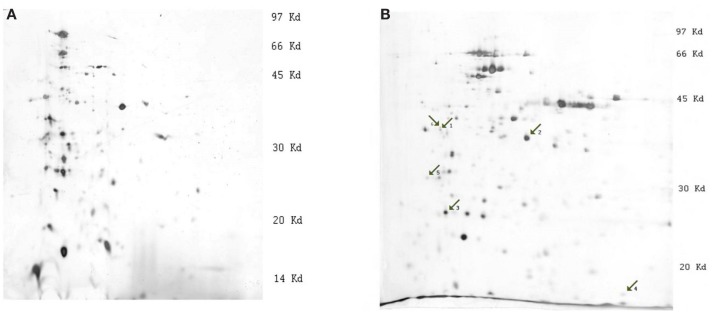
The 2-D gel electrophoresis of Y. pestis protein extracts was carried out on IEF strips with two pH gradients: 3.0–10.0 and 3.0–5.6 (Figures 1A,B). The majority of the proteins on the 3.0–10.0 pH gradient gels were concentrated between pH 3.0 and 6.0 (Figure 1A), so 2-D gel electrophoresis was repeated using strips with a pH gradient from 3.0 to 5.6 for better resolution of protein bands (Figure 1B).
Figure 1.
Images of silver stained 2-D gels of Y. pestis protein extracts (strain 1853, condition 28°C). (A) 2-D electrophoresis on strips with pH linear gradient 3.0–10.0, (B) 2-D electrophoresis on strips with pH linear gradient 3.0–5.6. The numbered arrows indicate the positions of differentially expressed proteins: 1, outer membrane protein C, porin; 2, outer membrane protein C2 porin; 3, Tellurium-resistance protein; 4, DNA-binding protein H-NS; and 5, F1 capsule antigen.
The comparison of images of different strains revealed consistent differential expression of bands. The most significant differences were observed at 28°C. Strains were divided into two groups according to their differences: (1) 1853 and 1390 and (2) 2944 and 8787. Differential expression of certain proteins was significantly higher in the first group of bacterial strains. MS analysis was used to determine the identity of the excised proteins (Table 1). To validate the significant differences between the proteins, quantitative Western blotting was used to study four of the five identified proteins. The criteria for the selection of proteins that we studied were the following reasons: (i) availability of the commercial antibodies, (ii) our ability to produce custom antibodies, and (iii) differential expression profiles of these proteins under the various growth conditions.
Table 1.
Differentially expressed proteins identified by 2-D electrophoresis by condition and strain.
| Protein | Condition/strain |
|---|---|
| Outer membrane protein C, porin | 28°C, 1853 and 1390 > 2944, 8787 |
| Tellurium-resistance protein | 28°C, 1853 and 1390 > 2944, 8787 |
| DNA-binding protein H-NS | 28°C, 1853 and 1390 > 2944, 8787 |
| F1 antigen | 37°C, 37°C + Ca2+ and 28°C, 1853 and 1390 > 2944, 8787 |
| Outer membrane protein-C2, porin | 28°C, 1853 and 1390 > 2944, 8787 |
Immunostaining
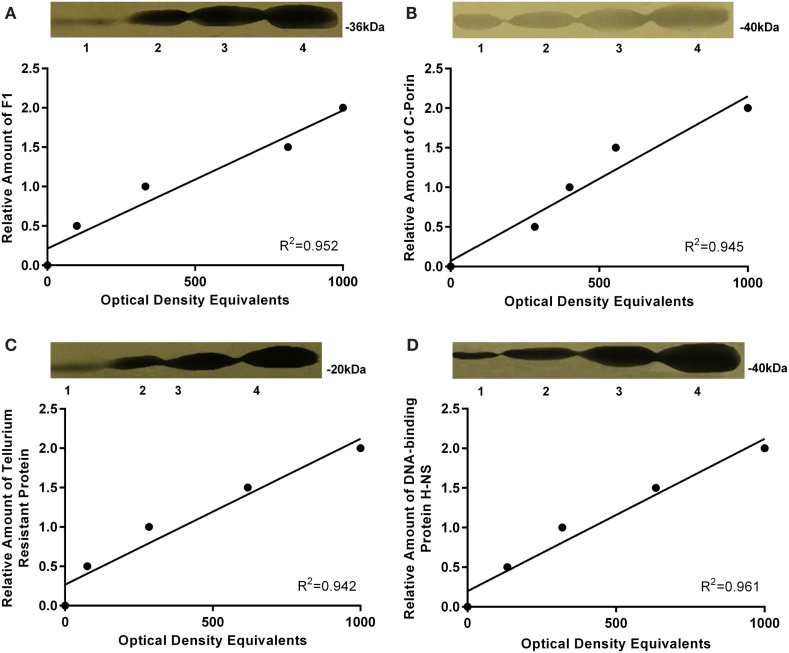
Standards were prepared as described above (15, 30, 45, and 60 μg total protein). For these standards, the optical densities of the bands immunostained for F1, outer membrane protein C, porin, tellurium-resistance protein, and DNA-binding protein H-NS were linearly related to the amounts of proteins in the bands (Figures 2A–D).
Figure 2.
Western blot autoradiographs and calibration plots. Western blot autoradiographs for single standard sample of the mixture of all Y. pestis strains protein extracts containing 15, 30, 45, and 60 μg of protein, respectively. Calibration plots are shown below each autoradiograph and reflect the linear response of immunostaining with the corresponding amount of loaded protein for (A) F1 antigen, (B) outer membrane protein C, porin, (C) Tellurium, and (D) DNA-binding protein H-NS.
Antibodies against F1 reacted with a protein band of molecular weight of approximately 36–38 kDa (Figure 2A). The molecular weight of F1 antigen is 17.7 kDa, but it migrates as a dimer in SDS electrophoresis (41). Outer membrane protein C, porin antibodies reacted with a protein band of apparent molecular weight approximately 38 kDa, which corresponds to the expected size of the target protein (Figure 2B). Tellurium-resistance protein antibodies reacted with a protein band of 20 kDa (Figure 2C) and DNA-binding protein H-NS antibodies reacted with a protein of apparent molecular weight 15–16 kDa (Figure 2D). Both of these weights corresponded to the expected size of the target proteins.
F1 Antigen
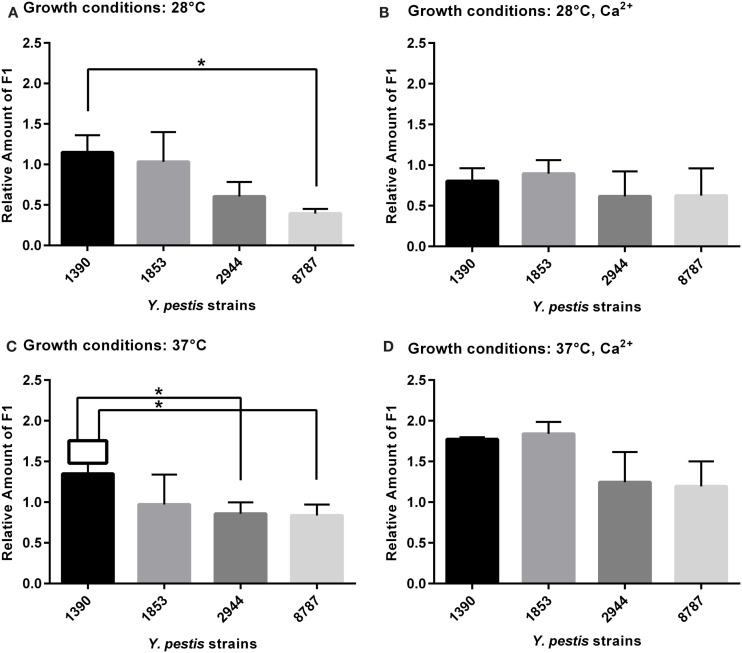
Y. pestis strain and growth condition were both found to be significant factors in the expression of F1 when analyzed by two-way ANOVA (respectively F3, 63 = 5.40, p = 0.002; F3, 63 = 9.95, p = 0.0001). As expected from 2-D electrophoresis data, the most significant changes were observed at 37°C (Figure 3C). The mean amount of F1 antigen was significantly higher in strain 1390 than in strains 2944 (t = 2.65, p = 0.038) and 8787 (t = 2.85, p = 0.029). The mean amount of F1 was also higher in strain 1390 than strain 8787 at 28°C (t = 3.47, p = 0.013; Figure 3A). At 28°C with Ca2+, and 37°C with Ca2+, mean amounts of F1 antigen in 1390 and 1853 were not significantly higher than mean amounts in 8787 and 2944 (Figures 3B,D, respectively).
Figure 3.
Relative amount of F1 antigen expressed by Y. pestis strains 1390, 1853, 2944, and 8787 at physiological growth conditions (A) 28°C; (B) 28°C, Ca2+; (C) 37°C; and (D) 37°C, Ca2+. The relative amount of protein expressed was calculated from immunostained bands, by dividing the optical density of the band by the optical density of the corresponding 30 μg total protein standard. Data represents the mean + SEM. *Indicates significant differences (p < 0.05).
Outer Membrane Protein C, Porin
Two-way ANOVA revealed significant differences in expression of outer membrane protein C, porin, between strains (F3, 63 = 8.53, p = 0.0001). The largest significant differences were observed at 28°C (Figure 4A). The mean amount of outer membrane protein C, porin was significantly higher in strain 1853 than strains 2944 (p = 0.02) and 8787 (p = 0.002), and in strain 1390 than strain 8787 (p = 0.041). The amount of outer membrane protein C, porin was significantly higher in strain 1853 than 8787 at 28°C with Ca2+ (p = 0.025, Figure 4B). Statistically significant differences were not observed between strains grown under other conditions (Figures 4C,D).
Figure 4.
Relative amount of outer membrane protein C, porin expressed by Y. pestis strains 1390, 1853, 2944, and 8787 at physiological growth conditions (A) 28°C; (B) 28°C, Ca2+; (C) 37°C; and (D) 37°C, Ca2+. The relative amount of protein expressed was calculated from immunostained bands, by dividing the optical density of the band by the optical density of the corresponding 30 μg total protein standard. Data represent the mean + SEM. *Indicates significant differences (p < 0.05).
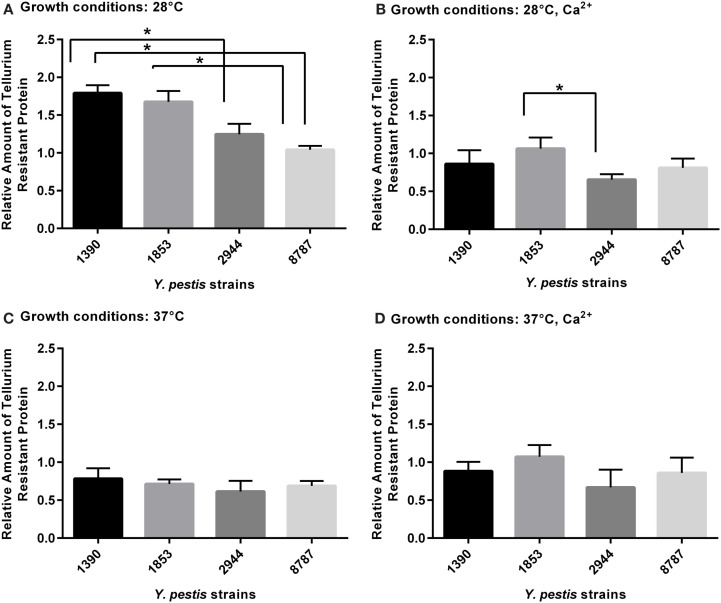
Tellurium-Resistance Protein
For the tellurium-resistance protein, the effects of strain and growth conditions on expression (respectively F3, 63 = 5.49, p = 0.002; F3, 63 = 21.03, p = 0.0001) were significant by two-way ANOVA. The mean amount of protein at 28°C was significantly higher in strain 1390 than strains 2944 (p = 0.019) and 8787 (p = 0.001), and the amount in strain 1390 was significantly higher than in strain 8787 (p = 0.006). At 28°C with Ca2+, the mean value of protein in 1853 was significantly higher than 2944 (p = 0.044) (Figures 5A–D); no other significant differences were observed.
Figure 5.
Relative amount of tellurium-resistance protein expressed by Y. pestis strains 1390, 1853, 2944, and 8787 at physiological growth conditions (A) 28°C; (B) 28°C, Ca2+; (C) 37°C; and (D) 37°C, Ca2+. The relative amount of protein expressed was calculated from immunostained bands, by dividing the optical density of the band by the optical density of the corresponding 30 μg total protein standard. Data represent the mean + SEM. *Indicates significant differences (p < 0.05).
DNA-Binding Protein H-NS
None of the differences in expression of DNA-binding protein H-NS between strains or under different growth conditions were significant by ANOVA (Figures 6A–D).
Figure 6.
Relative amount of DNA-binding protein H-NS expressed by Y. pestis strains 1390, 1853, 2944, and 8787 at physiological growth conditions (A) 28°C; (B) 28°C, Ca2+; (C) 37°C; and (D) 37°C, Ca2+. The relative amount of protein expressed was calculated from immunostained bands by dividing the optical density of the band by the optical density of the corresponding 30 μg total protein standard. Data represent the mean + SEM. *Indicates significant differences (p < 0.05).
In vitro Cytotoxicity Assay
Caspase-3 activity of Y. pestis strains was compared using one-way ANOVA; activity was significantly different between strains (F4, 19 = 22.15, p = 0.0001; Figure 7). The highest activity of caspase-3 was observed in strain 1853, which significantly exceeded the activity in strains 1390 (t = 3.24, p = 0.018), 8787 (t = 9.35, p = 0.0001), and EV (t = 11.20, p < 0.0001). Caspase-3 activity in strain 2944 was significantly higher than in strains 8787 and EV (t = 5.69, p = 0.001). Caspase-3 activity was also significantly higher in strain 1390 than strains 8787 (t = 3.55, p = 0.012) and EV (t = 4.48, p = 0.004) (Figure 7). No other significant differences were observed (by two-tailed t-test).
Figure 7.
Macrophage caspase-3 activity under the influence of Y. pestis strains 1390, 1853, 2944, 8787, and EV. In vitro cytotoxicity of Y. pestis strains and the vaccine strain EV was evaluated based on their ability to induce apoptosis in macrophage cultures. Data represent the mean + SEM. *Indicates significant differences (p < 0.05).
Discussion
Genetic polymorphisms are often associated with various Y. pestis strains isolated from natural foci in the Republic of Georgia, but it is not known if these polymorphisms affect gene expression or if there are differences between the proteomes of various strains (42, 43). The results of this study demonstrate that strains of Y. pestis isolated from natural foci in the Republic of Georgia differ at the proteomic level as well as the genetic level.
The main goal of this research was to identify a set of candidate proteins that are differentially expressed across the different Georgian Y. pestis isolates. A combination of 2-D electrophoresis and MS was used to identify several candidate proteins. However, 2-D electrophoresis with silver staining is not a strong quantitative approach, even when analyzed with even the most sophisticated software; therefore, further validation was required. To this end, we used quantitative Western blotting to measure the levels of proteins (the final products of gene expression). From candidate proteins, the following proteins were chosen as the focus of the experiments: F1 antigen; tellurium-resistance protein; outer membrane protein C, porin, and DNA-binding protein H-NS.
F1 antigen is widely accepted as a virulence factor of Y. pestis. F1 is encoded by the caf1 gene located on the large 100-kb pFra plasmid, which is unique to Y. pestis. F1 antigen is synthesized as a 15–16 kDa monomer and forms a large homopolymer (>200 kDa) on the bacterial cell surface in a stacked ring structure composed of heptamers (44, 45). The secretion and assembly of F1 require the caf1M and caf1A genes, which are homologous to the chaperone and usher protein families required for biogenesis of pili. F1 may be involved in the ability of Y. pestis to prevent uptake by macrophages, thus protecting Y. pestis from the host’s innate immune response (45). F1-antigen is also known to inhibit bacterial adhesion to epithelial cells (45).
As a differentially expressed protein, F1 antigen was detected on gels with bacterial extracts grown at 37°C and at 37°C with Ca2+. Western blotting analysis verified that the mean amount of F1 antigen was significantly higher in strain 1390 than in strain 2944 and 8787 at 37°C. The mean amounts of F1 antigen expressed at 37°C with Ca2+ were not significantly higher in strains 1390 and 1853 than strains 2944 and 8787. These results indicate that expression of F1 antigen differs from strain to strain. Further, the F1 antigen has been well-documented to be temperature regulated. Prior work demonstrates enhanced F1 expression at >35°C. Considering that our findings revealed enhanced expression at 37°C relative to 28°C (most evident when grown in the presence of calcium), this provides additional authenticity to our results (46).
Tellurium is a trace element that belongs to the same chemical group as selenium, sulfur, and oxygen. Tellurite oxyanions are highly toxic for most forms of life even at micromolar levels (47). Toxicity of tellurium may be mediated partly by reactive oxygen species that are generated as by-products of tellurite reduction (48). The primary functions of tellurium-resistance proteins are not known, although some sources suggest that they are involved in detoxification of antimicrobial compounds produced by host macrophages (47). Y. pestis cells in natural rodent hosts multiply initially in macrophage phagolysosomes. Survival and multiplication of Y. pestis in this new environment likely requires compensatory mechanisms involving expression of specific proteins compared to those expressed during extracellular growth. Indeed, 2-D electrophoresis and MS analysis has shown that intracellular and extracellular Y. pestis proteomes differ from each other in the expression of 12 proteins (49). Differentially expressed proteins that were upregulated inside the macrophages include tellurium-resistance proteins and also DNA-binding protein H-NS (49). Since the survival and multiplication of Y. pestis inside macrophages may be enhanced tellurium-resistance proteins, the strain-specific variations in tellurium-resistance protein expression might influence virulence during mammalian infection. The results of these experiments suggest that strains 1390 and 1853 are expressing tellurium-resistance protein at higher levels than strains 2944 and 8787.
Iron is an essential element for the survival of many microorganisms and its acquisition by bacteria can be central to the outcome of an infection (45). Recent data indicate that outer membrane proteins, including protein C, are transferrin binding proteins and could be involved in the acquisition of iron for growth within their host (47). Differences in the expression of outer membrane protein C, porin, between the strains were generally analogous to differences in expression of tellurium-resistance protein. Significant differences between the strains were observed at 28°C; the mean amount of protein was higher in strains 1390 and 1853 than strains 2944 and 8787.
DNA-binding protein H-NS belongs to a group of nucleoid-associated proteins, which are associated with the chromosome. They possess substantial non-specific DNA-binding affinity and have two major functions: gene regulation and chromosome organization (50). As mentioned above, this protein in is upregulated in Y. pestis inside of macrophages (49). DNA-binding protein H-NS was identified as a significantly differentially expressed protein when analyzed on silver-stained 2-D gels, but further quantitative analysis with Western blotting indicated that differences in expression between strains were not significant. The greatest difference, which was observed between strains 1390 and 8787, was only significant in a one-tailed t-test.
It is well documented that cell death plays a central role in host-pathogen interactions by eliminating the pathogen’s replicative niche and/or by eliminating immune cells and evading antimicrobial effector mechanisms (51). In these experiments, the ability to induce apoptosis in macrophage cell cultures was used to determine the toxicity of Y. pestis isolates. Macrophages are responsible for detecting, engulfing, and destroying pathogens and their apoptosis terminates the host’s immune response. Caspase-3 is a mediator of the pathogenic effect of Y. pestis in the livers of C57BL/6 mice (52); therefore the apoptosis activity, as indicated by caspase-3 expression, could be a measure of the virulence of Y. pestis isolates. Caspase-3 activity differed significantly among the study strains: the lowest significant expression was observed in strains EV and 8787, and the highest was observed in 1853. The apoptosis-inducing activities in strains 2944 and 1390 were also significantly higher than in strains 8787 and EV.
The overall comparative analysis of studied strains indicates that strain 8787 is distinct from other strains due to a low expression of virulence factors and cytotoxic activities. According to the neighbor-joining tree generated for 46 Y. pestis strains from the NCDCPH, strain 8787 is different from strains 1390, 1853, and 2944. Strains 1853 and 1390 are in close proximity to each other. The levels of expression of F1, tellurium-resistance protein, and outer membrane protein C, porin suggest that strains 1853 and 1390 are close to each other. According to the cytotoxicity assay, strains 1853 and 1390 are significantly different from each other and are both different from 8787 and 2944. Strain 2944 is characterized by a lower expression of the studied proteins and high cytotoxic activity, which is likely driven by factors other than those studied. Thus, the results obtained confirm both relatedness and differences between the strains.
Plasminogen activator, Pla is encoded by the pPCP plasmid and is one of the major virulence factors in Y. pestis (53). According to one study, the pPCP plasmid is present in all strains of Y. pestis from the NCDCPH collection (25), but in another series of experiments, the plasmid was found only in three strains (2944, 2614, and 790) (25, 30). If the plasmid is only found in three strains, the high cytotoxic activity of strain 2944 could be explained by the presence of the pPCP plasmid. According to the blotting data, the Pla is expressed in all of these strains (data not shown). We also consider the possibility that Pla gene from pPCP plasmid was integrated in to the bacterial chromosome, which has been reported in previous studies.
The proteome of Y. pestis as a function of changes in temperature and calcium provides information regarding the expression levels of virulence-associated factors, putative virulence factors, and metabolic and housekeeping proteins, as well as potential novel virulence determinants. Future studies using altered gel formulations to examine lower-molecular-weight proteins, and experiments to control the proteolytic activity of Pla will provide more information about these Y. pestis strains. Although, we can infer that there are differences between strains and the functions of the identified differentially expressed proteins, future studies using mutant strains of Y. pestis are needed. The proteomic analysis of these strains reported here allows for future comparisons of clinical and environmental isolates across the Caucasus.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was made possible by financial support provided by the U.S. Department of Defense (DoD), Defense Threat Reduction Agency (DTRA), through the Biological Threat Reduction Program’s (BTRP) TADR Activity Project TAP-6, in Georgia. The findings, opinions, and views expressed herein belong to the authors and do not reflect an official position of any organization listed nor does it reflect the official policy or position of the Department of the Navy, DoD, nor the U.S. Government. The authors thank Attimo Research and Development and Sonya Narodny for editorial assistance.
References
- 1.Anisimov AP, Lindler LE, Pier GB. Intraspecific diversity of Yersinia pestis. Clin Microbiol Rev (2004) 17(2):434–64. 10.1128/CMR.17.3.695.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inglesby TV, Dennis DT, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, et al. Plague as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA (2000) 283(17):2281–90. 10.1001/jama.283.17.2281 [DOI] [PubMed] [Google Scholar]
- 3.Cui Y, Yang X, Xiao X, Anisimov AP, Li D, Yan Y, et al. Genetic variations of live attenuated plague vaccine strains (Yersinia pestis EV76 lineage) during laboratory passages in different countries. Infect Genet Evol (2014) 26:172–9. 10.1016/j.meegid.2014.05.023 [DOI] [PubMed] [Google Scholar]
- 4.Hinnebusch BJ. The evolution of flea-borne transmission in Yersinia pestis. Curr Issues Mol Biol (2005) 7(2):197–212. [PubMed] [Google Scholar]
- 5.Perry RD, Fetherston JD. Yersinia pestis – etiologic agent of plague. Clin Microbiol Rev (1997) 10(1):35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wimsatt J, Biggins DE. A review of plague persistence with special emphasis on fleas. J Vector Borne Dis (2009) 46(2):85–99. [PubMed] [Google Scholar]
- 7.Acha PN, Szyfres B. Zoonoses and Communicable Diseases Common to Man and Animals. Washington, DC: Pan American Health Organization; (2001). p. 218–26. [Google Scholar]
- 8.Bacon M, Drake CH. Comparative susceptibility of various species of mice native to Washington to inoculation with virulent strains of Pasteurella pestis. J Infect Dis (1958) 102(1):14–22. 10.1093/infdis/102.1.14 [DOI] [PubMed] [Google Scholar]
- 9.Baltazard M, Karimi Y, Eftekhari M, Chamsa H, Mollaret HH. Interepizootic Conservation of Plague in its Inveterate Foci: Working Hypotheses. Frederick, MD: Army Biological Labs; (1964). [Google Scholar]
- 10.Hubbert WT, Goldenberg MI. Natural resistance to plague: genetic basis in the vole (Microtus californicus). Am J Trop Med Hyg (1970) 19(6):1015–9. [DOI] [PubMed] [Google Scholar]
- 11.Azizi MH, Azizi F. A history of the human plague in Iran. Arch Iran Med (2010) 13(6):563–9.010136/AIM.0022 [PubMed] [Google Scholar]
- 12.Biggins DE, Kosoy MY. Influences of introduced plague on North American mammals: implications from ecology of plague in Asia. J Mammal (2001) 82(4):906–16. [DOI] [Google Scholar]
- 13.Mostafavi E, Asadmanesh K, Nadaf S, Hanifi H, Hanifi M, Behzadi-yousefi M. Study of the plague infection in rodents and carnivores in Western Iran. The First Conference on Integrative Zoology. Iran: Ferdowsi University of Mashhad; (2012). [Google Scholar]
- 14.Wanrong Z. The succession of rodent populations in Erdosi desert-grasslands and the multiplicity of the reservoirs in plague natural foci. Chin J Vector Biol Control (1991) 2:257–60. [Google Scholar]
- 15.Cully JF, Williams ES. Interspecific comparisons of sylvatic plague in prairie dogs. J Mammal (2001) 82:894–905. [DOI] [Google Scholar]
- 16.Bearden SW, Brubaker RR. Recent findings regarding maintenance of enzootic variants of Yersinia pestis in sylvatic reservoirs and their significance in the evolution of epidemic plague. Vector Borne Zoonotic Dis (2010) 10(1):85–92. 10.1089/vbz.2009.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorange EA, Race BL, Sebbane F, Hinnebusch BJ. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J Infect Dis (2005) 191(11):1907–12. 10.1086/429931 [DOI] [PubMed] [Google Scholar]
- 18.Brubaker RR. Factors promoting acute and chronic diseases caused by yersiniae. Clin Microbiol Rev (1991) 4(3):309–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duplantier JM, Duchemin JB, Chanteau S, Carniel E. From the recent lessons of the Malagasy foci towards a global understanding of the factors involved in plague reemergence. Vet Res (2005) 36(3):437–53. 10.1051/vetres:2005007 [DOI] [PubMed] [Google Scholar]
- 20.World Health Organization. Interregional Meeting on Prevention and Control of Plague. Antananarivo: World Health Organization; (2010). [Google Scholar]
- 21.Ben-Ari T, Neerinckx S, Gage KL, Kreppel K, Laudisoit A, Leirs H, et al. Plague and climate: scales matter. PLoS Pathog (2011) 7(9):e1002160. 10.1371/journal.ppat.1002160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisen RJ, Gage KL. Adaptive strategies of Yersinia pestis to persist during inter-epizootic and epizootic periods. Vet Res (2009) 40(2):1. 10.1051/vetres:2008039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A (1999) 96(24):14043–8. 10.1073/pnas.96.24.14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, Neyt C, et al. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev (1998) 62(4):1315–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Revazishvili T, Rajanna C, Bakanidze L, Tsertsvadze N, Imnadze P, O’Connell K, et al. Characterisation of Yersinia pestis isolates from natural foci of plague in the Republic of Georgia, and their relationship to Y. pestis isolates from other countries. Clin Microbiol Infect (2008) 14(5):429–36. 10.1111/j.1469-0691.2008.01953.x [DOI] [PubMed] [Google Scholar]
- 26.Beridze L, Chubinidze S, Tsertsvadze N, Tsintsadze L, Gaphrindashyili M, Katsitadze G. Characteristics of natural foci of plague existing on territory of Georgia. Sakartvelos Sameditsino Moambe (2004) 524:22–8. [Google Scholar]
- 27.Chubinidze S, Katsitadze G, Tsertsvadze N, Eloshvili M, Tsanava S. [Dynamics of epizootic activity of natural foci of plague in Georgia]. Georgian Med News (2008) 163:47–53. [PubMed] [Google Scholar]
- 28.O’Connel KP, Skowronski EW, Sulakvelidez A, Bakanidze L. Emerging and Endemic Pathogens: Advances in Surveillance, Detection and Identification. 2010 ed Netherlands: Springer; (2010). 175 p. [Google Scholar]
- 29.Cummings CA, Bormann Chung CA, Fang R, Barker M, Brzoska P, Williamson PC, et al. Accurate, rapid and high-throughput detection of strain-specific polymorphisms in Bacillus anthracis and Yersinia pestis by next-generation sequencing. Investig Genet (2010) 1(1):5. 10.1186/2041-2223-1-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajanna C, Revazishvili T, Rashid MH, Chubinidze S, Bakanidze L, Tsanava S, et al. Characterization of pPCP1 plasmids in Yersinia pestis strains isolated from the former soviet union. Int J Microbiol (2010) 2010:760819. 10.1155/2010/760819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong ZZ, Zhou DS, Song YJ, Zhang L, Pei D, Han YP, et al. Genetic variations in the pgm locus among natural isolates of Yersinia pestis. J Gen Appl Microbiol (2005) 51(1):11–9. 10.2323/jgam.51.11 [DOI] [PubMed] [Google Scholar]
- 32.You YH, Wang P, Wang YH, Zhang MJ, Song ZZ, Hai R, et al. Comparative genomic analysis of gene variations of two Chinese Yersinia pestis isolates from vaccine strain EV76. Biomed Environ Sci (2012) 25(4):440–8. 10.3967/0895-3988.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 33.Rudolph AE, Stuckey JA, Zhao Y, Matthews HR, Patton WA, Moss J, et al. Expression, characterization, and mutagenesis of the Yersinia pestis murine toxin, a phospholipase D superfamily member. J Biol Chem (1999) 274(17):11824–31. 10.1074/jbc.274.17.11824 [DOI] [PubMed] [Google Scholar]
- 34.Cornelis GR. Yersinia type III secretion: send in the effectors. J Cell Biol (2002) 158(3):401–8. 10.1083/jcb.200205077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry RD, Harmon PA, Bowmer WS, Straley SC. A low-Ca2+ response operon encodes the V antigen of Yersinia pestis. Infect Immun (1986) 54(2):428–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price SB, Straley SC. lcrH, a gene necessary for virulence of Yersinia pestis and for the normal response of Y. pestis to ATP and calcium. Infect Immun (1989) 57(5):1491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeannin P, Bottazzi B, Sironi M, Doni A, Rusnati M, Presta M, et al. Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity (2005) 22(5):551–60. 10.1016/j.immuni.2005.03.008 [DOI] [PubMed] [Google Scholar]
- 38.Solomonia R, Nozadze M, Mikeladze E, Zhgenti E, Chanturia G, Kekeklidze M, et al. Characterization of atypical Y. pestis strains isolated in natural foci of Georgia. In Abstracts of 112th ASM General Meeting. San-Francisco: (2012). p. 99. [Google Scholar]
- 39.Knowlton KU, Jeon ES, Berkley N, Wessely R, Huber S. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J Virol (1996) 70(11):7811–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zauberman A, Cohen S, Mamroud E, Flashner Y, Tidhar A, Ber R, et al. Interaction of Yersinia pestis with macrophages: limitations in YopJ-dependent apoptosis. Infect Immun (2006) 74(6):3239–50. 10.1128/iai.00097-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chromy BA, Choi MW, Murphy GA, Gonzales AD, Corzett CH, Chang BC, et al. Proteomic characterization of Yersinia pestis virulence. J Bacteriol (2005) 187(23):8172–80. 10.1128/JB.187.23.8172-8180.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajanna C, Ouellette G, Rashid M, Zemla A, Karavis M, Zhou C, et al. A strain of Yersinia pestis with a mutator phenotype from the Republic of Georgia. FEMS Microbiol Lett (2013) 343(2):113–20. 10.1111/1574-6968.12137 [DOI] [PubMed] [Google Scholar]
- 43.Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci U S A (2004) 101(51):17837–42. 10.1073/pnas.0408026101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knight SD. Structure and assembly of Yersinia pestis F1 antigen. Adv Exp Med Biol (2007) 603:74–87. 10.1007/978-0-387-72124-8_6 [DOI] [PubMed] [Google Scholar]
- 45.Williamson ED, Oyston PC. Protecting against plague: towards a next-generation vaccine. Clin Exp Immunol (2013) 172(1):1–8. 10.1111/cei.12044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sebbane F, Jarrett C, Gardner D, Long D, Hinnebusch BJ. The Yersinia pestis caf1M1A1 fimbrial capsule operon promotes transmission by flea bite in a mouse model of bubonic plague. Infect Immun (2009) 77(3):1222–9. 10.1128/IAI.00950-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor DE. Bacterial tellurite resistance. Trends Microbiol (1999) 7(3):111–5. 10.1016/S0966-842X(99)01454-7 [DOI] [PubMed] [Google Scholar]
- 48.Calderon IL, Arenas FA, Perez JM, Fuentes DE, Araya MA, Saavedra CP, et al. Catalases are NAD(P)H-dependent tellurite reductases. PLoS One (2006) 1:e70. 10.1371/journal.pone.0000070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ponnusamy D, Hartson SD, Clinkenbeard KD. Intracellular Yersinia pestis expresses general stress response and tellurite resistance proteins in mouse macrophages. Vet Microbiol (2011) 150(1–2):146–51. 10.1016/j.vetmic.2010.12.025 [DOI] [PubMed] [Google Scholar]
- 50.Wang W, Li GW, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science (2011) 333(6048):1445–9. 10.1126/science.1204697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Philip NH, Brodsky IE. Cell death programs in Yersinia immunity and pathogenesis. Front Cell Infect Microbiol (2012) 2:149. 10.3389/fcimb.2012.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye Z, Gorman AA, Uittenbogaard AM, Myers-Morales T, Kaplan AM, Cohen DA, et al. Caspase-3 mediates the pathogenic effect of Yersinia pestis YopM in liver of C57BL/6 mice and contributes to YopM’s function in spleen. PLoS One (2014) 9(11):e110956. 10.1371/journal.pone.0110956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bearden SW, Fetherston JD, Perry RD. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun (1997) 65(5):1659–68. [DOI] [PMC free article] [PubMed] [Google Scholar]